IMED1002 - Chromosome Replication (L13)
1/25
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
26 Terms
DNA and Chr replication is restricted to
S phase of the cell cycle and proceeds in two directions
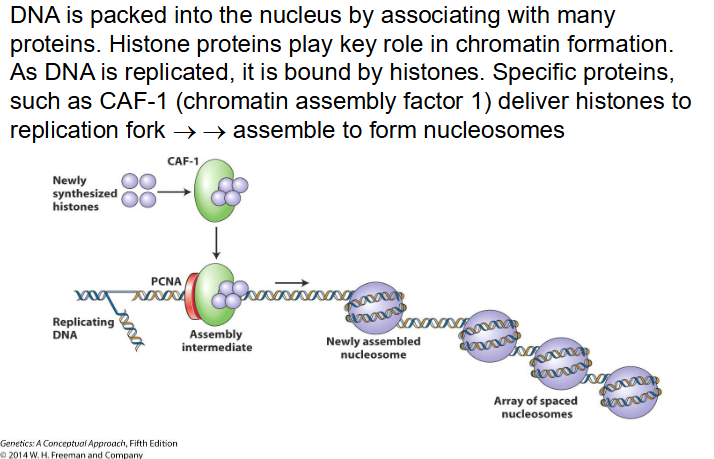
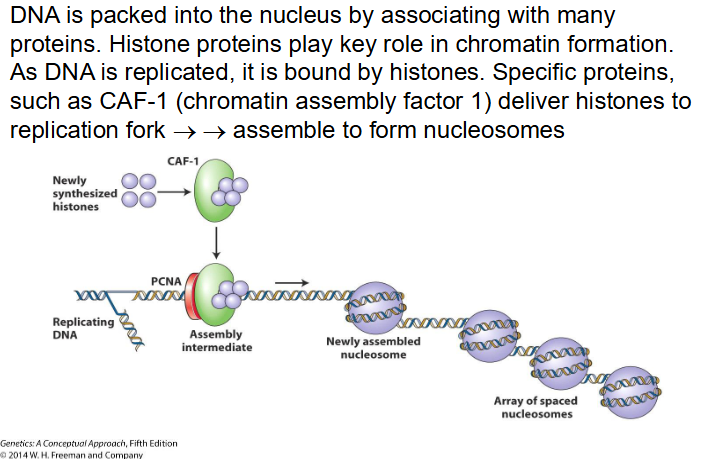
CAF-1
Chromatin Assembly Factor 1 deliver histones to replication fork
Chromatosome
nucleosome + H1 histone
Parental Histones and New Histones
- DNA can have old histones
- but during DNA replication new histones can also be made.
- final DNA has a mixture of new and old histones
Common amino acids in Histones
lysine, arginine, and histidine.
Multiple Origins of Replication
Speed up replication of eukaryotic chromosomes. Eukaryotic cells have several starting points.
E-coli vs Eukaryotes DNA replication
- E.coli have a fast rate of replication. Eukaryotes would take forever to replicate if it weren't for multiple origins of replication
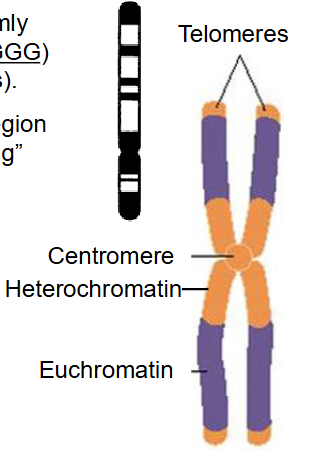
Simplified Structure of Nuclear Chromosome
- ends of linear chr have tandemly repeated G-rich sequences (TTAGGG) called telomeres (thousands of nts)
- the centromere is a constricted region along the chromosome, "separating" the two arms (p and q)
- For a chromosome to be copied it requires three elements: centromere, replication origins - initiation, telomeres (telos = end)
- Euchromatin and heterochromatin distributed across chromosomes

Replication Origin Number in organisms
- in bacterium: only one
- in eukaryotes: thousands
- condensed chromatin replicates late, while less condensed regions replicate earlier
Replicon
Unit of replication, consisting of DNA from the origin of replication to the point at which replication on either side of the origin ends.
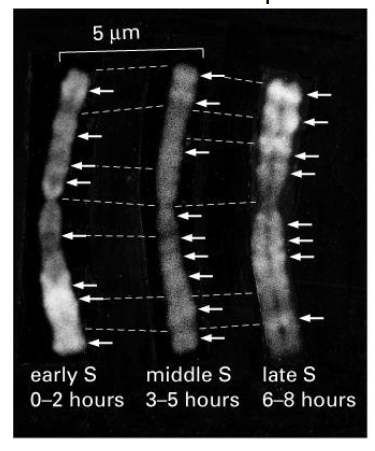
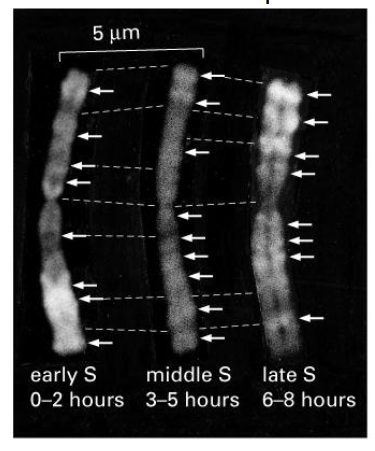
Regions of chromosomes replicate at different times
- you will notice that at different times during the S phase, different regions along the chromosome will be replicating
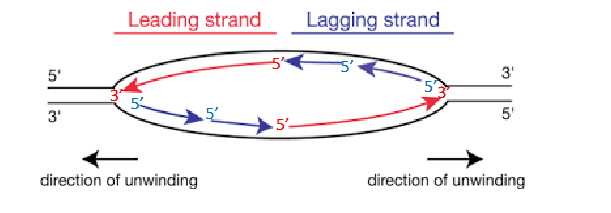
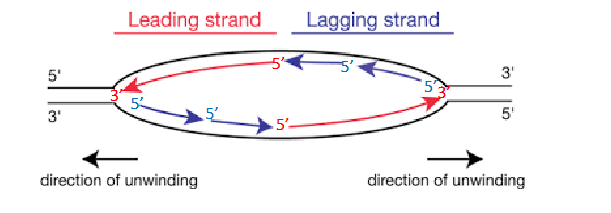
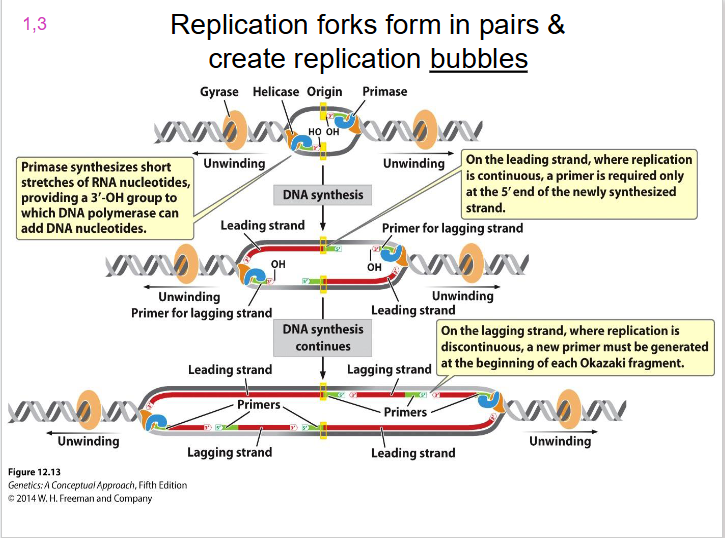
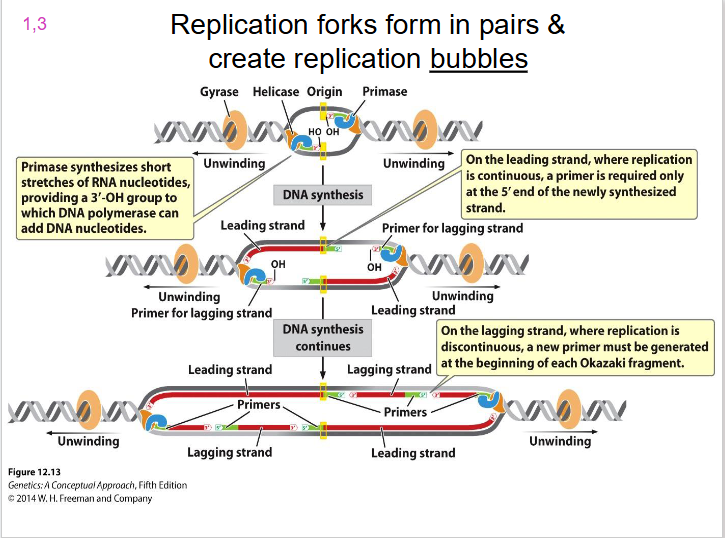
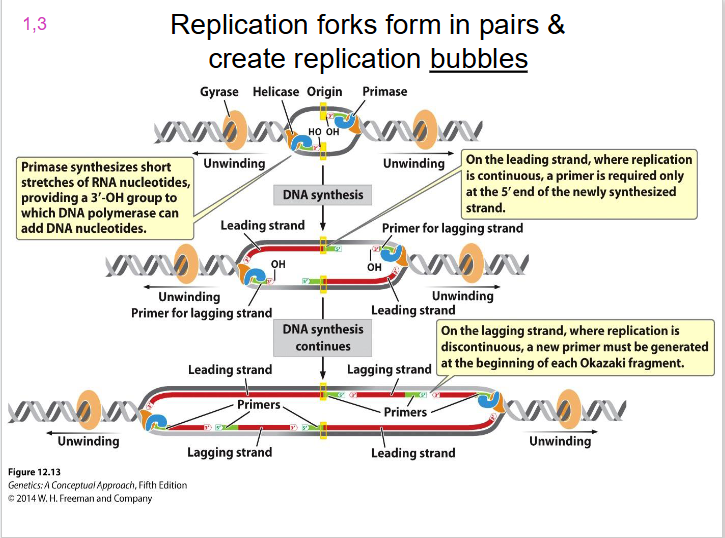
Two Replication Forks
each replication bubble has, they are on either side. Direction of unwinding is away from the centre of the bubble on both sides. both face the direction of the leading strand.
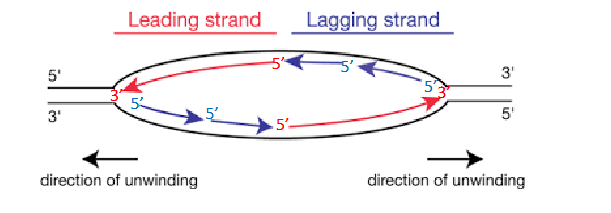
Regulation of Chromosome Replication
- regulated mainly through control of initiation
- initiation is related to sequences on DNA, packaging of DNA and activites of genes
- Timing when different origins are used is related to chromatin structure and cell cycle phase
- replication forks form in pairs and create replication bubbles moving in opposite directions
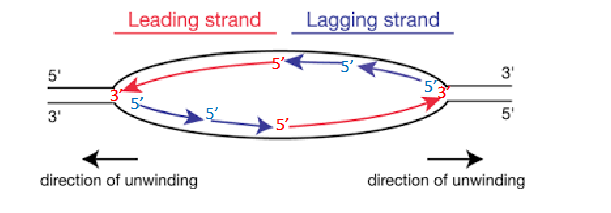
Where does initiation occur
at the origin of replication
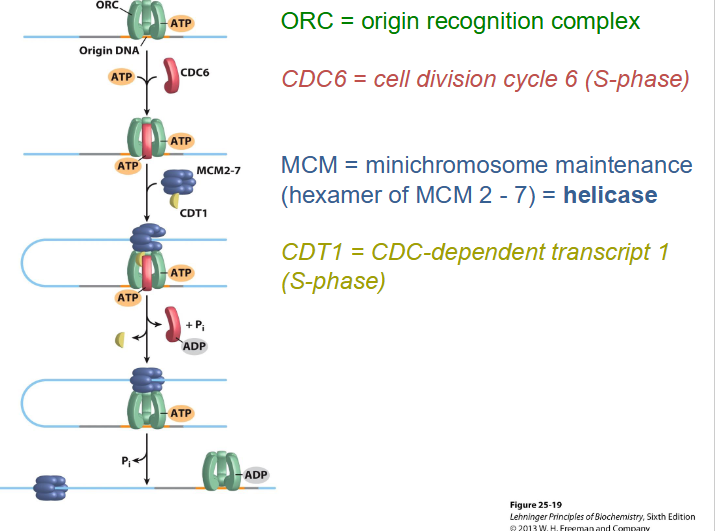
Initiation of Eukaryotic DNA replication
- Assembly of the replication proteins is ordered and begins at precise chromosomal locations = origins, length and sequences vary greatly between species
- helicase breaks apart DNA at origin
- since there are many origins on chromosmes, we need to coordinate replication
- this is done through ORC (origin recognition complex) (defo on next slide)
- it must ensure that all DNA is replicated in the S phase, so will have genes and chromosomes in new cells
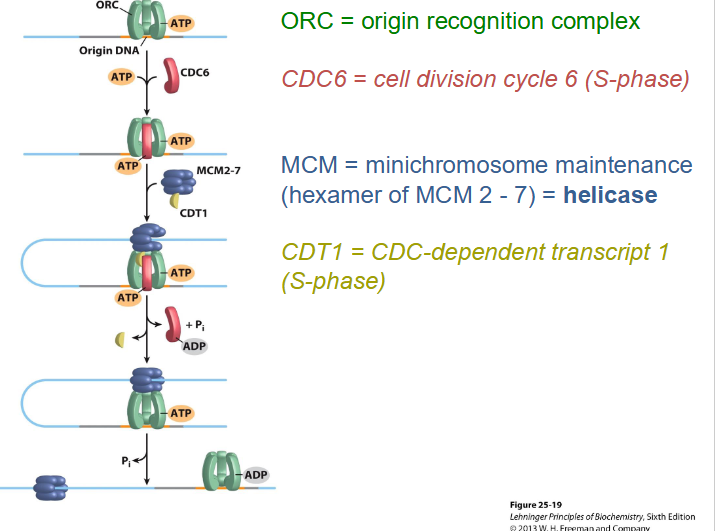
Origin Recognition Complex (ORC)
- multiprotein complex, binds to origin and separates DNA strands by recruiting replication proteins to ensure that each origin is only used once

What type of bases are origins rich in
A-T bases (Adenine, Thymine). Because they have least number of hydrogen bonds they are the easiest to break apart.
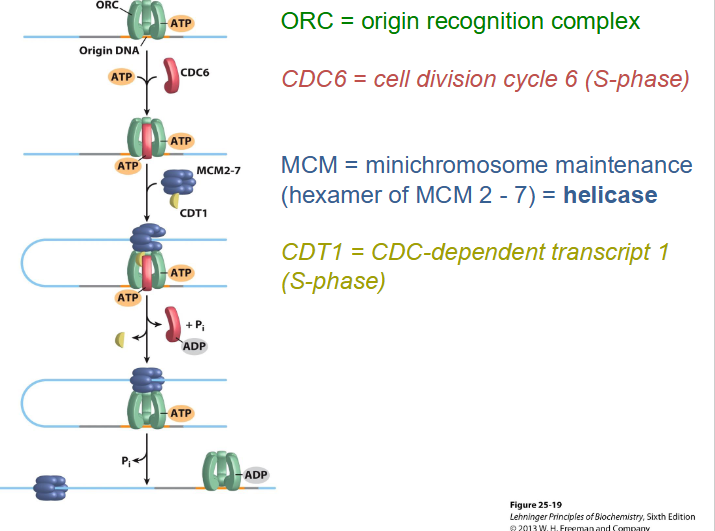
Licensing
- to initiate replication, DNA is "licensed" at origins and each origin is ONLY LICENSED ONCE, can't get re-initiation (only allowed to happen once)
- DNA helicase (heterohexameric protein, hetero = different, hexameric = 6, so heterohexameric means 6 different proteins) is recruited to the DNA cell-phase (S) specific proteins to ensure DNA replication occurs only in the S phase and once this happens it can not happen again
- then, get other proteins binding helicase (minichromosome maintenance = MCM), which prevents further initiation of replication at that origin. Ensure initiation is only once per cell cycle and only once per origin.
IMPORTANT SLIDE TO UNDERSTAND HOW LICENSING WORKS
SLIDE 14. not needed to know in detail but it helps find out how licensing works
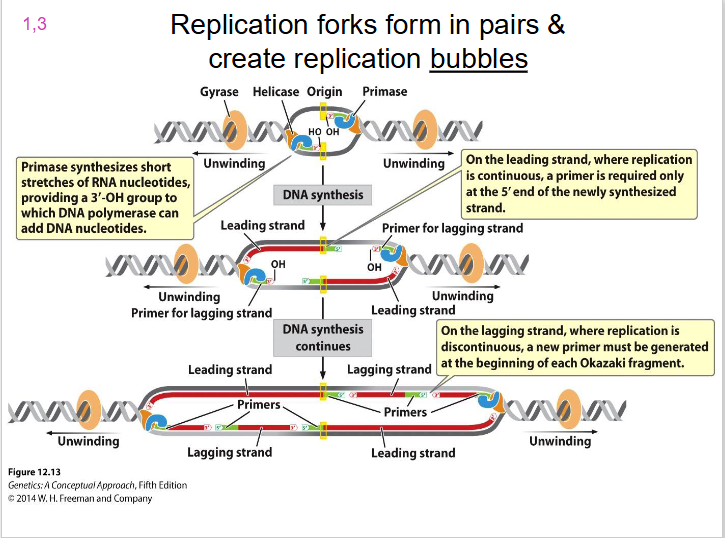
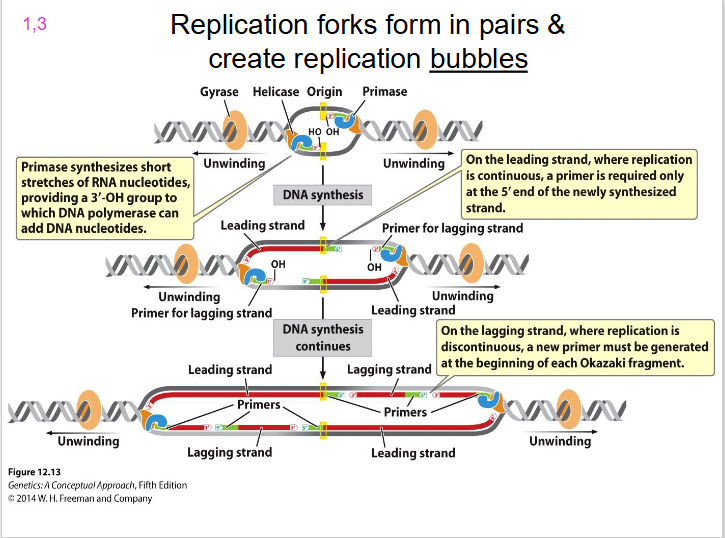
DIAGRAM SHOWING REPLICATION BUBBLE
SLIDE 15
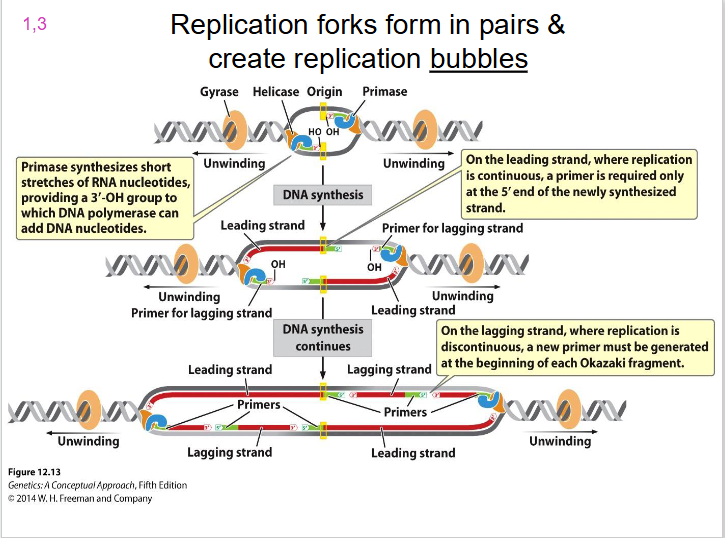
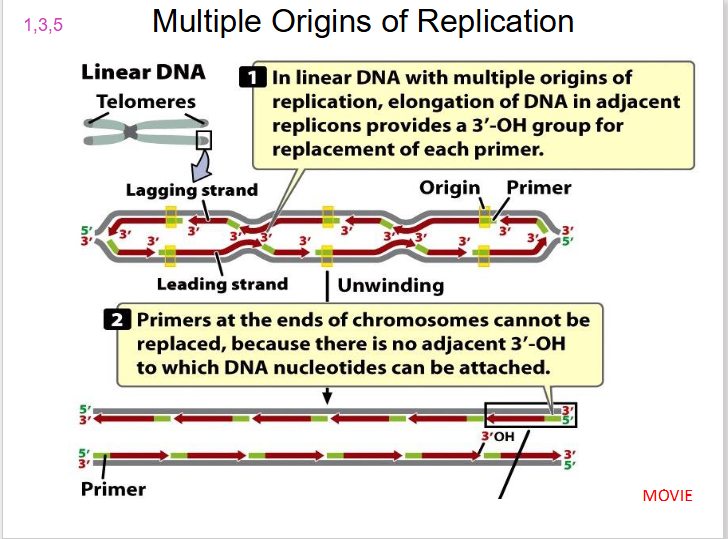
DIAGRAM SHOWING MULTIPLE ORIGINS OF REPLICATION
SLIDE 16
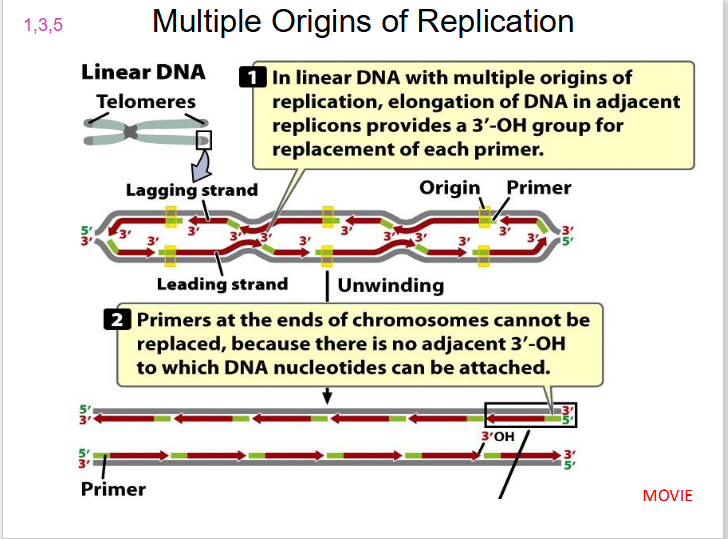
Chromosome Ending
- at the end, 3' group overhangs
- no 3' OH group to which DNA nucleotides can be attached, producing a gap
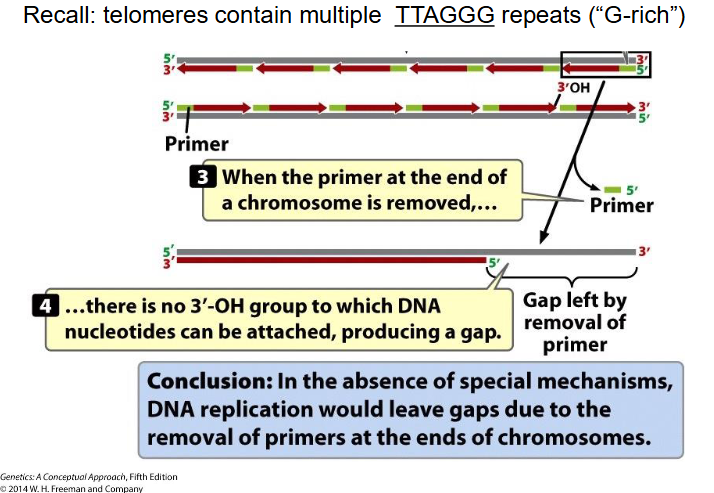
DIAGRAM SHOWING END REPLICATION PROBLEM
SLIDE 17
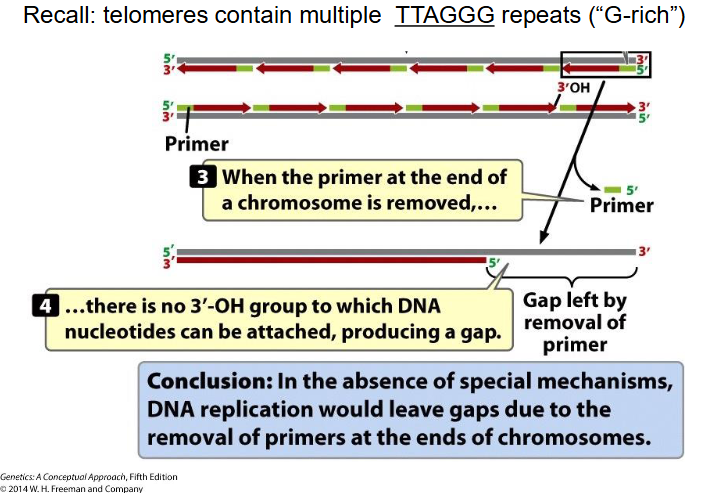
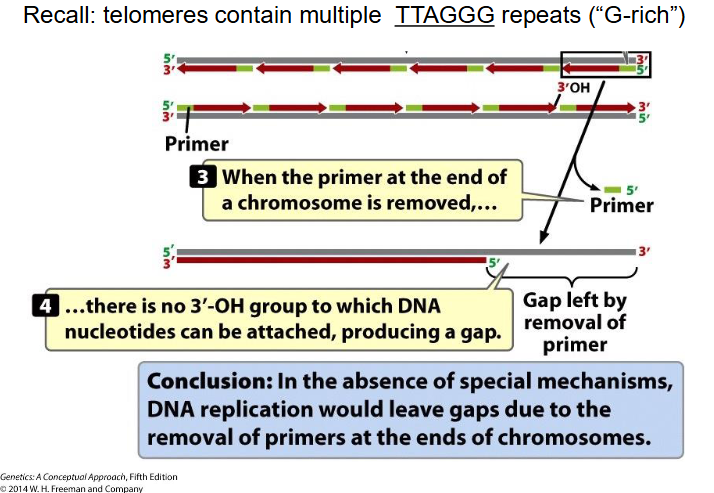
End Replication Problem
- At extreme end of newly synthesised DNA strand, no template for RNA primer to base-pair with, so chr would get shorter every replication round (there would be gaps) (lose TTAGGG repeats, since telomeres contain mulitple TTAGGG repeats (G rich))
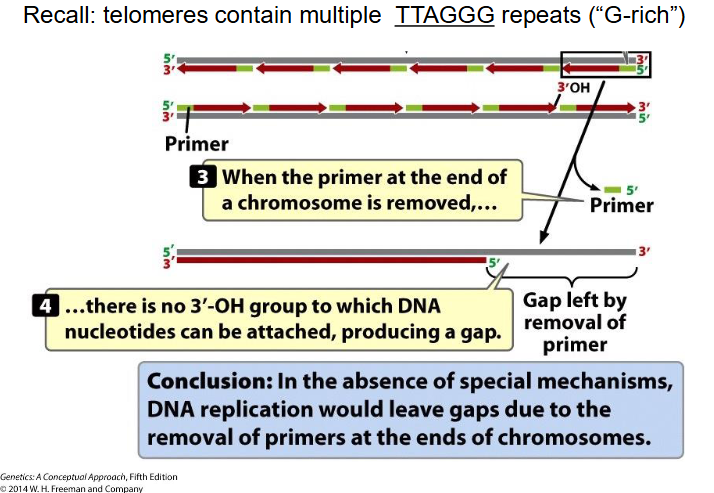
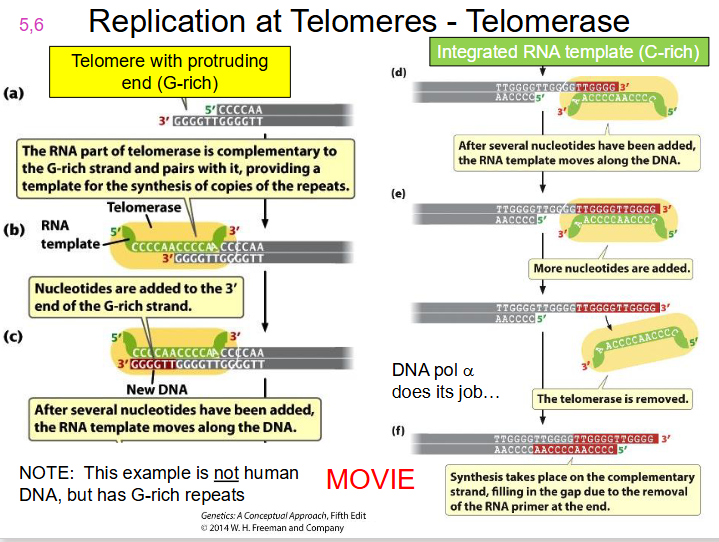
End Replication Problem Solution
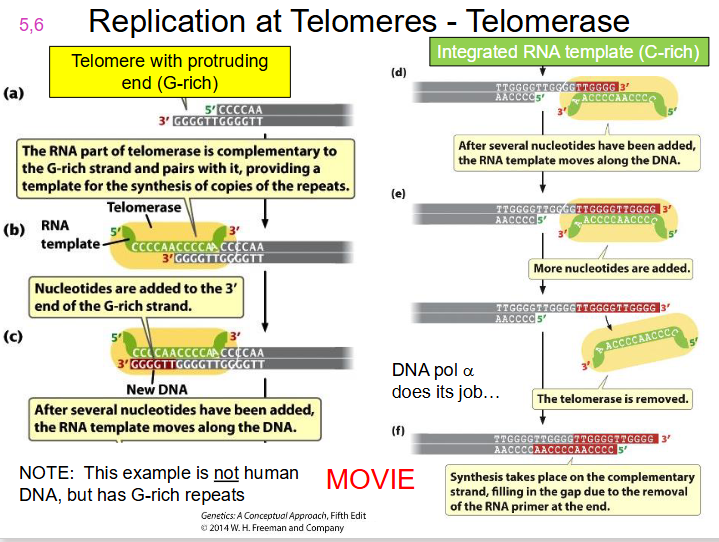
telomerase.
- A specialised enzyme, telomerase, with a catalytic subunit = Telomerase Reverse Transcriptase (TERT) along with RNA component (complementary to telomere repeat), completes the chromosome end to maintain chromosome length
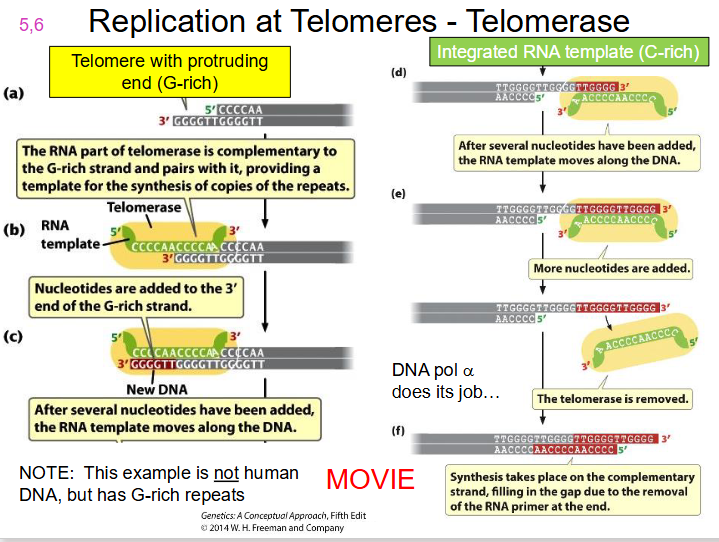
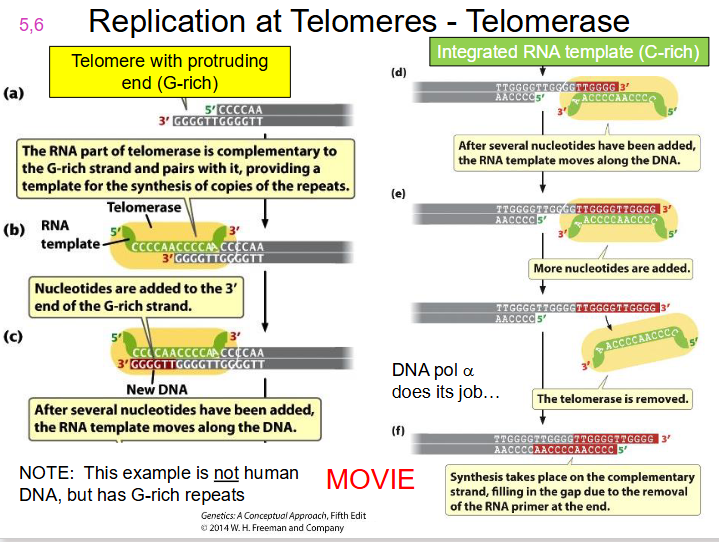
DIAGRAM OF TELOMERASE
SLIDE 19. WATCH TELOMERASE MOVIE ON LMS