Translational - Protein Synthesis in Prokaryotes & Eukaryotes
1/6
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
7 Terms
List the basic steps that must occur in protein synthesis, including details of each step.
Identify the structure of tRNA molecules and the impact of structure on function.
Describe the contribution of each step to the quality control of protein production.
Describe the structure of ribosomes and know the function of ribosomes in protein synthesis.
List the translation factors (other proteins) and their roles in initiation, elongation and termination.
Calculate the energy required for protein synthesis.
List the basic steps that must occur in protein synthesis, including details of each step.
Protein synthesis, also known as translation, is the process where the cell decodes mRNA to build a specific protein. It occurs in three main stages: Initiation, Elongation, and Termination.
Here are the basic steps that must occur, with details for each.
Overview of Translation
Goal: To accurately convert the nucleotide sequence of mRNA into the amino acid sequence of a polypeptide chain.
Key Players:
mRNA: The genetic blueprint.
Ribosome: The molecular machine that catalyzes protein synthesis.
tRNA: The adapter molecule that brings the correct amino acid.
Amino Acids: The building blocks of the protein.
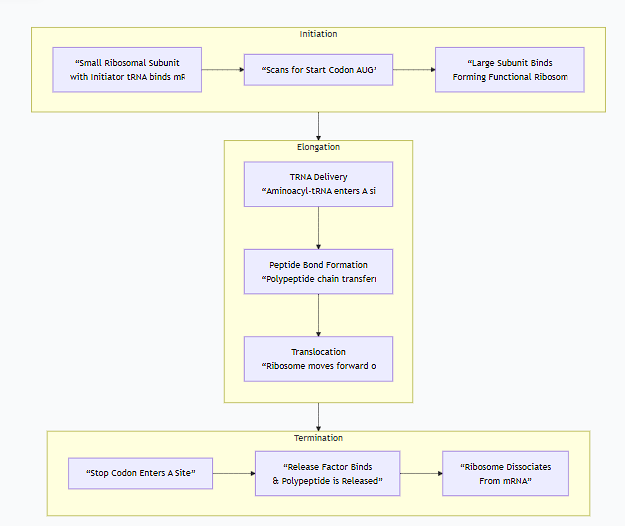
The following flowchart provides a high-level overview of the entire translation process, from initiation to a finished protein:
Stage 1: Initiation
Goal: To correctly assemble the ribosome on the mRNA start codon.
Detailed Steps (in Eukaryotes):
Small Subunit Binding: The small ribosomal subunit, loaded with a special initiator tRNA (carrying methionine, tRNA<sub>i</sub><sup>Met</sup>), binds to the 5' cap of the mRNA.
Scanning: The small subunit "scans" the mRNA in the 5' to 3' direction, moving codon by codon.
Start Codon Recognition: It stops scanning when the initiator tRNA's anticodon base-pairs with the AUG start codon on the mRNA. This sets the reading frame for the rest of the translation.
Large Subunit Joining: The large ribosomal subunit then binds, forming a functional ribosome. The initiator tRNA is sitting in the P (Peptidyl) site. The A (Aminoacyl) site is empty and ready for the next tRNA.
Stage 2: Elongation
Goal: To sequentially add amino acids to the growing polypeptide chain in the correct order.
This is a cyclic, three-step process that repeats for each new amino acid. The following diagram illustrates this codon-by-codon elongation cycle:
Detailed Steps (Elongation Cycle):
Codon Recognition (Delivery):
An incoming aminoacyl-tRNA (a tRNA with its specific amino acid attached) carrying the correct anticodon for the mRNA codon in the A site is delivered by elongation factor EF-Tu.
The ribosome checks for proper codon-anticodon pairing to ensure accuracy.
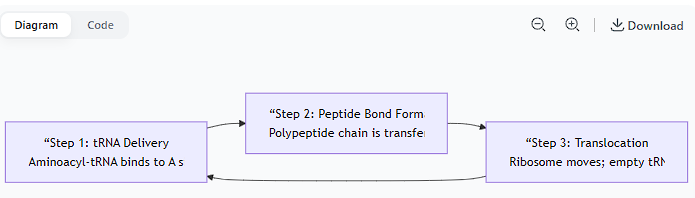
Peptide Bond Formation:
The ribosome's peptidyl transferase activity (catalyzed by rRNA in the large subunit) catalyzes the formation of a peptide bond between the amino acid in the P site and the amino acid in the A site.
This transfers the entire growing polypeptide chain from the tRNA in the P site to the aminoacyl-tRNA in the A site.
Translocation:
The ribosome moves exactly one codon downstream (3') along the mRNA. This shift is powered by the elongation factor EF-G.
This movement has three consequences:
The now empty tRNA is moved from the P site to the E (Exit) site.
The tRNA carrying the growing polypeptide chain is moved from the A site to the P site.
The A site is emptied and is now over the next mRNA codon, ready for the next aminoacyl-tRNA to bind.
The empty tRNA in the E site is released.
This cycle repeats for each codon in the mRNA.
Stage 3: Termination
Goal: To release the finished polypeptide chain and disassemble the translation machinery.
Detailed Steps:
Stop Codon Recognition: Elongation continues until a stop codon (UAA, UAG, or UGA) enters the A site. There is no tRNA with an anticodon for these codons.
Release Factor Binding: A protein called a release factor (RF) binds to the stop codon in the A site.
Polypeptide Release: The release factor triggers the peptidyl transferase activity to hydrolyze (cut) the bond between the completed polypeptide and the tRNA in the P site. This releases the finished protein.
Ribosome Recycling: The ribosome dissociates into its large and small subunits, releasing the mRNA and the final tRNA. These components can now be used for another round of translation.
Summary Table of Key Steps
Stage | Key Event | Molecular Action |
|---|---|---|
Initiation | Ribosome assembles on start codon. | Small subunit + initiator tRNA (Met) find AUG; large subunit joins. |
Elongation | Codon Recognition | Correct aminoacyl-tRNA binds to A site. |
Peptide Bond Formation | Polypeptide chain is transferred to new amino acid. | |
Translocation | Ribosome moves; tRNAs shift sites (A→P→E); empty tRNA ejected. | |
Termination | Stop codon is reached. | Release factor binds; protein is released; ribosome dissociates. |
Identify the structure of tRNA molecules and the impact of structure on function.
The structure of a tRNA molecule is a classic example of how form dictates function in molecular biology. Its unique shape is perfectly designed for its essential role as the molecular "adapter" in translation.
The Structure of tRNA Molecules
The structure of tRNA can be understood at four levels, but its final functional form is its three-dimensional shape.
1. Primary Structure (The Sequence)
This is the linear sequence of 73-93 ribonucleotides. Some of these bases are highly modified post-transcriptionally (see below).
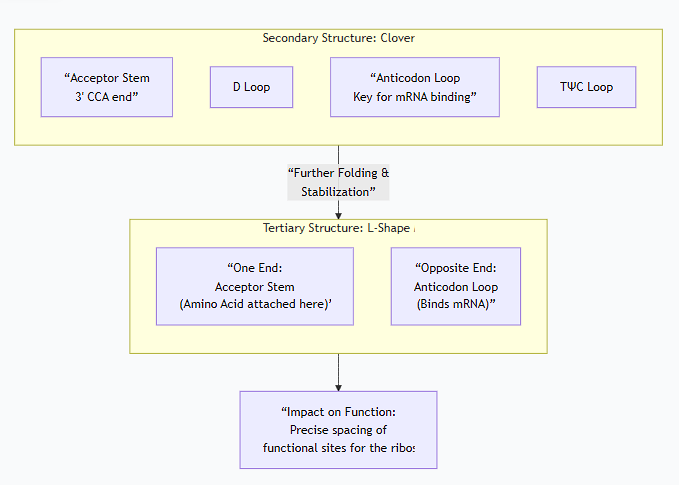
2. Secondary Structure (The 2D "Cloverleaf")
The molecule folds into a characteristic "cloverleaf" pattern held together by hydrogen bonds between complementary base pairs. This structure consists of four (sometimes five) double-helical stems and three loops.
Acceptor Stem: The 3' end of the tRNA. It always ends with the sequence CCA-3'OH. The amino acid is covalently attached to the terminal adenosine (A) of this stem. This is the "loading dock" for the amino acid.
D Loop (Dihydrouridine Loop): Contains the modified base dihydrouridine. It is important for the proper folding and stability of the tRNA.
Anticodon Loop: This is the most critical loop for function. It contains the anticodon, a sequence of three nucleotides that base-pairs with the complementary codon on the mRNA during translation.
TΨC Loop: Contains a conserved sequence of ribothymidine (T), pseudouridine (Ψ), and cytidine (C). This loop is crucial for binding the tRNA to the ribosome.
3. Tertiary Structure (The 3D "L-Shape")
The 2D cloverleaf folds further into a compact, inverted L-shaped molecule. This is the functional form of tRNA.
This folding is stabilized by hydrogen bonds between nucleotides in the D and TΨC loops.
In this L-shape:
The Acceptor Stem (with its amino acid) is at one end.
The Anticodon Loop is at the opposite end.
The following diagram illustrates the relationship between the secondary cloverleaf structure and the final, functional tertiary L-shape:
The Impact of Structure on Function
The unique L-shaped structure is absolutely essential for tRNA's role as an adapter. It optimizes the molecule for interaction with the ribosome and its partners.
1.Precise Positioning in the Ribosome
The ribosome has three binding sites: A, P, and E. The L-shape allows a single tRNA to span two of these sites simultaneously.
The acceptor stem (one end of the 'L') interacts with the peptidyl transferase center in the large ribosomal subunit, where the peptide bond is formed.
The anticodon loop (the other end of the 'L') interacts with the mRNA codon in the small ribosomal subunit.
This precise distance and orientation are critical for the ribosome to hold the mRNA, the growing peptide chain, and the incoming amino acid in perfect alignment for catalysis.
2.Accurate Codon Recognition
The structure of the anticodon loop presents the three-nucleotide anticodon in a way that allows it to "read" the mRNA codon via Watson-Crick base pairing.
This ensures the correct amino acid is incorporated into the growing protein chain according to the genetic code.
3.Interaction with Key Enzymes
Aminoacyl-tRNA Synthetases: Each of these 20 enzymes recognizes and attaches a specific amino acid to its correct tRNA. They do this by binding to specific structural features of the tRNA, including the anticodon loop and the D loop. This ensures the high fidelity of the genetic code; the right amino acid is matched to the right tRNA (and thus the right mRNA codon).
Elongation Factors (e.g., EF-Tu): These proteins that deliver aminoacyl-tRNA to the ribosome also recognize the overall L-shape and specific structural elements of the tRNA.
4.Role of Modified Bases
The numerous post-transcriptional base modifications (e.g., Dihydrouridine, Pseudouridine, Inosine) are not just decorative. They are crucial for function:
Stability and Folding: They facilitate the precise folding of the tRNA into its L-shape and stabilize the structure.
Wobble Base Pairing: Modifications in the anticodon (especially the first position, I-34) allow a single tRNA to recognize more than one codon, explaining the degeneracy (redundancy) of the genetic code. This is known as the wobble hypothesis.
Accurate Reading: They prevent misreading of the codon and ensure the tRNA is correctly positioned on the ribosome.
Summary Table: Structure-Function Relationship
Structural Feature | Functional Impact |
|---|---|
Acceptor Stem (3' CCA end) | Site of amino acid attachment. The amino acid is held far from the anticodon, preventing steric interference. |
Anticodon Loop | Binds to the mRNA codon. Ensures the correct genetic information is read. |
L-Shaped 3D Structure | Fits precisely into the ribosome's A, P, and E sites. Allows the acceptor end and anticodon end to interact simultaneously with the large and small ribosomal subunits. |
D Loop and TΨC Loop | Critical for proper folding, stability, and recognition by aminoacyl-tRNA synthetases and elongation factors. |
Modified Bases | Promote folding, stability, and accurate codon recognition via wobble pairing. |
In conclusion, the elegant L-shaped structure of tRNA is a masterpiece of molecular evolution. It perfectly spaces its two critical business ends—the amino acid attachment site and the anticodon—allowing this small molecule to serve as the essential physical link between the language of nucleic acids and the language of proteins.
Describe the contribution of each step to the quality control of protein production.
Quality control in protein production is not a single step but a series of rigorous checks integrated throughout the entire process, from gene to finished protein. Each step contributes to ensuring the correct protein is made in the right amount, at the right time, and in the right form.
Here is a description of the contribution of each key step to the quality control of protein production.
1. Transcription & RNA Processing: Controlling the Blueprint
The first line of defense is ensuring the mRNA template is correct and stable.
Transcription Fidelity: While RNA polymerase has a higher error rate than DNA polymerase, mistakes are less permanently damaging because mRNA is short-lived. However, accurate transcription initiation ensures the correct gene is expressed.
Splicing Accuracy: The spliceosome must precisely remove introns and join exons. Errors can lead to frameshifts or nonfunctional proteins. Alternative splicing is a regulated form of this, generating different protein isoforms from a single gene as needed.
RNA Surveillance (Nonsense-Mediated Decay - NMD): This is a major QC checkpoint. If a splicing error or mutation creates a premature stop codon (nonsense codon), the NMD pathway detects and destroys the faulty mRNA before it can produce a truncated, potentially toxic protein.
Contribution: This stage ensures that only a correct, full-length, and stable mRNA blueprint is sent to the cytoplasm for translation.
2. tRNA Charging: Ensuring the Correct Adapter
This is arguably the most critical step for the accuracy of the genetic code itself.
Aminoacyl-tRNA Synthetases: Each of these 20 enzymes is responsible for covalently attaching a specific amino acid to its correct tRNA.
Double-Sieve Mechanism: Many synthetases have a double-check system:
Coarse Sieve: Rejects amino acids that are larger than the correct one.
Fine Sieve: Hydrolyzes (destroys) any incorrectly attached amino acid that is similar in size but not identical to the correct one.
Proofreading: The enzyme actively checks the amino acid in its active site and will remove it if it is incorrect.
Contribution: This step guarantees that each tRNA adapter is carrying its cognate (correct) amino acid. A mistake here would directly incorporate the wrong amino acid into the protein, as the ribosome only reads the tRNA's anticodon.
3. Translation (Ribosome Function): Accurate Decoding and Elongation
The ribosome itself is a sophisticated QC machine during the elongation phase.
Codon-Anticodon Proofreading: When an aminoacyl-tRNA enters the A site, the ribosome checks if its anticodon matches the mRNA codon. The initial binding is loose; only correct pairing induces a conformational change that tightly grips the tRNA and catalyzes peptide bond formation.
Kinetic Proofreading: Incorrect tRNAs bind more weakly and dissociate from the A site more quickly than correct ones. The ribosome uses this time delay (requiring GTP hydrolysis by EF-Tu) as a window of opportunity to reject mismatched tRNAs before the peptide bond is formed.
Contribution: The ribosome ensures that only tRNAs with the correct anticodon are used for protein synthesis, adding the amino acid specified by the mRNA sequence.
4. Cotranslational Folding and Chaperone Assistance
Quality control continues as the protein is being synthesized.
Ribosome as a Chaperone: The ribosome tunnel and exit site help prevent the nascent polypeptide from aggregating or misfolding prematurely.
Molecular Chaperones (e.g., Hsp70): These proteins bind to the hydrophobic regions of the nascent chain as it exits the ribosome, preventing improper interactions and allowing the protein to fold into its correct native structure.
Signal Recognition Particle (SRP): For secreted or membrane proteins, SRP recognizes a signal sequence and halts translation until the ribosome is targeted to the endoplasmic reticulum (ER). This ensures these proteins are folded in the correct compartment.
Contribution: Prevents the production of misfolded and aggregated proteins, which are often nonfunctional and can be toxic to the cell.
5. Post-Translational Quality Control
The final checks occur after the protein is fully synthesized.
ER Quality Control: In the ER, proteins are scrutinized. Correctly folded proteins are allowed to proceed to the Golgi. Misfolded proteins are identified by chaperones like Calnexin/Calreticulin and given chances to refold.
The Ubiquitin-Proteasome System (UPS): Proteins that fail to fold correctly in the cytosol or ER are tagged with a chain of ubiquitin molecules. This tag marks them for destruction by a large protease complex called the proteasome, which degrades them into small peptides.
Autophagy: Large protein aggregates or damaged organelles can be engulfed by a double-membrane vesicle (autophagosome) and delivered to the lysosome for degradation.
Contribution: This is the "final exam." It eliminates irreversibly misfolded proteins, preventing the accumulation of cellular "junk" that can disrupt function and lead to disease (e.g., Alzheimer's, Cystic Fibrosis).
Summary Table of Quality Control Steps
Stage | Key Quality Control Mechanism | Consequence of Failure |
|---|---|---|
mRNA Production | Nonsense-Mediated Decay (NMD) | Production of truncated, dysfunctional proteins. |
tRNA Charging | Aminoacyl-tRNA Synthetase Proofreading | Mis-incorporation of wrong amino acids into the protein sequence. |
Translation | Ribosomal Kinetic Proofreading | Misreading of the genetic code; incorrect protein sequence. |
Folding | Molecular Chaperones (Hsp70, etc.) | Protein misfolding, aggregation, and toxicity. |
Final Check | Ubiquitin-Proteasome System & ERAD | Accumulation of toxic protein aggregates and cellular dysfunction. |
In summary, quality control in protein production is a multi-layered, energy-intensive process. From verifying the mRNA blueprint to destroying faulty final products, each step adds a crucial layer of scrutiny to ensure the proteome remains functional and the cell remains healthy.
Describe the structure of ribosomes and know the function of ribosomes in protein synthesis.
The ribosome is one of the most complex and ancient molecular machines in the cell, responsible for the crucial task of protein synthesis (translation).
The Structure of Ribosomes
Ribosomes are not membrane-bound organelles. They are massive complexes of ribosomal RNA (rRNA) and proteins. Their structure is organized around a core of catalytic RNA, with proteins stabilizing the complex.
1. Composition: The "Ribozyme"
Made of rRNA and Protein: In eukaryotes, a ribosome is about 60% rRNA and 40% protein. The rRNA is not just structural; it is directly responsible for the key catalytic activity.
Two Subunits: Ribosomes consist of a large subunit and a small subunit that fit together. These subunits are assembled in the nucleus (in eukaryotes) and join in the cytoplasm during protein synthesis.
Ribosome Type | Prokaryotic (e.g., E. coli) | Eukaryotic (e.g., Human) |
|---|---|---|
Overall Size | 70S | 80S |
Large Subunit | 50S (contains 23S & 5S rRNA + proteins) | 60S (contains 28S, 5.8S, & 5S rRNA + proteins) |
Small Subunit | 30S (contains 16S rRNA + proteins) | 40S (contains 18S rRNA + proteins) |
(Note: The "S" stands for Svedberg unit, a measure of sedimentation rate that depends on both mass and shape, which is why the subunit values don't add up to the whole.)
2. Key Functional Sites
The ribosome has three primary binding sites for tRNA molecules, which are essential for its function. These sites are formed at the interface between the large and small subunits.
A Site (Aminoacyl-tRNA site): This is the entry site for the incoming Aminoacyl-tRNA carrying the next amino acid to be added to the chain.
P Site (Peptidyl-tRNA site): This site holds the Peptidyl-tRNA—the tRNA that is carrying the growing polypeptide chain.
E Site (Exit site): This is the Exit site for the deacylated tRNA (the empty tRNA) after it has donated its amino acid to the chain.
Additionally, the large subunit contains the peptidyl transferase center, which is the actual site of peptide bond formation.
The Function of Ribosomes in Protein Synthesis
The ribosome's function is to accurately and efficiently decode an mRNA molecule to synthesize a polypeptide chain. It acts as the stage where mRNA, tRNA, and amino acids come together.
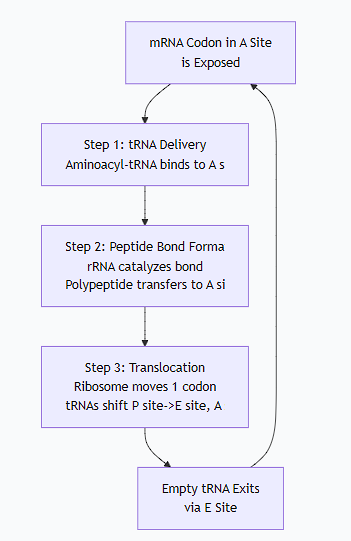
The following diagram illustrates the core functional cycle of the ribosome during the elongation phase of translation:
Detailed Ribosome Functions:
mRNA Binding and Reading Frame Maintenance:
The small ribosomal subunit binds to the mRNA and ensures it is read in the correct groups of three nucleotides (codons). The interaction between the 16S/18S rRNA and the mRNA's Shine-Dalgarno sequence (in prokaryotes) or 5' cap (in eukaryotes) ensures proper start site selection.
tRNA Binding and Decoding:
The ribosome provides the A, P, and E sites that hold the tRNAs in precise orientation. It facilitates the base-pairing between the mRNA codon in the A site and the tRNA anticodon, ensuring the correct amino acid is added.
Catalysis of Peptide Bond Formation:
This is the ribosome's most critical chemical function. The peptidyl transferase activity is catalyzed not by a protein, but by the 23S/28S rRNA in the large subunit. This makes the ribosome a ribozyme. It catalyzes the nucleophilic attack by the amino group in the A site on the carbonyl carbon of the amino acid in the P site, forming a new peptide bond.
Translocation:
After the peptide bond is formed, the ribosome must move to the next codon. Using energy from GTP hydrolysis (via elongation factor EF-G), the ribosome shifts precisely one codon downstream. This moves the tRNAs from the A site to the P site, and from the P site to the E site, ejecting the deacylated tRNA from the E site.
Ensuring Fidelity:
The ribosome has built-in proofreading mechanisms. It can detect incorrect codon-anticodon pairs and reject the wrong tRNA before the peptide bond is formed, ensuring the accuracy of the protein sequence.
Interaction with Factors:
The ribosome has specific binding sites for initiation, elongation, and release factors. These protein factors are essential for the ribosome to start, elongate efficiently, and correctly terminate the polypeptide chain.
In summary, the ribosome is a magnificently engineered machine. Its structure—composed of two subunits with specific rRNA cores and functional sites—is perfectly designed for its function: to read the genetic code in mRNA and use it to catalyze the assembly of amino acids into a protein chain with high speed and accuracy.
List the translation factors (other proteins) and their roles in initiation, elongation and termination.
Translation factors are essential proteins that guide the ribosome through the initiation, elongation, and termination phases of protein synthesis. They ensure the process is accurate, efficient, and properly regulated.
Here is a list of the key translation factors and their roles, categorized by the stage in which they act.
1. Initiation Factors
Initiation factors are responsible for assembling the ribosomal complex at the correct start codon on the mRNA. This process is more complex in eukaryotes, which is reflected in the greater number of factors.
Prokaryotic (Bacterial) Initiation Factors
Factor | Role |
|---|---|
IF1 | Binds to the A site of the small ribosomal subunit, blocking it and ensuring the initiator tRNA starts in the P site. |
IF2 | A GTP-binding protein that recruits the initiator tRNA (fMet-tRNA<sup>fMet</sup>) to the P site of the small subunit. |
IF3 | Binds to the small subunit, preventing the large subunit from binding prematurely. It also helps check the accuracy of the initiator tRNA binding. |
Eukaryotic Initiation Factors (eIFs)
Eukaryotic initiation involves many more factors to handle the 5' cap, scan for the start codon, and deal with a more complex cellular environment.
Factor | Role |
|---|---|
eIF1 & eIF1A | Promote scanning and ensure the ribosome starts at the correct AUG codon. eIF1 helps in start codon selection. |
eIF2 | Crucial GTP-binding protein. In its active GTP-bound form, it delivers the initiator tRNA (Met-tRNA<sup>Met</sup>) to the small ribosomal subunit. This is a major regulatory point for global protein synthesis. |
eIF3 | The largest factor. Binds to the small subunit, preventing large subunit association, and promotes the binding of other factors. |
eIF4F Complex | The "Cap-Binding Complex." Binds to the 5' cap of the mRNA and prepares it for recruitment to the ribosome. It consists of: |
→ eIF4E | Directly binds to the 5' cap. |
→ eIF4G | Acts as a scaffolding protein, bridging the mRNA, the ribosome, and other factors. |
→ eIF4A | An RNA helicase that unwinds secondary structures in the 5' UTR of the mRNA, allowing the ribosome to scan. |
eIF5 | A GAP (GTPase-Activating Protein) for eIF2. It triggers the hydrolysis of eIF2-bound GTP to release initiation factors after the start codon is found. |
eIF5B | A GTPase that catalyzes the final joining of the large (60S) ribosomal subunit to form the complete 80S ribosome. |
2. Elongation Factors
Elongation factors are highly conserved between prokaryotes and eukaryotes. They facilitate the cyclic process of adding amino acids to the growing polypeptide chain.
Factor (Prokaryotic / Eukaryotic) | Role |
|---|---|
EF-Tu / eEF1A | The "Delivery Driver." This is a GTP-binding protein that forms a complex with aminoacyl-tRNA. It delivers the correct aminoacyl-tRNA to the A site of the ribosome. Its GTPase activity provides a proofreading step; incorrect tRNAs dissociate before GTP is hydrolyzed. |
EF-Ts / eEF1B | The "Recycler." Acts as a Guanine Nucleotide Exchange Factor (GEF) for EF-Tu/eEF1A. It promotes the exchange of spent GDP for fresh GTP, reactivating EF-Tu/eEF1A for another round of delivery. |
EF-G / eEF2 | The "Motor." This GTP-binding protein catalyzes translocation. After peptide bond formation, EF-G binds to the ribosome and, using GTP hydrolysis as an energy source, pushes the ribosome one codon forward. This moves the tRNAs from the A→P site and P→E site. |
3. Termination Factors (Release Factors)
Termination factors recognize the stop codons and trigger the release of the finished polypeptide chain.
Factor (Prokaryotic / Eukaryotic) | Role |
|---|---|
RF1 / eRF1 | Recognizes the stop codons UAA and UAG and triggers polypeptide chain release. |
RF2 / eRF1 | Recognizes the stop codons UAA and UGA and triggers polypeptide chain release. |
RF3 / eRF3 | A GTP-binding protein that stimulates the activity of RF1/RF2 (in bacteria) or eRF1 (in eukaryotes). It helps release the class-1 release factor (RF1/eRF1) from the ribosome after termination. |
- / Rli1 (ABCE1) | In eukaryotes, this ATP-binding protein is essential for ribosome recycling, working alongside eRF1 to split the ribosome into its subunits. |
Ribosome Recycling Factor (RRF)
Prokaryotes: After termination, RRF and EF-G bind to the post-termination ribosome to split the large and small subunits apart, releasing the mRNA and deacylated tRNA. This allows all components to be reused.
Eukaryotes: The process is less defined but involves eRF1, eRF3, and Rli1 (ABCE1).
Summary Table
Stage | Key Factor(s) | Primary Function |
|---|---|---|
Initiation | eIF2 / IF2 | Deliver the initiator tRNA to the small ribosomal subunit. |
eIF4F Complex | Bind the 5' cap of mRNA and unwind secondary structure. | |
Elongation | EF-Tu / eEF1A | Deliver aminoacyl-tRNA to the A site. |
EF-G / eEF2 | Catalyze translocation of the ribosome. | |
Termination | RF1, RF2 / eRF1 | Recognize stop codons and catalyze release of the polypeptide. |
Recycling | RRF & EF-G | Split the ribosome into its subunits for reuse. |
In essence, translation factors are the dedicated managers of the protein synthesis assembly line. They ensure that the ribosome—the production machine—is correctly set up, supplied with the right parts (tRNAs), moved along the instructions (mRNA), and properly shut down and reset once the product (protein) is complete.
Calculate the energy required for protein synthesis.
Calculating the energy required for protein synthesis is a fundamental concept in biochemistry. The process is highly energy-intensive, and the cost is almost always counted in molecules of ATP (and GTP, which is energetically equivalent to ATP).
The total energy required can be broken down into the costs for each major step: amino acid activation, initiation, elongation, and termination.
Step 1: Amino Acid Activation (Charging the tRNA)
This is the first and most straightforward energy cost.
Process: Attaching an amino acid to its corresponding tRNA.
Reaction:
Amino Acid + tRNA + ATP → Aminoacyl-tRNA + AMP + PPᵢEnergy Cost: 2 ATP equivalents per amino acid.
1 ATP is hydrolyzed to AMP, which is equivalent to 2 ATP because it takes 2 high-energy phosphate bonds to convert AMP back to ATP (AMP → ADP → ATP).
The pyrophosphate (PPᵢ) is rapidly hydrolyzed by pyrophosphatase into two inorganic phosphates (2Pᵢ), releasing heat and making this step irreversible.
Cost so far: 2 ATP per amino acid in the protein.
Step 2: Initiation
The energy cost here differs between prokaryotes and eukaryotes.
In Prokaryotes (Bacteria):
Process: Assembling the ribosome at the start codon with the initiator tRNA.
Energy Cost: 1 GTP is hydrolyzed by initiation factor IF2.
Total: 1 ATP equivalent.
In Eukaryotes:
Process: A more complex process involving scanning the mRNA for the start codon.
Energy Cost: The hydrolysis of 1 ATP by the eIF4A helicase (to unwind mRNA secondary structure) is often included in the minimal count, though other GTP hydrolysis events (eIF2, eIF5B) also occur.
Total (Minimal Estimate): 1 ATP equivalent.
Cost so far: +1 ATP for initiation (regardless of protein length).
Step 3: Elongation (The Cyclic, Repetitive Cost)
This is where the bulk of the energy is spent, as it occurs for every single amino acid added after the first one.
For each elongation cycle (adding one amino acid), two key GTP hydrolysis events occur:
Delivery of Aminoacyl-tRNA (by EF-Tu in prokaryotes / eEF1A in eukaryotes):
Cost: 1 GTP per amino acid added.
Translocation (by EF-G in prokaryotes / eEF2 in eukaryotes):
Cost: 1 GTP per amino acid added.
Cost per amino acid during elongation: 2 ATP equivalents.
Step 4: Termination
Process: Releasing the finished polypeptide chain.
Energy Cost: In prokaryotes, termination factor RF3 hydrolyzes 1 GTP. In eukaryotes, eRF3 hydrolyzes 1 GTP.
Total: 1 ATP equivalent per protein synthesized.
The Calculation: Putting It All Together
Let's calculate the cost to synthesize a protein with n amino acids.
Total Energy = Cost of Activation + Cost of Initiation + Cost of Elongation + Cost of Termination
Activation:
namino acids × 2 ATP/aa =2nATPInitiation:
1ATPElongation: We add
(n - 1)amino acids during elongation (the first amino acid is already placed during initiation).(n - 1)amino acids × 2 ATP/aa =2(n - 1)ATP
Termination:
1ATP
Total ATP = 2n + 1 + 2(n - 1) + 1
Let's simplify this equation:Total ATP = 2n + 1 + 2n - 2 + 1Total ATP = 4n + 0
The Final Formula: Total Energy Cost ≈ 4n ATP Equivalents
Where n is the number of amino acids in the protein.
Example Calculation
How much energy is required to synthesize a 300-amino acid protein?
Using the formula:
4 × 300 = 1200 ATP equivalents.
Breakdown:
Activation: 300 aa × 2 ATP = 600 ATP
Initiation: 1 ATP
Elongation: 299 aa × 2 ATP = 598 ATP
Termination: 1 ATP
Total: 600 + 1 + 598 + 1 = 1200 ATP
Summary Table
Step | Energy Cost (in ATP equivalents) |
|---|---|
Amino Acid Activation | 2 per amino acid ( |
Initiation | 1 |
Elongation | 2 per amino acid added during elongation ( |
Termination | 1 |
TOTAL | ~4 per amino acid in the final protein ( |
Important Considerations
This is a Minimal Estimate: This calculation (
4n) is the widely accepted textbook figure. In reality, additional energy might be spent on proofreading (extra GTP hydrolysis by EF-Tu for incorrect tRNAs), mRNA unwinding, and error correction systems, which could push the cost slightly higher.GTP = ATP: For energy accounting, GTP is considered equivalent to ATP because the cell can easily convert one to the other (e.g., NDPK: GDP + ATP ⇌ GTP + ADP).
High Energy Investment: This massive energy expenditure (e.g., 1200 ATP for a small protein) underscores the huge metabolic commitment of protein synthesis. It explains why this process is so tightly regulated by the cell.