Immunology - Lecture 4 - Innate Immunity: Pattern recognition receptors
1/146
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
147 Terms
What are PAMPS
Pathogen associated molecular patterns (PAMPs)
What are the receptors for PAMPs called?
Pattern recognition receptors (PRRs)
Where are PRRs encoded in?
PRRs are genetically encoded and inherited through the germline
Contrast to the random generation of antigen receptors of the adaptive immune system
The specificity of PRRs have been shaped via evolution to recognize PAMPS
What are DAMPs?
Damage associated molecular patterns (DAMPs)
While the cellular perturbations and DAMPs are often the result of infection, they can also be induced in other contexts
Tissue damage or neoplastic transformation
What happens to many microbes after entering tissues?
They are recognized, ingested, and killed by phagocytes.
What cells recognize, ingest, and kill microbes if they cross epithelial barriers?
Resident phagocytic cells such as macrophages and monocytes, granulocytes, and dendritic cells.
What are the main classes of phagocytic cells in the innate immune system?
Macrophages, monocytes, granulocytes, and dendritic cells.
What are macrophages and where are they found?
Macrophages are major phagocytic cells found in most tissues at homeostasis, often in connective tissues
How do macrophages arise?
Macrophages arise from progenitor cells that enter tissues during embryonic development or from circulating monocytes.
What are embryonic progenitors of macrophages and where do they arise from?
Embryonic progenitors arise from the fetal liver, yolk sac, or aorta-gonad-mesonephros (AGM).
What are the names of macrophages in different tissues?
Microglia in neural tissue, Kupffer cells in the liver.
What receptors do human macrophages express, and what is their function?
They express CD14 and CD16 (FcγRIII) and survey for injury to the endothelium.
What are granulocytes and what is their role?
Granulocytes include neutrophils, eosinophils, and basophils, and are involved in pathogen killing.
Which granulocyte has the greatest phagocytic activity and is involved in innate immunity?
Neutrophils have the greatest phagocytic activity and are involved in innate immunity.
Why are macrophages and granulocytes important in innate immunity?
They can recognize, ingest, and destroy pathogens without an adaptive immune response.
What is the third class of phagocytic cells?
Immature dendritic cells.
What are the main types of dendritic cells and what is their function?
Conventional dendritic cells (cDCs) and plasmacytoid dendritic cells (pDCs); both are involved in immune response activation.
What role do cDCs play in immunity?
cDCs generate peptide antigens to activate T cells and induce an adaptive immune response.
What is the role of pDCs?
pDCs produce large amounts of type I interferon, an antiviral interferon.
How do macrophages and neutrophils recognize pathogens?
By means of cell surface pattern recognition receptors (PRRs) that bind pathogen-associated molecular patterns (PAMPs).
What is phagocytosis and how does it occur?
The process by which a cell engulfs a pathogen using its plasma membrane to form a phagosome.
What happens in a phagolysosome?
The phagosome fuses with lysosomes to form a phagolysosome, which becomes acidified and kills the microbe.
How do neutrophils kill microbes?
Neutrophils use cytoplasmic granules containing antimicrobial peptides and enzymes to kill microbes.
What is receptor-mediated endocytosis?
A process by which extracellular material is taken into a cell and degraded.
What is macropinocytosis?
The nonspecific ingestion of large amounts of extracellular fluid and its contents.
What is Dectin-1 and what does it recognize?
Dectin-1 is a C-type lectin expressed by macrophages and neutrophils; it recognizes β(1,3)-linked glycans found in fungal cell walls.
What is the mannose receptor and what does it recognize?
A C-type lectin expressed by macrophages and dendritic cells; recognizes mannosylated ligands on fungi, bacteria, and viruses.
What are scavenger receptors and their types?
Scavenger receptors recognize anionic polymers and lipoproteins; Class A (e.g., SR-A I, SR-A II, MARCO) and Class B (e.g., CD36).
What is CD36 and what does it bind?
CD36 is a Class B scavenger receptor that binds long fatty acid chains.
What receptors are important for macrophage and neutrophil phagocytosis?
Complement receptors and Fc receptors, which facilitate phagocytosis of complement-coated microbes or antibody-coated microbes.
What does G protein-coupled receptor (GPCR) signaling involve?
GPCR signaling involves the following steps:
1. In the inactive state, the G protein's α subunit binds GDP and is associated with the β and γ subunits.
2. Ligand binding to the receptor induces a conformational change, allowing the receptor to bind the G protein, replacing GDP with GTP.
3. GTP binding triggers the dissociation of the G protein into the Gα subunit and the Gβγ complex, each of which activates intracellular proteins.
4. The Gα subunit activates GTPases Rac and Rho, while the Gβγ complex activates GTPase Cdc42, leading to NADPH oxidase assembly and reactive oxygen species (ROS) production.
5. Chemokine signaling similarly activates chemotaxis, leading cells to migrate toward chemokine sources.
6. The response ends when the intrinsic GTPase activity of the Gα subunit hydrolyzes GTP to GDP, and the G protein subunits reassociate.
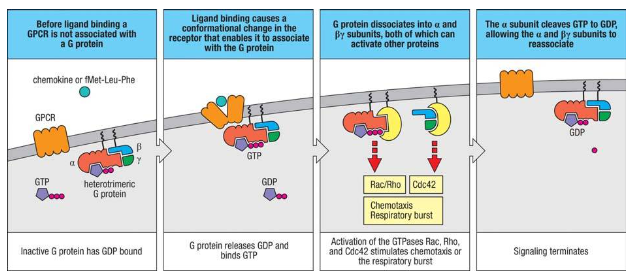
Give me an example of a G protein coupled receptor
fMet-Leu-Phe (fMLF)
What is the target and location of TLR-1:TLR-2 heterodimer?
TLR-1:TLR-2 heterodimer targets lipomannans (Mycobacteria), diacyl and triacyl lipopeptides (bacteria), lipoteichoic acids (gram-positive bacteria), and cell wall beta glucans (fungi). It is located on the cell surface.
What is the target and location of TLR-2:TLR-6 heterodimer?
TLR-2:TLR-6 heterodimer targets lipomannans (Mycobacteria), diacyl and triacyl lipopeptides (bacteria), lipoteichoic acids (gram-positive bacteria), and cell wall beta glucans (fungi). It is located on the cell surface.
What is the target and location of TLR-3?
TLR-3 targets double-stranded RNA (viruses) and poly I:C. It is located in the endosome.
What is the target and location of TLR-4?
TLR-4 targets lipopolysaccharide (LPS) from bacteria. It is located on the cell surface.
What is the target and location of TLR-5?
TLR-5 targets flagellin from bacteria. It is located on the cell surface.
What is the target and location of TLR-7?
TLR-7 targets single-stranded RNA (viruses). It is located in the endosome.
What is the target and location of TLR-8?
TLR-8 targets single-stranded RNA (viruses). It is located in the endosome.
What is the target and location of TLR-9?
TLR-9 targets DNA with unmethylated CpG motifs from bacteria and DNA viruses. It is located in the endosome.
Which cell types express Toll-Like Receptors (TLRs)?
TLRs are expressed on macrophages, dendritic cells, B cells, stromal cells, and epithelial cells.
Which cell types express Toll-Like Receptors (TLRs)?
TLRs are expressed on macrophages, dendritic cells, B cells, stromal cells, and epithelial cells.
What are TLR-1, TLR-2, and TLR-6, and what do they recognize?
TLR-1, TLR-2, and TLR-6 are cell surface receptors that recognize lipoteichoic acid, diacyl, and triacyl lipoproteins found in both gram-positive and gram-negative bacteria.
What induces the formation of TLR-2 heterodimers?
Ligand binding induces the formation of heterodimers of TLR-2 with TLR-1 or TLR-2 with TLR-6.
Where is TLR-5 expressed, and what does it recognize?
TLR-5 is expressed on the cell surface of certain macrophages, dendritic cells, and intestinal epithelial cells, and it recognizes flagellin.
What activates TLR-5?
TLR-5 is activated by monomeric flagellin, which is shed during flagellar assembly or when flagella are degraded.
Which TLRs recognize nucleic acids, and where are they located?
TLR-3, TLR-7, TLR-8, and TLR-9 recognize nucleic acids and are located in the endosome.
What role does the acidic endosome lumen play in TLR recognition?
The acidic endosome lumen contains proteases that degrade internalized microbes, releasing nucleic acids for recognition by TLRs.
What are the similarities between TLR-7 and TLR-8?
TLR-7 and TLR-8 share very similar amino acid sequences and are both activated by degradation products of single-stranded RNA.
What type of nucleic acids do TLR-7 and TLR-8 bind?
TLR-7 and TLR-8 bind to small RNA fragments released when viruses are degraded in the endosome.
What does TLR-9 recognize, and why is unmethylated CpG important?
TLR-9 recognizes single-stranded DNA with unmethylated CpG dinucleotides, which limits the potential for recognizing self DNA.
What kind of DNA does TLR-9 recognize, and what must happen for recognition?
TLR-9 recognizes single-stranded DNA, which must be melted from double-stranded form for recognition.
Which cells express TLR-3, and what does it recognize?
TLR-3 is expressed by macrophages, conventional dendritic cells, and intestinal epithelial cells, and it recognizes double-stranded RNA (dsRNA).
How is TLR-3 activated by double-stranded RNA (dsRNA)?
TLR-3 is activated by dsRNA after direct endocytosis of viruses with dsRNA genomes or by phagocytosis of dying cells in which viruses are replicating.
What is required for TLR-3, TLR-7, TLR-8, and TLR-9 to reach the endosome?
TLR-3, TLR-7, TLR-8, and TLR-9 must interact with a specific protein, UNC93B1, to reach the endosome.
What does TLR-4 recognize, and which accessory proteins are involved?
TLR-4 recognizes bacterial lipopolysaccharide (LPS) in association with host accessory proteins MD-2 and CD14.
What role do MD-2 and CD14 play in TLR-4 signaling?
MD-2 binds to TLR-4 within the cell and is necessary for the correct trafficking of TLR-4 to the cell surface and for recognition of LPS, while CD14 assists in binding LPS.
How does TLR-4 recognize lipopolysaccharide (LPS)?
TLR-4 recognizes LPS using the ectodomain and accessory protein MD-2, which binds five lipid chains of LPS.
What happens when TLR-4:MD-2 complex encounters LPS?
When TLR-4:MD-2 complex encounters LPS, five lipid chains bind to MD-2, and the sixth chain binds to a second TLR-4 ectodomain, inducing dimerization.
What transcription factors do TLRs activate?
TLRs activate NF-kB, AP-1, and IRF transcription factors.
What are the roles of NF-kB, AP-1, and IRF in TLR signaling?
NF-kB and AP-1 primarily induce the expression of proinflammatory cytokines and chemotactic factors, while IRFs induce type 1 interferon production.
What is the role of NF-kB in TLR signaling?
NF-kB generates proinflammatory cytokines like TNF-alpha, IL-1b, and IL-6, and recruits neutrophils, monocytes, and dendritic cells to the site of infection.
What is the role of IRF in TLR signaling?
IRFs, especially IRF3 and IRF7, are important for inducing antiviral type 1 interferon, while IRF5 is involved in proinflammatory cytokine production.
Which transcription factors are involved in inducing proinflammatory cytokines?
NF-kB and AP-1 are involved in inducing proinflammatory cytokines.
Which IRFs are important for inducing type 1 interferon, and what is their role?
IRF3 and IRF7 are important for inducing antiviral type 1 interferon, which helps the body respond to viral infections.
What is the function of IRF5?
IRF5 is involved in the production of proinflammatory cytokines.
What cytokines are generated by NF-kB activation?
NF-kB generates cytokines such as TNF-alpha, IL-1b, and IL-6.
What role does NF-kB play in immune cell recruitment?
NF-kB recruits immune cells like neutrophils, monocytes, and dendritic cells to the site of infection.
How does TLR-3 activate IRFs?
TLR-3 uses its cytoplasmic TIR domain to activate IRFs.
How does TLR-4 activate IRFs, and how does it compare to TLR-3?
TLR-4 also uses the TRIF pathway to activate IRF3, but it is weaker compared to TLR-3.
Which TLRs use MyD88-dependent signaling to activate IRFs?
TLR-7, TLR-8, and TLR-9 use MyD88-dependent signaling to activate IRFs and induce type I interferon.
What is the collective ability of TLRs in activating IRFs and NF-kB?
The collective ability of TLRs to activate both IRFs and NF-kB allows them to stimulate either antiviral or antibacterial responses as needed.
What do RIG-I-like receptors (RLRs) detect, and what do they activate?
RLRs detect cytoplasmic viral RNAs and activate MAVS to induce type I interferon production and pro-inflammatory cytokines.
How do RLRs detect viral RNA?
RLRs bind to viral double-stranded RNA (dsRNA) using an RNA helicase-like domain.
What is the role of RIG-I in RNA detection?
RIG-I detects viral RNA in the cytoplasm by sensing an unmodified 5’-triphosphate group.
How does RIG-I discriminate between host and viral RNA?
RIG-I discriminates between host and viral RNA by sensing differences at the 5’ end of RNA transcripts; eukaryotic RNA is capped with 7-methylguanosine, whereas most viral RNA is not.
What feature of viral RNA does RIG-I detect?
RIG-I detects the lack of a 5’ cap and the presence of an unmodified 5’-triphosphate group.
What happens when RIG-I detects uncapped 5’-triphosphate dsRNA?
Detection of uncapped 5’-triphosphate dsRNA by RIG-I leads to assembly of RIG-I proteins, enabling aggregation of CARDs and interaction with MAVS.
Where is the adaptor protein MAVS located, and what is its role?
MAVS is attached to the mitochondrial outer membrane and interacts with RIG-I to activate downstream signaling.
What transcription factors are activated by RIG-I signaling?
RIG-I signaling activates IRF3 and NF-κB, producing type I interferon and pro-inflammatory cytokines.
What cytokines are produced by RIG-I signaling?
The cytokines produced include TNF-alpha, IL-1, and IL-6.
What is MDA-5, and how is it different from RIG-I?
MDA-5 is similar to RIG-I but senses longer dsRNA and does not require the 5’-triphosphate for recognition.
How do RIG-I and MDA-5 reduce the chance of activating RLRs by cellular RNA?
RIG-I and MDA-5 recognize specific features of dsRNA (5’ triphosphate for RIG-I and longer dsRNA for MDA-5) to reduce the chance of activation by cellular RNA.
What does the cGAS-STING pathway detect, and what does it induce?
The cGAS-STING pathway detects cytosolic DNA and induces the production of type I interferon.
What is the role of type I interferon in antiviral defense?
Type I interferon is antiviral; it inhibits viral replication, degrades viral RNA, prevents viral protein synthesis, enhances innate immunity, activates NK cells, promotes macrophage activity, and activates the JAK-STAT pathway.
What is the function of the JAK-STAT pathway in type I interferon signaling?
The JAK-STAT pathway leads to the transcription of various genes that help establish an antiviral state in infected and neighboring cells.
How do innate sensors discriminate between host and viral cytosolic RNA?
Innate sensors use specific modifications like the 5’cap to discriminate between host and viral cytosolic RNA.
Where is host DNA restricted, and what happens when DNA is introduced into the cytosol?
Host DNA is restricted to the nucleus, but viral and other foreign DNA may be located in the cytosol, where it induces the production of type I interferons.
What is the main sensor of cytosolic DNA, and how does it function?
The main sensor of cytosolic DNA is cGAS (cyclic GAMP synthase), which binds directly to cytosolic DNA and produces the second messenger cGAMP.
What molecule is produced by cGAS, and what does it activate?
cGAS produces cGAMP, which binds and activates the STING pathway.
What is STING, and how is it activated?
STING (Stimulator of Interferon Genes) is a protein localized in the endoplasmic reticulum membrane. It exists as an inactive homodimer until cGAMP binding induces a conformational change.
What happens after STING is activated by cGAMP?
After STING is activated, it recruits TBK1, which activates IRF3, leading to the production of type I interferon, similar to signaling by TLR3 and RLR.
Where does cGAS reside, and what does it detect?
cGAS resides in the cytoplasm and serves as a sensor of double-stranded DNA from viruses.
How does cGAMP activate STING, and what is the downstream effect?
cGAMP binds and activates the STING dimer on the endoplasmic reticulum, leading to the recruitment of TBK1 and activation of IRF3.
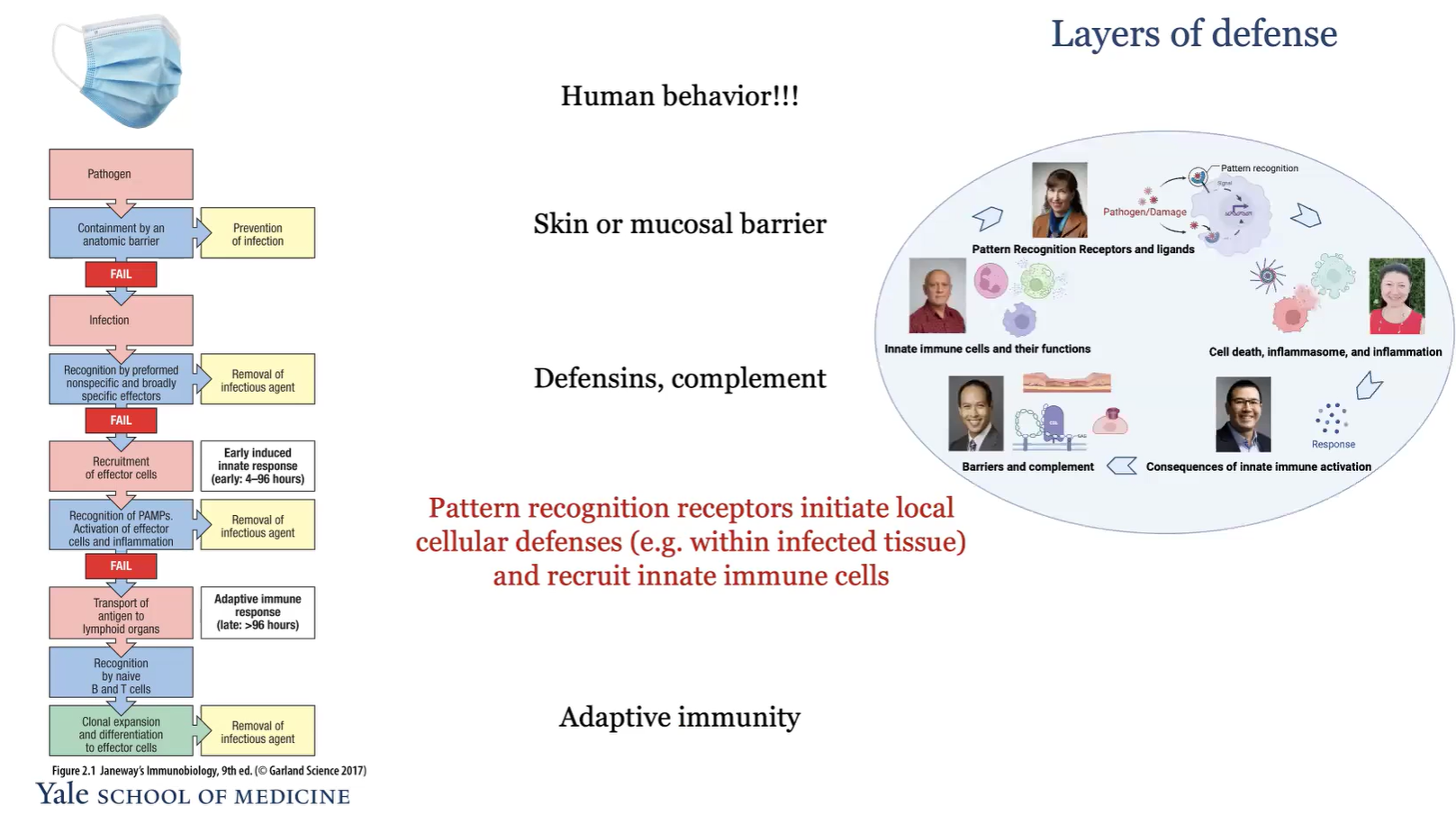
What are the layers of defense?
Human behavior
Skin or mucosal barrier
Defensins, complement
Pattern recognition recpetors initate local cellular defenses and recruit innate immune cells
adaptive immunity

Pathogen associated molecular patterns (PAMPs)
What is fMet-Leu-Phe (fMLF), and why is it important?
fMet-Leu-Phe (fMLF) is a tripeptide that serves as a signal for the presence of bacterial infection. It is recognized by a G protein-coupled receptor (fMLF receptor) on immune cells. fMLF is important because it contains an N-formylmethionine (fMet) residue, which is unique to bacterial polypeptides, allowing immune cells to distinguish bacterial proteins from host proteins. Binding of fMLF to its receptor activates intracellular signaling pathways that enhance the immune response, including the production of reactive oxygen species (ROS) and the migration of immune cells to the site of infection.
What does fMet-Leu-Phe (fMLF) bind to?
fMet-Leu-Phe (fMLF) binds to a G protein-coupled receptor (fMLF receptor) on immune cells.

Nucleic acids, viral nucleic acids are very different
Abnormal RNA structures
For DNA, location is important. DNA should be in the nucleus
Also, human DNA is methylated at CpG, but virus are not
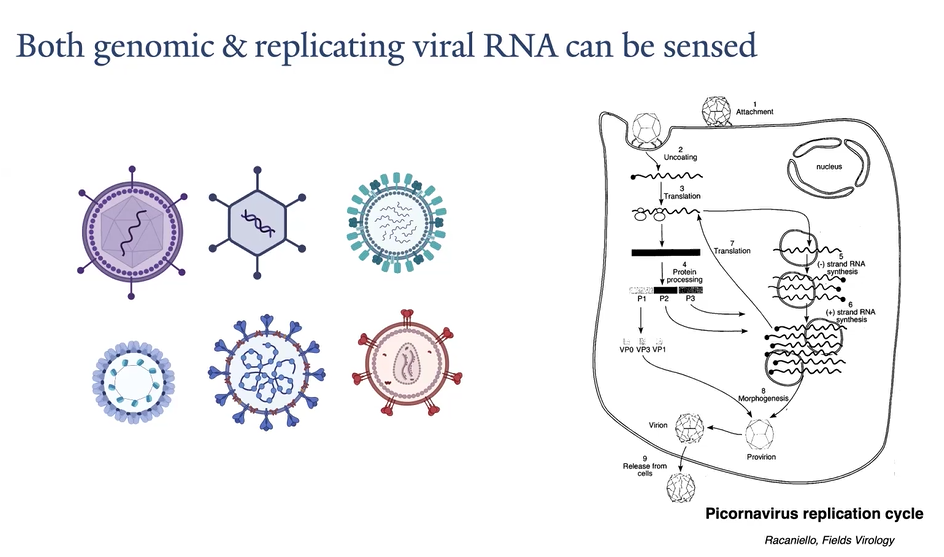
What is the key difference between the 5' end of eukaryotic cell RNA and viral RNA?
Eukaryotic cell RNA is capped on the 5' end to allow translation and prevent degradation, whereas viral RNA may or may not have a conventional cap.