Lec 15 - RNA Splicing
1/108
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
109 Terms
eukaryotes, introns, exons
Splicing is a critical step in gene expression in _____ (organisms with cells that have a nucleus), where non-coding sections (____) are removed from the newly transcribed RNA, and the remaining coding sections (_____) are joined together to form a mature messenger RNA (mRNA)
encode, removed
genes are organized in the DNA of eukaryotes, consisting of exons (segments that ___ protein) and introns (non-coding segments that must be __)
isoforms, cells, times, transcription
Alternative splicing is a mechanism that allows a single gene to code for multiple, distinct proteins (called ____) by treating different segments as exons or introns in different ___ or at different ___. This is a powerful way to regulate gene expression after ____ (post-transcriptional
diseases
how errors or changes (mutations) in the splicing process can lead to human ____
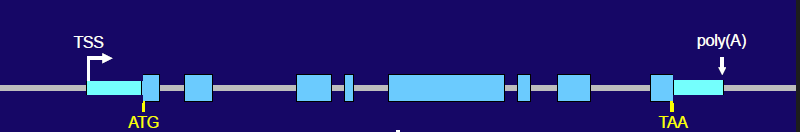
TSS, ATG, TAA, last
Gene Structure (Top Strand):
The gene starts at the Transcription Start Site (____) and includes segments called exons (light blue blocks) and segments called introns (lines between exons).
Key codons are marked:
___ is the start codon for translation (marks the beginning of the protein-coding sequence),
___ is a stop codon (marks the end of the protein-coding sequence).
The region after the ___ exon is involved in adding a poly(A) tail
pre, 5’, introns
Transcription, Capping (Second Strand): The DNA is first transcribed into a __-mRNA (precursor mRNA). A protective cap (represented by the green circle) is added to the __ end of the molecule. The pre-mRNA still contains both exons and _
3’, introns
3'-end processing (Third Strand): The __ end of the pre-mRNA is processed, and a long tail of Adenine nucleotides, called the poly(A) tail (AAAAA), is added. This tail helps with nuclear export, translation, and mRNA stability. This molecule now has the cap on the 5' end and the poly(A) tail on the 3' end, but still contains __. The thin lines with loops indicate the sections that will be removed (the introns)
5’ UTR, 3’ UTR, leave
Splicing (Bottom Strand): The introns are removed, and the exons are joined together. The resulting mature mRNA is much shorter.
The region before the ATG start codon is the __ __ (Untranslated Region).
The region after the TAA stop codon but before the poly(A) tail is the __ _(Untranslated Region).
This final, spliced mRNA is ready to __ the nucleus and be translated into protein.

nucleus, DNA, RNA, leaves, cytoplasm, ribosome, transfer, protein
classic, complete overview of gene expression—the process by which information from a gene is used to synthesize a functional gene product, like a protein.
Location: Eukaryotic cells have a __ surrounded by a Cytoplasm (the cell interior) and enclosed by a Cell membrane.
Transcription: This process takes place in the Nucleus. The genetic information stored in the __ is copied (transcribed) into a molecule of m__ (messenger RNA) .
mRNA Transport: The mRNA ___ the nucleus and moves into the Cytoplasm.
Translation: This process occurs in the Cytoplasm , specifically on a structure called the __, which reads the code in the mRNA.
tRNA (_____ RNA) molecules bring the correct amino acids to the ribosome based on the mRNA code.
The ribosome joins these amino acids together to form a chain, which folds into a functional ___
pre-mRNA, spliceosome, mature
gene expression model from page 4 by inserting the crucial step of splicing.
Transcription: In the Nucleus , DNA is transcribed into a __-__ that contains both coding segments (Exons ) and non-coding segments (Introns ).
Splicing: The pre-mRNA then undergoes Splicing , also in the Nucleus. This process is carried out by a large complex of proteins and RNA molecules called the ____, which removes the Introns and joins the Exons to produce the mature mRNA.
mRNA Transport & Translation: The ___ mRNA then moves to the Cytoplasm to be read by the Ribosome (with the help of tRNA ), a process called Translation , which synthesizes the Protein
initiating, highest, product, intermediate, lariat
This page presents an experimental result from a historic in vitro (in a test tube) experiment on beta-globin pre-mRNA splicing
Experiment: The left side shows an image that resembles a gel electrophoresis, which separates nucleic acids (like RNA) by size.
The lanes represent different time points after ___ the splicing reaction: 0 (start), 15' (15 minutes), 30' (30 minutes), 1h (1 hour), 2h, 3h, and 4h
The bands on the gel indicate molecules of different sizes.
Interpretation (Right Side): The diagrams on the right correspond to the RNA molecules observed at different sizes in the gel lanes over time.
The __ band represents the initial pre-mRNA containing Exon 1 and Exon 2
As time progresses (15' and beyond), the higher band decreases, and new, lower bands appear, which represent the products and intermediates of splicing:
A ___ representing Exons 1 and 2 spliced together (mature mRNA). This is the desired final product and is the lowest major band.
An ____ where Exon 1 has been cut from the intron, and the intron (containing Exon 2) forms a loop called a ___ (the loop with 2 attached)
The excised intron (a lariat structure, appearing further down).
A molecule representing Exon 1 joined to the lariat (Exon 1-lariat intermediate).
transesterification
This page introduces the two-step chemical pathway of pre-mRNA splicing. This process involves two consecutive _____ reactions, which are swaps of chemical bonds without adding or consuming energy.
GU, A, AG, 5’ splice, 3’ splice, branch
Two-Step Pre-mRNA Splicing Pathway
Starting Material (Top): Shows a pre-mRNA segment: Exon 1 followed by the intron, which has a specific consensus sequence __..__...__, and then Exon 2.
The intron starts with GU (__ __ site) and ends with AG (__ __ site).
An internal Adenosine (A) is the ___ point.
2’-OH, phosphate, breaks, lariat, Exon 1, Exon 2
Two-Step Pre-mRNA Splicing Pathway
Step 1 (Middle):
The __-__ (hydroxyl group) of the Adenosine (A) at the branch point attacks the ___ at the 5' splice site (GU).
This _____ the bond at the 5' splice site, freeing the 5' end of the intron.
The intron's 5' end then covalently joins to the branch point A, forming a three-way junction called an RNA _____(the loop).
The product of this step is: a free __ _ with a 3'-OH group, and the lariat-intron attached to __ __
3’-OH, phosphate, breaks, mature, degraded
Two-Step Pre-mRNA Splicing Pathway
Step 2 (Bottom):
The newly freed __-__ of Exon 1 attacks the __ at the 3' splice site (AG).
This __ the bond at the 3' splice site, freeing the lariat-intron.
This action joins Exon 1 to Exon 2 to form the ____mRNA.
The excised intron is released as a lariat structure, which is subsequently ___
three, 2’, 5’
chemical view of the unique junction that is formed in the RNA lariat intermediate.
The Lariat Junction:
Branch Point Detail: The diagram highlights an Adenosine (A) nucleotide, which is the branch point in the intron.
This Adenosine is connected to _____ other nucleotides, forming a branch.
It is connected via its regular 3' carbon to a neighboring nucleotide (part of the intron strand continuing 3' downstream) and via its 5' carbon to another neighboring nucleotide (part of the intron strand continuing 5' upstream).
Crucially, a bond is also formed from the 2' carbon of the branch point Adenosine to the phosphate group of the Guanosine (G) at the 5' end of the intron. This forms a rare __ to __ phosphodiester bond, which is the signature feature of the lariat branch point
5, snRNAs
key molecular sequences that define the boundaries of the intron, and the molecular machinery that recognizes them.
The Spliceosome: This is the massive molecular machine responsible for pre-mRNA splicing. It consists of __ small nuclear RNAs (__) and over 170 proteins.
beginning, internal, end, U, C
Consenus Splicing Signals anf the Spliceosome
Key Recognition Sites: An intron (the region between Exon 1 and Exon 2) has three essential signals:
5' splice site: The _____ of the intron, typically with the consensus sequence GU.
Branch site: An _____ Adenosine (A) within the intron, which is critical for the first step of splicing.
3' splice site: The _____ of the intron, typically with the consensus sequence YYYYYYY-AG, where "Y" is a pyrimidine (__ or __). This region is known as the polypyrimidine tract.
U1 snRNP, U2 snRNP, U2AF
Consensus Splicing signals and the Spliceosome
Initial Recognition
The __ __ (small nuclear ribonucleoprotein) , along with its associated proteins (like the 70K protein ), recognizes and binds to the 5' splice site.
The __ __ recognizes and binds to the branch site.
The __ (U2 Auxiliary Factor) 65/35 complex recognizes and binds to the polypyrimidine tract and 3' splice site
snRNAs
This page focuses on the RNA components of the spliceosome, known as _____ (small nuclear RNAs).
Image Interpretation: The image shows a gel (likely an autoradiograph) used to separate the small RNA molecules present in the nucleus based on their size (S values, a measure of sedimentation rate).
Major snRNAs of the Spliceosome: The distinct bands represent the core snRNAs essential for the function of the major spliceosome: U1, U2, U4, U5, U6
These five snRNAs are the catalytic and structural core of the spliceosome. They are responsible for base-pairing with the pre-mRNA and with each other to orchestrate the two chemical steps of splicing.
one, many
An snRNP is a complex of ___ snRNA molecule and ____ associated proteins.
snRNA, Sm, Sm, unique
Components of U1 snRNP:
U1 ___: The RNA component, shown on the right with its internal secondary structure (loops and stems).
____ Site and _____ Proteins: The U1 snRNA has a specific sequence (the Sm site ) where a ring of seven core proteins, called Sm proteins , bind.
U1-specific proteins: These proteins are ___ to the U1 snRNP and include 70K , A , and C (with their corresponding molecular weights like 52K, 31K, 17.5K) . The 70K, A, and C proteins are shown as circles on the left, demonstrating their relative sizes
RBD, beta, alpha, RNP1, RNP2
This page illustrates a common protein domain found in many splicing factors and other RNA-binding proteins.
RNA-recognition motif (RRM): This is one of the most common domains in proteins that bind RNA. It is often referred to as an ____ (RNA-binding domain)
Structure: The RRM is a conserved structure characterized by:
A four-stranded ___-sheet (represented by the broad red arrows beta1, beta2, beta3, beta4). The strands are typically antiparallel.
Two __-helices (represented by the green/red ribbons).
Key RNA-Binding Loops: The beta-sheet contains two characteristic sequences, ___ and ___, which are involved in directly binding to the RNA molecule. The right-hand image shows a molecular model where the protein surface (blue/white) interacts with the RNA backbone (yellow and red)
stem loops
Model: This image is a structural model of the U1 snRNP. The yellow strands represent the U1 snRNA, which has several ___ ___ (SL) —regions of the RNA that fold back and pair with themselves.
core
Sm Core: The seven core Sm proteins form a ring-like structure, known as the Sm __, around the U1 snRNA. They are individually labeled (D1 , D2 , D3 , B , F, etc.)
70K, A
U1-specific proteins: The proteins unique to U1 are seen binding to the outer parts of the RNA:
U1-__ is shown as a large white mass.
U1-__ is shown as a gray mass
E, polyprimidine, 3’, splicing
Spliceosome Assembly on Pre-mRNA
1. __ Complex (Early or Commitment Complex)
Binding: The U1 snRNP binds to the 5' splice site (GU). Splicing factors (like U2AF) recognize the ___ tract (Py) and the __ splice site (AG).
Result: This initial binding commits the transcript to ___.
A, branch, polypyrimidine, pre
Spliceosome Assembly on Pre-mRNA
2. __ Complex (Pre-spliceosome Complex)
Binding: The U2 snRNP binds to the ____ point Adenosine (A) and the ___ tract. This binding forces the branch point Adenosine to bulge out, which is necessary for its nucleophilic attack in the first splicing step.
Result: The __-spliceosome is formed.
B, Exon, Exon,catalytic
Spliceosome Assembly on Pre-mRNA
__Complex (Pre-catalytic Complex)
Binding: The large U4/U6.U5 tri-snRNP complex joins the assembly.
U5 snRNP binds to both ___ 1 and ___- 2, aligning them for the splicing reactions.
U4 and U6 snRNAs are held together by extensive base-pairing, and U6 has a complex base-pairing with U4 and U2.
Rearrangement and Activation: U1 is displaced, and U4 dissociates from U6. U6 then base-pairs with U2, forming the active ___ center.
C, lariat, ligation
4. __ Complex (Catalytic Complex)
Catalysis: This complex is the activated spliceosome where the two transesterification reactions occur.
The first C complex performs the first attack (___ formation).
The second C complex performs the second attack (__ ligation).
Result: The intron (lariat) is excised, and the two exons are joined together to form the mature exon 1 | exon 2 mRNA
U1, U6, U5
molecular recognition between the pre-mRNA and the snRNAs at the 5' splice site.
The key is complementary base-pairing.
The 3' end of the __ snRNA (sequence: GUCCAPsiPsiCAUApppG) base-pairs with the 5' splice site of the pre-mRNA (sequence: CAG GURAGU), where the 'GU' is the start of the intron. This pairing is crucial for the initial recognition and commitment of the transcript (E Complex formation)
The diagram also shows the __ snRNA base-pairing with a sequence near the 5' splice site (GURAGU ), indicating its role in the final catalytic phase (C Complex), where it takes over from U1 snRNA
__ snRNA is shown interacting with the sequences flanking the 5' splice site, which is essential for aligning the two exons before they are joined.
5’ splice, nucleophilic
This page reinforces the two crucial base-pairing interactions that define the two most important sites in the intron.
The U1 snRNA (specifically its 3' end) forms a base-pairing complex with the ___ __ site (CAGGUAAGU) at the beginning of the intron on the Pre-mRNA. This confirms the initial recognition (E Complex).
The U2 snRNA base-pairs with the sequence around the Branch point (UACUAC) on the pre-mRNA.
Crucially, the binding of U2 to this region forces the branch point Adenosine (A) to bulge out (form a loop) from the double-stranded region, where its 2'-OH group is exposed. This exposed hydroxyl is what performs the ____ attack to begin the first splicing step.
splicing
This massive diagram provides a detailed inventory of the many protein factors involved in the spliceosome assembly at different stages.
Spliceosome Components: The major spliceosome is incredibly complex, involving numerous components. This chart groups the proteins by the complex they belong to and the snRNA they associate with.
Complex A Proteins (~10 proteins): These are the factors involved in the initial commitment and U2 snRNP binding. Key groups include:
U1 RNA-associated proteins (Sm, 70K, A, C).
U2 RNA-associated proteins (Sm, SF3 family of proteins).
Other factors like hnRNPs (heterogeneous nuclear ribonucleoproteins) and SR proteins (Serine/Arginine-rich proteins
Complex B Proteins (~65 proteins): This is where the U4/U6.U5 tri-snRNP joins, bringing in a huge number of additional factors. The number of proteins jumps from ~10 to ~65
This complex includes the Prp19 complex (a set of proteins critical for spliceosome activation) and the many proteins associated with the U4/U6.U5 tri-snRNP (e.g., Lsm proteins, U5-specific, U4-specific proteins).
Complex C Proteins (Further Activation/Catalytic): After the two splicing reactions, the spliceosome disassembles, and various components are released or stay bound to the mRNA. This stage involves factors that aid in the final step (Step II factors ) and the release/recycling of the components.
Key takeaway: The transition from Complex A to Complex B involves a major recruitment of factors, and the entire ______ machinery is one of the largest and most intricate molecular machines in the cell.
ATP, RNA-RNA, RNA-protein
This page focuses on a specific class of proteins—the ATP-dependent RNA helicases—that are critical for the dynamic and energy-intensive process of splicing.
Function: These enzymes use the energy from ____ hydrolysis (breaking down ATP) to change the shape (conformation) of the spliceosome by unwinding __-__ or __-__ interactions (they are RNA-dependent ATPases/RNA helicases ). These rearrangements are necessary for the spliceosome to transition between its assembly (E, A, B, C) and catalytic states
stages
Key Helicases in the Splicing Cycle: The diagram shows how different Prp (Pre-mRNA Processing) proteins act at specific ___ of the cycle:
Prp5: Involved in the formation of the A complex (U2 recruitment).
Prp28: Involved in the transition from E/A complex to B complex (U1 removal).
Brr2: Acts at B complex to unwind U4/U6, allowing U6 to pair with U2 for catalysis.
Prp2: Involved in the first catalytic step (lariat formation).
Prp16: Involved in the second catalytic step (exon ligation).
Prp22 : Involved in the final step, releasing the mature mRNA and the lariat-intron.
Prp43: Involved in the disassembly of the spliceosome.
helicases
Splicing is not a passive process; it is an ATP-dependent, dynamic cycle driven by a series of RNA ___ that perform sequential unwinding and rearrangement steps to ensure fidelity and completion.
Prp18
This page shows an experiment demonstrating the function of one specific protein, __, which is a splicing factor required for the second catalytic step of splicing (joining the two exons).
Experiment (Similar to Page 6): This is another time-course splicing assay using gel electrophoresis, but it compares different conditions:
NE (Nuclear Extract): A complete extract of the nucleus, which should perform splicing normally. The final product (1 | 2 151) appears over time.
Pre-immune depleted: A control extract, where the extract has been treated with antibodies that don't recognize Prp18. Splicing should still occur normally152.
alpha-hPrp18 depleted: The extract has been treated with an antibody that specifically removes the human Prp18 protein.
Result: The final product (1 | 2) is significantly reduced or absent, but the first-step intermediate (lariat with Exon 2 or the Exon 1 product of the first step 154) accumulates. This shows that the first step happens, but the second step is blocked.
alpha-hPrp18 depleted + hPRP18: The depleted extract is rescued by adding back purified human Prp18 protein.
Result: The final product (1 | 2) is restored, confirming that Prp18 is necessary and sufficient for the second step of splicing to proceed.
second
This experiment proves the necessity of Prp18 as a ____-step splicing factor, acting after the lariat has formed to facilitate the final exon-joining reaction
GU, A, polypyrimidine, AG
This page provides a summary of the canonical (standard) consensus sequences required for splicing in both yeast and humans.
Canonical Splicing Signals: These are the highly conserved sequences found at the three key sites in an intron.
5' splice site : The consensus is a sequence highly enriched (specifically __ at the intron boundary).
Branch site : This region contains the critical _____. The consensus sequences vary between yeast and human (e.g., in yeast, the sequence is highly specific: UACUAAC).
3' splice site : Characterized by the ___ tract (a stretch of C's and U's) followed immediately by the required __ sequence
acessory
Comparison (Yeast vs. Human):
Yeast: Has a small number of genes (6,000) and very few introns (235). The splicing signals are extremely highly conserved and predictable (e.g., UACUAAC branch site).
Human: Has many more genes (>20,000) and vastly more introns (>200,000). The consensus sequences are more degenerate (less strict/repeate) compared to yeast, indicating that human splicing relies more on ____ proteins (splicing factors) to find and define the correct sites
-1, +1, +2
This page focuses specifically on the 5' splice site and the base-pairing with U1 snRNA.
U1 snRNP Recognition 166: The top line shows the 3' end of the U1 snRNA 167base-pairing with the 5' splice site sequence (CAG GUAAGU)168.
Nucleotide Numbering: The sequence of the 5' splice site is typically numbered relative to the exon-intron junction:
The last nucleotide of the exon is __ (e.g., 'G' in CAG).
The first two nucleotides of the intron are __ and __ (always GU), which are the most critical for recognition.
Subsequent intron nucleotides are +3, +4, +5, etc.169169169169.
GU
The __ at the +1/+2 position is the most critical and universally conserved element of the 5' splice site, and its complementary base-pairing with U1 snRNA is the initial step of spliceosome assembly.
alternative splicing
The vast majority of human genes (>75%) use ______ ____ to create multiple versions of protein
three
On average, a single human gene produces at least _____different protein versions (isoforms).
human
___ protein-coding genes
gene number ~25,000
largest gene 2.4 Mb (avg = 27 kb)
introns per gene 1 - 316 (avg = 8)
largest exon 17 kb (avg = 150 bp)
largest intron 800 kb (avg = 3 kb)
alternatively spliced > 75%
isoforms per gene avg ≥3
long, shot
Human genes are characterized by very ____ introns and many ___ exons, and the majority of genes utilize alternative splicing to generate significant protein diversity from a limited number of genes.
specific
Alternative Splicing Patterns
Exon Inclusion/Skipping (Cassette): A __ exon (the red/orange block) is either included in the mature mRNA (included) or entirely skipped (spliced out with the introns)
mutually
Mutually Exclusive Exons: A region contains two possible exons, but only one of the two is incorporated into the final mRNA. The choice is ____ exclusive.(two or more events, possibilities, or things cannot occur or exist at the same time. If one event happens, it makes the other(s) impossible)
length
Alternative 3' Splice Sites: The spliceosome can recognize and cut at one of two different 3' splice sites (AG), changing the ____ of the exon
length
Alternative 5' Splice Sites: The spliceosome can recognize and cut at one of two different 5' splice sites (GU), also changing the __ of the exon
non-functional
Retained Intron: An entire intron segment is left in the mature mRNA, which can introduce a frameshift or a premature stop codon, often leading to a __-__ protein
first
Alternative Promoters: The gene can start at different Transcription Start Sites, leading to an mRNA with different ___ exons.
length
Alternative Polyadenylation Sites: The gene can end at different points, leading to a mature mRNA with a different ____ of its 3' UTR.
TP53
This page uses the TP53 gene (a major tumor suppressor gene) as an example to show the biological reality of alternative splicing.
TP53 Gene Locus: The diagram is a view from the Ensembl genome browser, showing the different mRNA molecules (transcripts) produced from the single TP53 gene.
Multiple Isoforms: Instead of just one product, the TP53 gene produces many different splice variants, or isoforms, which are named (e.g., TP53-203, TP53-229, TP53-202, etc.).
Functional Classes: These isoforms fall into different functional categories:
Protein coding: Transcripts that can be successfully translated into a protein.
Nonsense mediated decay (NMD): Transcripts that contain a premature stop codon due to the way they were spliced, leading to their degradation by the NMD pathway.
Retained intron: Isoforms where an intron was left unspliced.
Key takeaway: The ___ example vividly illustrates the principle of alternative splicing: a single gene can generate a complex family of transcripts with different structures and fates, including functional proteins and transcripts destined for degradation (a form of gene regulation).
inactivates
This page provides a clinical example of a disease caused by a splicing mutation.
Adermatoglyphia: The top image shows the fingertips of a person lacking fingerprints191. This rare condition is known as "Immigration-delay disease" because it complicates identification documents192.
Genetic Cause: The condition is caused by a mutation in a skin-specific isoform of the SMARCAD1 gene193.
The Splicing Mutation: A single nucleotide change is responsible: IVS1 + 1g >t
IVS1 refers to Intron 1.
+1 means the first nucleotide of Intron 1.
g>t means the Guanine G at this position is mutated to a Thymine (T)
The Effect: The G at the +1 position is the most critical nucleotide for the 5' splice site (GU)197. Changing it to T ___ the authentic 5' splice site. This disrupts the normal splicing, resulting in the production of a non-functional protein specifically in the skin
diversity
Significance of Alternative Splicing
Major Source of Protein Isoform ___: More than 75% of human genes encode at least two isoforms.
Protein Isoform Functions: The different proteins produced from a single gene can have:
Related functions (e.g., in different tissues).
Distinct functions (e.g., binding different targets).
Antagonistic functions (e.g., one promotes a process, the other inhibits it).
Differential localization (e.g., one in the nucleus, one in the cytoplasm)
off, NMD
Regulation and Consequences
Post-transcriptional On/Off Switch: Alternative splicing can effectively turn a gene "__" by leading to a transcript that cannot produce a functional protein.
Frameshifting: When an exon is included or skipped, if its length is not a multiple of three nucleotides (nt), it shifts the reading frame for all subsequent codons. This nearly always leads to a premature stop codon.
Inclusion of alternative exons with in-frame stop codons: Some exons contain a stop codon, and their inclusion results in a truncated protein.
Consequences: These errors can result in inactive or dominant-negative truncated proteins , or the transcript is rapidly destroyed by Nonsense-mediated mRNA decay (__)
Dscam
This page provides an extreme example of the diversity generated by alternative splicing in the fruit fly (Drosophila).
The ___ (Down syndrome cell adhesion molecule) gene is involved in neuronal wiring and immune defense
Hyper-variability: The Dscam gene is a record holder for alternative splicing because it contains multiple sets of mutually exclusive exons
In a region designated Exon 4, there are 12 different possible exons, only one of which can be chosen.
In Exon 6, there are 48 different possible exons.
In Exon 9, there are 33 different possible exons.
In Exon 17, there are 2 different possible exons.
Total Isoforms: The total number of unique mRNA molecules (isoforms) that can be generated is found by multiplying the number of choices for each variable exon211:
12 \times 48 \times 33 \times 2 = \mathbf{38,016}
combinatorial
Key takeaway: The Dscam gene demonstrates the enormous ___ power of alternative splicing, allowing a single gene to encode tens of thousands of different protein isoforms.
chosen
two major classes of signals and factors (cis and trans) that regulate which splice sites are ___, thereby controlling alternative splicing.
within
Cis-acting Signals
These are the sequences __ the RNA molecule itself (the pre-mRNA) that govern splicing choice
consensus, skipped
Cis
Strength of Splice Sites: How closely a 5' or 3' splice site matches the ___ sequence (GU, AG, etc.) determines its "strength." Weaker sites are often ____ unless aided by other factors
activator
Cis
Splicing Enhancers (Exonic and Intronic): Sequences in the exons (ESE - Exonic Splicing Enhancer) or introns (ISE - Intronic Splicing Enhancer) that recruit _____ proteins to promote the inclusion of the adjacent exon.
repressor
Cis
Splicing Silencers (Exonic and Intronic): Sequences in the exons (ESS - Exonic Splicing Silencer) or introns (ISS - Intronic Splicing Silencer) that recruit _____ proteins to inhibit the inclusion or promote the skipping of the adjacent exon.
folding
Cis
RNA secondary structure: The ____ of the pre-mRNA can physically mask or expose splice sites
rate
Cis
Chromatin features: Features of the DNA and its packaging can influence the transcription __, which can indirectly affect splicing (see Page 33).
splicing factors, cis
Trans-acting Factors
These are the protein molecules (___ __) that bind to the _-acting sequences to determine the splicing outcome
enhancers, silencers
Trans
Activators and/or Repressors: Proteins that recognize and bind to the ____ and _____. Examples include:
SR proteins (Serine/Arginine-rich): Typically promote exon inclusion (activators).
hnRNP A/B proteins (Heterogeneous Nuclear Ribonucleoproteins): Often function as repressors.
PTB, Nova, Fox-1, etc.
assist
Trans
Co-activators and Co-repressors: Proteins that __ the main activators/repressors (e.g., Srm160
exonic, intronic, splicesome
This diagram illustrates the mechanism of how trans-acting factors (Activators and Repressors) bind to the cis-acting signals (Enhancers and Silencers) to regulate splice site choice.
Enhancer Mechanism (Activation):
A splicing Activator (A) (e.g., an SR protein with an RS domain ) binds to an ____ Splicing Enhancer (ESE) or an ___ Splicing Enhancer (ISE).
The Activator can then interact with the core spliceosome components (like U1 snRNP or the U2AF proteins ) and a Co-Activator, helping to stabilize the binding of the ____ to weak adjacent splice sites, promoting the inclusion of the exon.
repressor, skipped
This diagram illustrates the mechanism of how trans-acting factors (Activators and Repressors) bind to the cis-acting signals (Enhancers and Silencers) to regulate splice site choice.
Silencer Mechanism (Repression):
A splicing ___ (R) (e.g., hnRNPA1 with an RRM domain ) binds to an Exonic Splicing Silencer (ESS) or an Intronic Splicing Silencer (ISS).
The Repressor can block the spliceosome from binding to the adjacent splice sites, either by physically hindering it or by competing with an activator, thereby causing the exon to be ___.
regulate
Splicing factors ____ alternative splicing by binding to enhancers or silencers on the pre-mRNA and either recruiting (activators) or blocking/competing with (repressors) the core spliceosome machinery.
female
The sxl gene (Sex-lethal): This gene is the master switch for _____ development.
activated, repressor, inactivated, nonfunctional
Female Embryo (Left Side):
(a) Early in embryogenesis: A special promoter (PE) is briefly ____, and the pre-mRNA is transcribed and spliced into a functional Early Sxl protein232232232232.
(b) Later in embryogenesis: The early Sxl protein acts as a splicing ___. It represses the splicing of the sxl pre-mRNA itself. This repression causes the skipping of exon 3233, The resulting mRNA (Exons 1-2-4) 234is translated into a functional Late Sxl protein235.
Male Embryo (Right Side):
(a) Early in embryogenesis: The special early promoter (PE) is _____, so no Early Sxl protein is made.
(b) Later in embryogenesis: With no early Sxl repressor, the sxl pre-mRNA is spliced by default. This includes exon 3237. Exon 3 contains a stop codon, resulting in a ___ Sxl protein238
repressor, autoregulation
Key takeaway: The Sex-lethal protein acts as its own splicing _____ to ensure its continued production only in females. If no Sxl protein is made initially (in males), the default splicing pathway leads to a non-functional protein. This is an example of _____ via alternative splicing.
active
Sxl Protein Action (Female Only):
The functional Sxl protein (made only in females, as shown on Page 30) acts as a repressor on the transformer (tra) pre-mRNA.
In Females: Sxl represses the inclusion of an exon with a stop codon in the tra pre-mRNA, leading to an ___ protein.
In Males: With no Sxl protein, the default splicing produces a truncated, non-functional Tra protein.
alternative, default
Tra Protein Action (Female Only):
The functional Tra protein (made only in females) acts as an activator of ____ splicing on the Double-sex (Dsx) pre-mRNA.
In Females: Tra protein promotes the inclusion of a female-specific exon, leading to the female Dsx protein.
In Males: With no Tra protein, the ____ splicing leads to the male Dsx protein
transcription
Final Outcome: The female Dsx protein and the male Dsx protein are different __ factors that activate or repress different sets of genes, ultimately controlling the development of male or female somatic tissues
X
Alternative splicing creates an irreversible cascade (a sex-determination cascade ) where the initial production of Sxl protein (determined by the number of __ chromosomes) controls the subsequent splicing events of other genes to determine the final sex of the fly.
Pol II
Transcription: The process of making RNA from DNA, carried out by ___ _____
idependent, concurent, coupled
Three Models of Transcription Splicing Interaction:
_______ reactions: Transcription finishes first, and then the full-length pre-mRNA is spliced afterward.
_______ reactions: Splicing begins while transcription is still happening (the spliceosome works on the RNA "co-transcriptionally," as the RNA is being made).
_____ reactions: Splicing is directly linked to or dependent on the speed or pausing of the RNA polymerase.
supports
The evidence overwhelmingly ___ the models of Concurrent and Coupled reactions in eukaryotes, meaning that splicing does not wait for the entire pre-mRNA to be finished; it begins while the RNA Polymerase is still transcribing the gene.
skipping
Scenario 1: Fast Elongation/No Pauses:
Pol II transcribes rapidly.
The spliceosome components (like U1 ) have less time to assemble on a Weak 3' Splice Site (Weak 3'SS) before the polymerase passes.
Because the weak site is skipped, the downstream splice site is used instead, leading to Exon ____
weak, included
Scenario 2: Slow Elongation/with Pauses:
Pol II slows down or pauses near the alternative exon.
This "pause time" is enough to allow splicing factors (like U2AF ) to fully assemble on the __ 3'SS.
The complex is stabilized, the weak site is recognized, and the exon is therefore ___.
inclusion
The rate at which the RNA Polymerase II transcribes the DNA can directly influence alternative splicing choice. Slower transcription gives the spliceosome more time to assemble on weak splice sites, often leading to exon ___.
inactive
Splicing mutations can lead to…
______ authentic splice sites: A mutation makes a normal (authentic) 5' (GU) or 3' (AG) splice site non-functional, causing the entire exon to be skipped or the intron to be retained. (Example: Adermatoglyphia on Page 25).
Create
Splicing mutations can lead to…
____ ectopic splice sites in introns or exons: A mutation creates a new, functional splice site where there shouldn't be one, leading to the insertion of an extra, abnormal segment into the mature mRNA
cryptic
Splicing mutations can lead to…
Activate ___ splice sites in introns or exons: A mutation makes a previously ignored, weak, or cryptic (silent but can be revealved) splice site suddenly functional, causing a portion of an intron to be retained or an exon to be shortened
enhancers
Splicing mutations can lead to…
Create or destroy exonic or intronic splicing ____: A mutation either removes a sequence that recruits an activator or creates a new one that causes incorrect inclusion
silencers
Splicing mutations can lead to…
Create or destroy exonic or intronic splicing _____: A mutation either removes a sequence that recruits a repressor or creates a new one that causes incorrect skipping
intron, skipping, activation
A mutation that disrupts a splice site (lightning bolt over the exon/intron boundary) can result in:
____ retention (1): The spliceosome fails to recognize the splice site, so the adjacent intron is not removed and remains in the mature mRNA. This often leads to a premature stop codon and NMD (Nonsense-Mediated mRNA Decay).
Exon ____(2): The spliceosome ignores the entire exon because its boundaries are too weak or incorrect. The flanking exons are spliced directly together, resulting in the exclusion of the central exon.
_____ of cryptic splice site (3): The mutation causes the spliceosome to use an alternative, normally ignored splice site in the intron or a nearby exon. This results in an mRNA that is shorter or longer than normal.
10%
Clinical Relevance: More than ___ of disease-causing mutations disrupt splice sites. This highlights the importance of precise splicing for human health
mutation
This page describes an experimental method, the minigene splicing assay, used to study the effect of a specific ____ on splicing.
shorter
Minigene: Scientists create an artificial, ___ version of a gene (a minigene) that contains the exons and introns of interest (e.g., Exons 7 and 8 of the GTF2H1 gene).
transfected
Transfection: The minigene is introduced (______) into cells.
point
Mutations Studied: The experiment tests the normal (Wild-Type, WT) gene and two different __ mutations: +3a and +6u269269269269. These labels refer to the position and type of mutation near the splice site
mRNA
RT-PCR (Reverse Transcription-Polymerase Chain Reaction): After the minigene is transcribed and spliced by the cell's machinery, the resulting ____ is isolated and analyzed using RT-PCR. RT-PCR distinguishes between the different spliced isoforms (e.g., exon included vs. exon skipped) based on their size.
site, abolishes
Results:
WT: Shows 100% inclusion of Exon 7 (the normal product).
+3a: Shows 9 inclusion, meaning the mutation severely disrupts the splice __ and causes the exon to be mostly skipped.
+6u: Shows 0% inclusion, meaning the mutation completely ____ the correct splicing.
RNA
This page introduces Spinal Muscular Atrophy (SMA), a severe, common, and often fatal genetic disease, which serves as a major clinical example of a splicing-related disorder.
Spinal Muscular Atrophy (SMA):
It is the leading genetic cause of infant mortality.
It is a devastating disease: half of children diagnosed before age 2 die before their 2nd birthday.
Key takeaway: SMA is a serious, life-threatening genetic disease, whose cause is directly related to a defect in ___ splicing, as described on subsequent pages.
SMN1, SMN, snRNP, partial
Spinal Muscular Atrophy.
Clinical Features: SMA is a pediatric neuromuscular disorder characterized by the degeneration of lower motor neurons.
Genetics:
It is an autosomal recessive disorder (meaning both copies of the causative gene must be mutated).
Incidence: Affects about 1 in 6,000 live births.
The disease is caused by the loss of function of the ___ gene.
The SMN1 gene codes for the essential Survival of Motor Neuron (____) protein.
SMN is vital for promoting the assembly of the ____-complex (the building blocks of the spliceosome).
It localizes to the cytoplasm and nuclear "gems".
SMN2 Paralog: Humans have a second, very similar gene called SMN2, which is a paralog (a similar gene arising from duplication). SMN2 provides only ____ function because its splicing is defective. The severity of SMA is often inversely correlated with the number of SMN2 copies a patient has