Regulation of Eukaryotic Gene Expression
1/42
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
43 Terms
What does differential gene expression mean in eukaryotes? By what three methods do we see differential gene expression?
Almost all the cells in an organism are genetically identical. Differences between cell types results from differential gene expression, due to differences in the epigenome, differences in chromatin structure, and covalent modifications of the DNA. This is regulated at many stages.
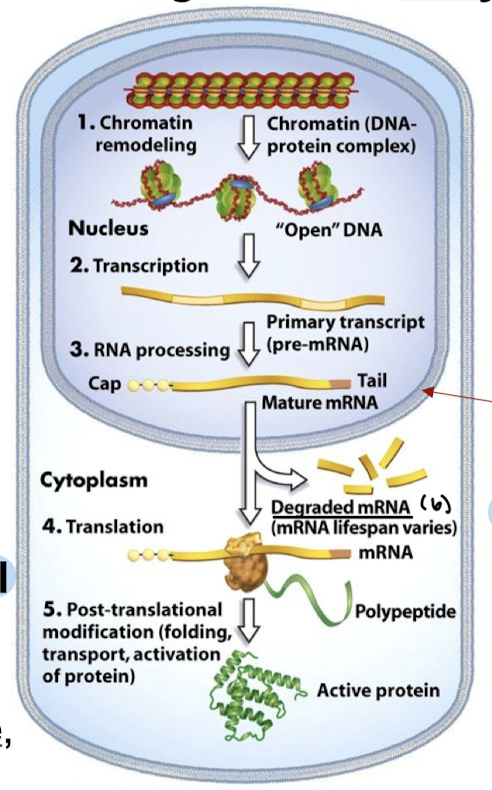
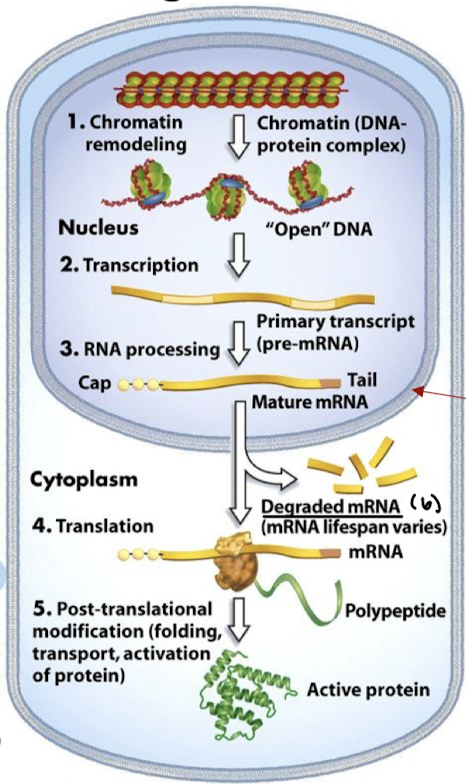
In what six stages can differential gene expression occur?
Chromatin remodeling
Transcription
RNA processing
mRNA stability/degradation
Translation
Post-translational modification (folding, transport, activation of protein)
How is gene expression regulated in RNA processing/post-transcriptional regulation?
Alternative RNA splicing
How do noncoding RNAs affect differential gene expression/regulation?
microRNAs (miRNA) and small interfering RNAs (siRNA) affect mRNA translation, chromatin configuration, and mRNA stability.
What is different about eukaryotic gene expression from prokaryotic gene expression? Operons? Mono or polycistronic?
Each structural gene has its promoter and is transcribed separately. No operons, polycistronic.
Where are control elements found? What are coordinately controlled genes?
Control elements are found per gene. Coordinately controlled genes are unlinked, but may have similar sequences within their promoters and be bound by some of the same transcription factors.
What do TFs intearct with in eukaryotes?
Some TFs interact directly with RNA polymerase, some interact with other proteins associated with RNA pol.
What is required for access of eukaryotic promoters to RNA polymerase?
Chromatin remodeling is hindered because RNAP is hindered by chromatin structure.
What is the default gene expression for prokaryotes and eukaryotes?
Eukaryotic gene expression requires a complicated set of proteins, and by default is “off” until transcription factors bind. RNAPII cannot bind on its own. (Prokaryotes are default “on”)
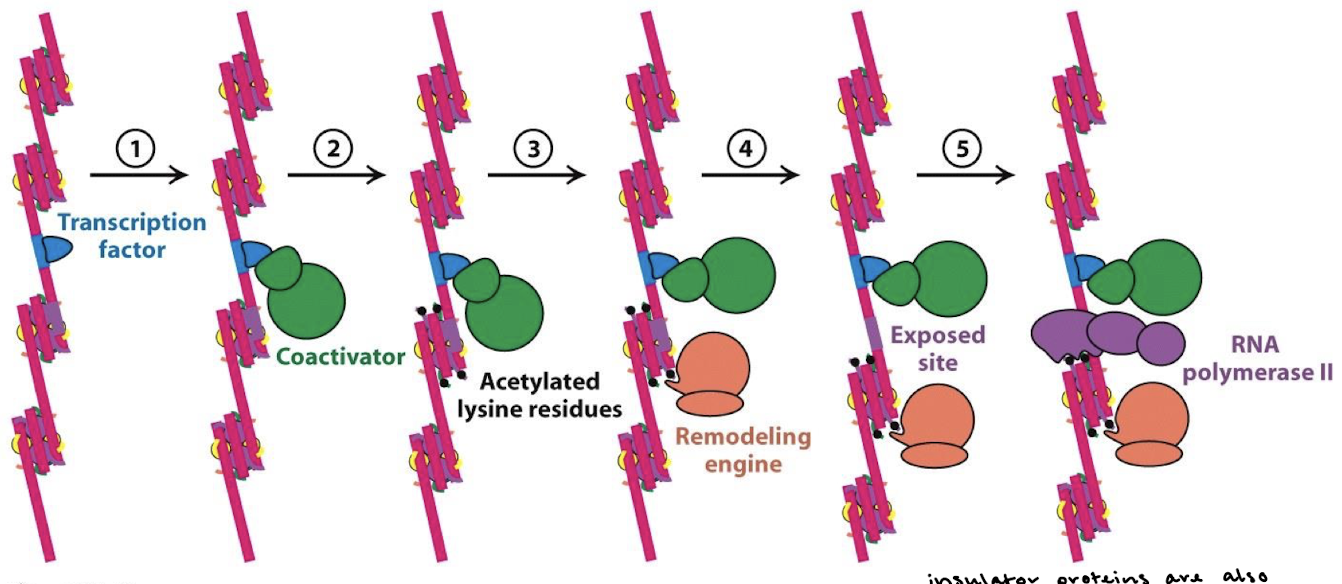
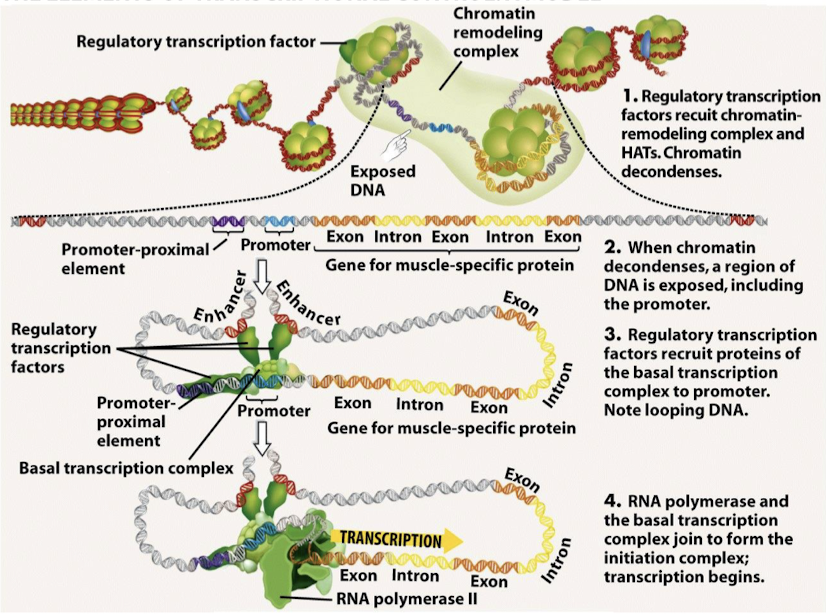
Explain the model of transcriptional control starting with condensed chromatin to initiation.
Regulatory transcription factors recruit chromatin-remodeling complexes and HATs. Chromatin decondenses.
When chromatin decondenses, a region of DNA is exposed, including the promoter.
Regulatory transcription factors recruit proteins of the basal transcription complex to the promoter. Note looping DNA.
RNA polymerase and the basal transcription complex join to form the initation complex; transcription begins.
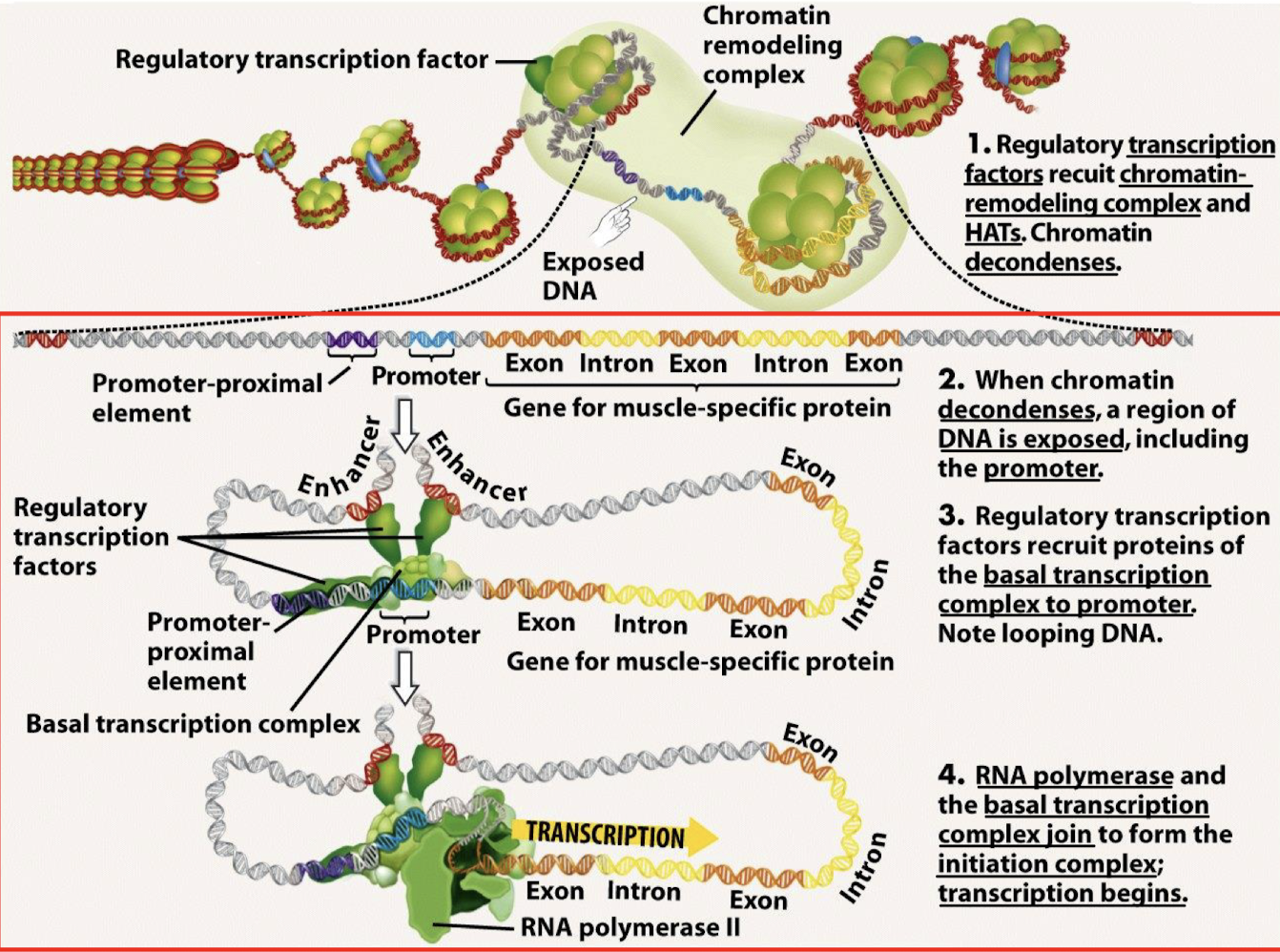
What is the role of the basal transcription complex?
Initiates transcription at a low frequency.
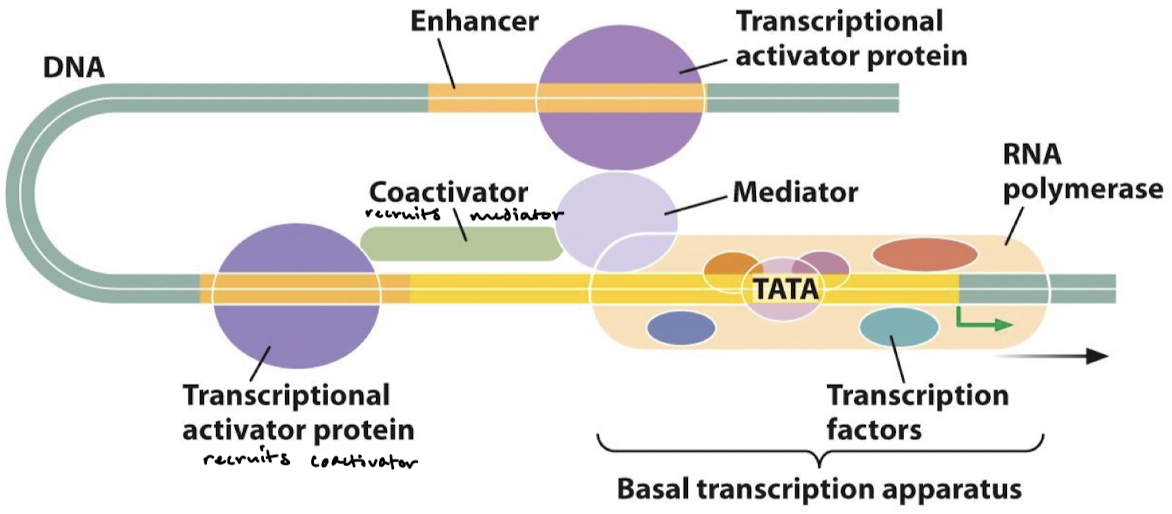
What is the function of the TF activation domain (AD)?
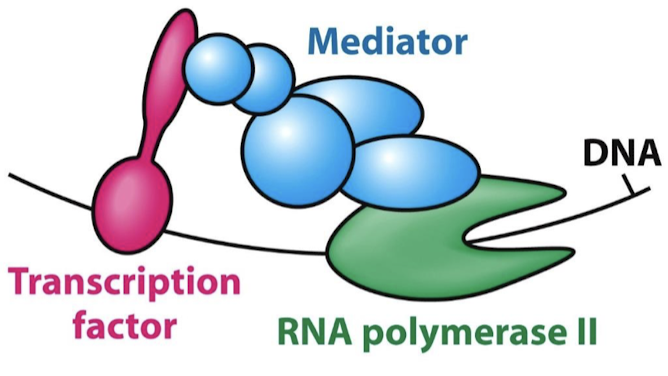
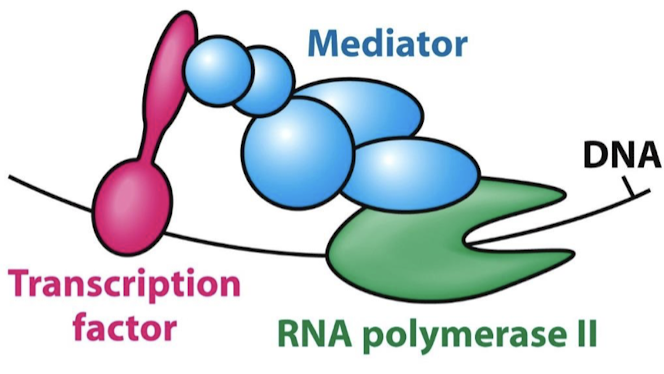
TF AD generally recruit other proteins that promote transcription. They interact directly with Pol II or with closely associated proteins through intermediary proteins (mediators).
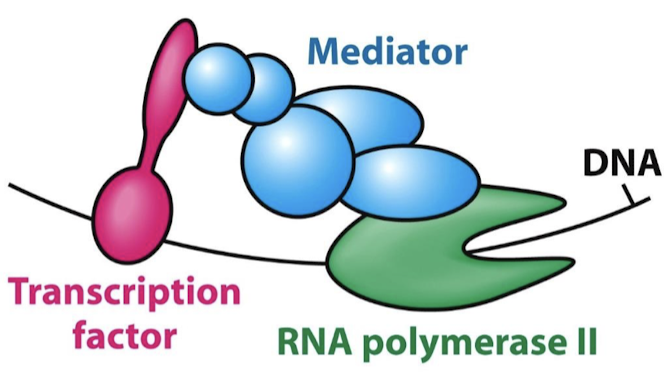
What is the function of mediator proteins?
They are complexes of 25-30 subunits that act as a bridge between TF ADs and RNA Pol II.
What are three common features of TF ADs?
Redundant: Part of AD can be deleted without loss of function
Modular: Can activate transcription when paired with a variety of DNA-binding domains.
Act Synergistically: Two activation domains acting together create stronger effect than either acting separately.
What are enhancers?
Regulatory DNA sequences, when bound by transcription factors, enhance the transcription of an associated gene.
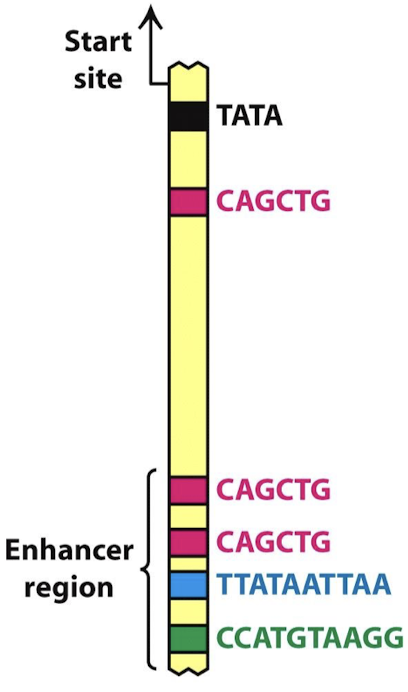
What are two common features of enhancers?
Modular: one gene may have multiple enhancers, each meant for a particular tissue or developmental (temporal) expression
Combinatorial: two genes may have the same enhancers for a factor, yet they do not show identical tissue-specific expression (other tissue-specific factors combine with the factor and modulate specific expression).
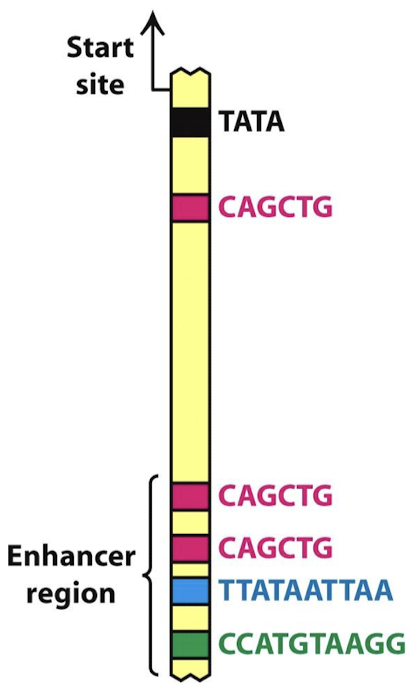
If an enhancer inhibits transcription, it is called a ________.
Silencer
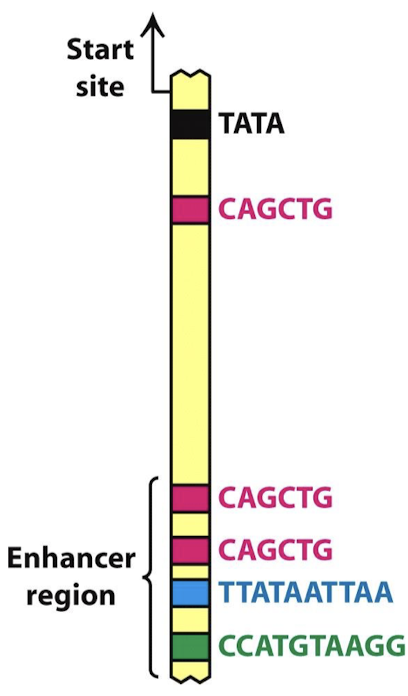
Explain a model for combination of control elements in liver and lens cells.
A particular combination of control elements can activate transcription only when the appropriate activator proteins are present.
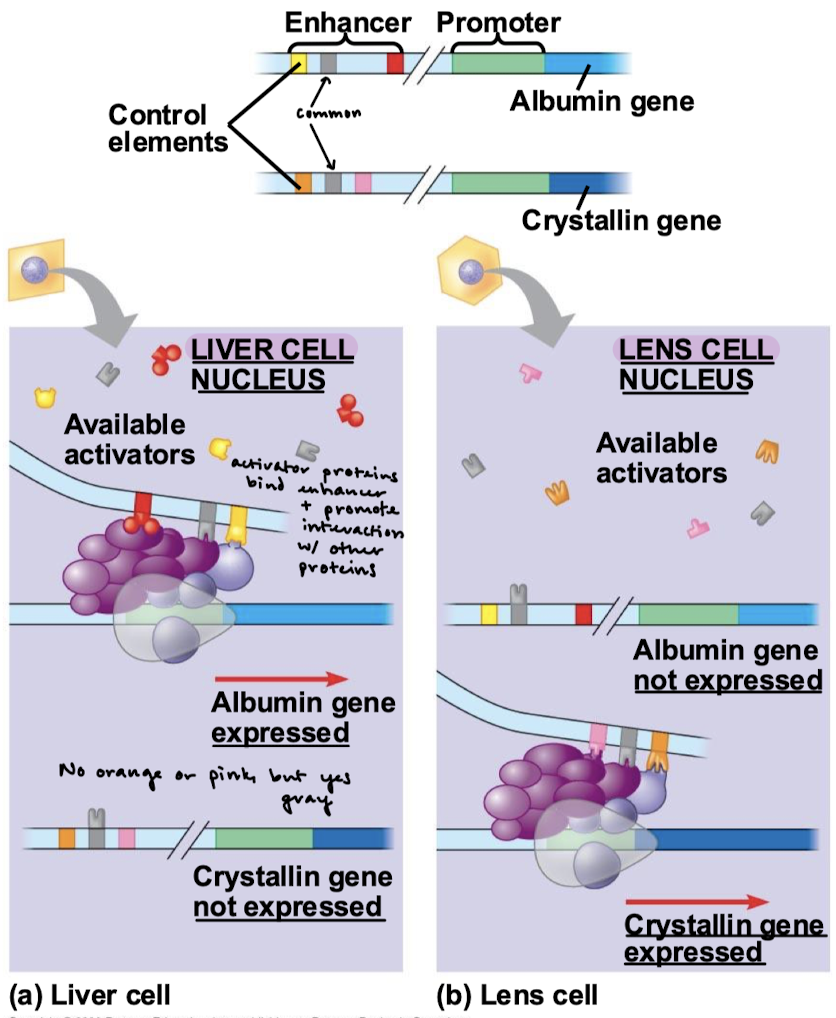
What is the function of an insulator protein? Where do they bind on DNA?
Insulator proteins are a genetic boundary element that blocks the interaction between enhancers and promoters. They bind stretches of DNA (as few as 42 bp) located between the enhancer/silencer(s) and promoter(s) and of neighboring genes or clusters of neighboring genes.
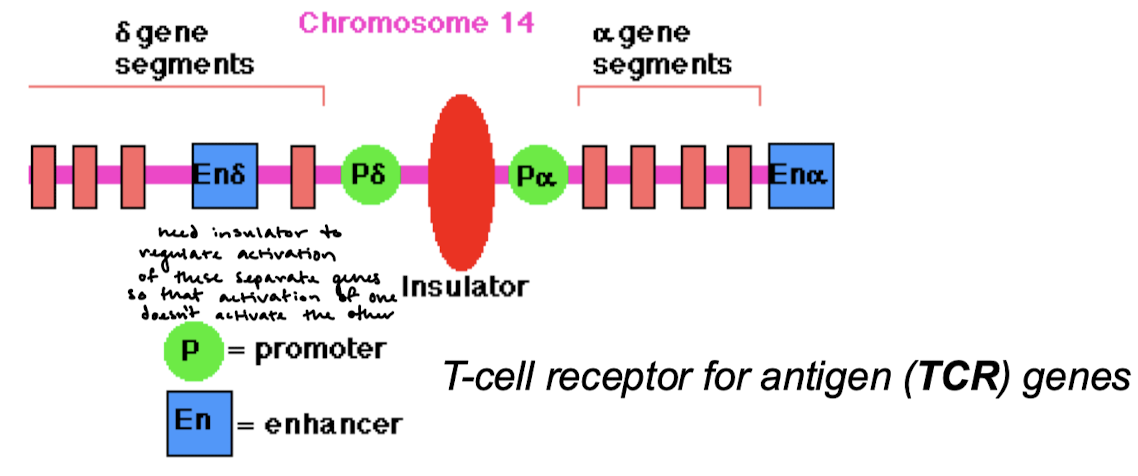
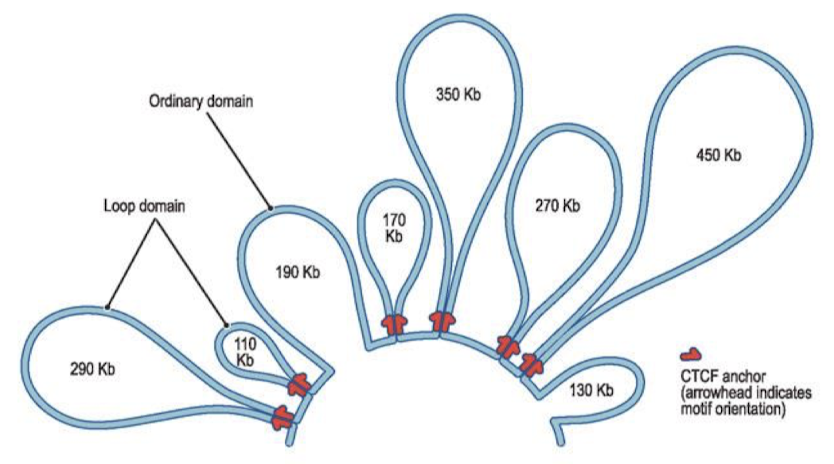
What forms at insulator sequences? What do insulator complexes form when they interact with neighboring insulators? Why is this important?
Protein complexes; chromosome loops. Enhancers cannot affect promoters outside of their loop.
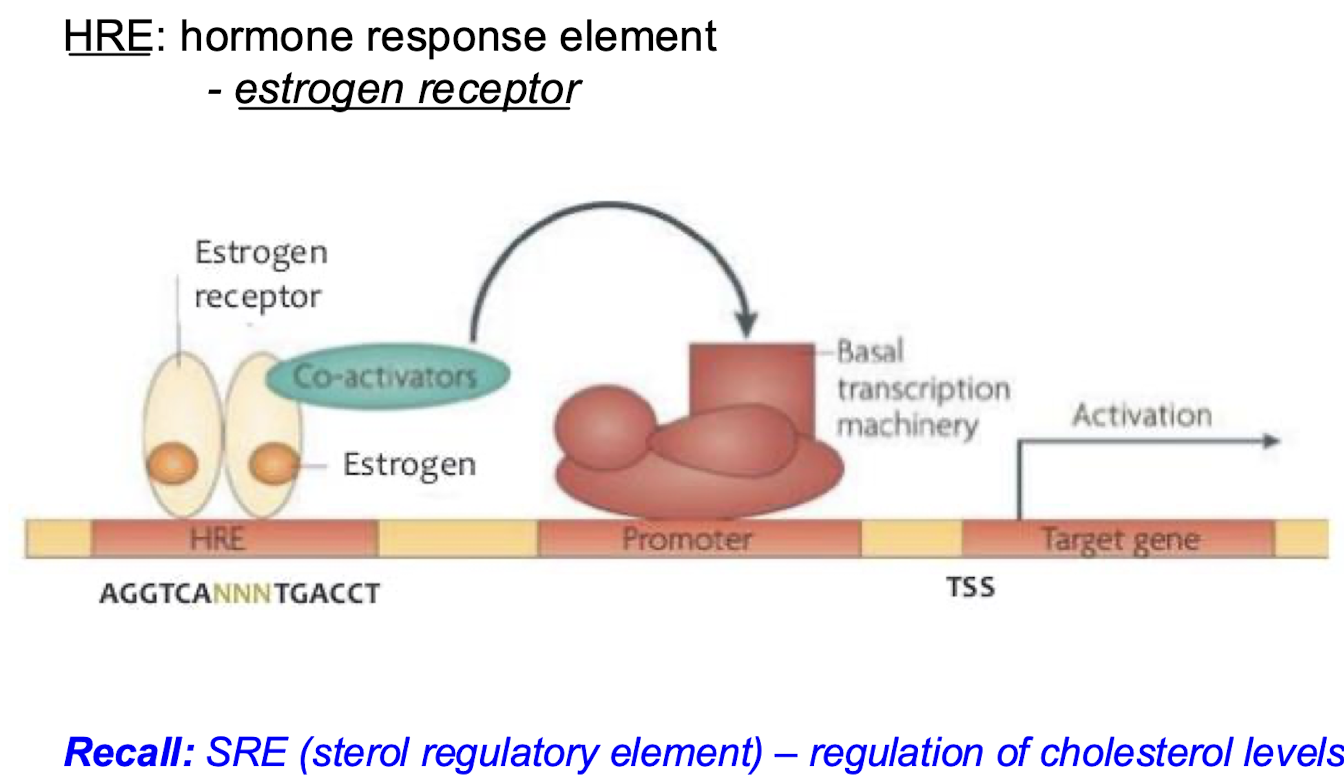
What is a response element? What is an example?
DNA sequences that are bound by the complex of specific factors (e.g. the steroid bound to its receptor) that identify genes regulated in response to particular physiological challenges. The hormone response element is an example because it binds the estrogen receptor when it is bound to estrogen. It will recruit coactivators and promote transcription of the target gene.
What are two changes that can be made to chromatin structure that affects the expression of genes? Explain what each does.
DNA methylation of cytosine bases adjacent to guanine nucleotides (CpG) — CpG islands. This decreases the expression of genes (silencing).
Chromatin-remodeling complexes bind directly to DNA sites and reposition nucleosomes (acetylation and deacetylation).
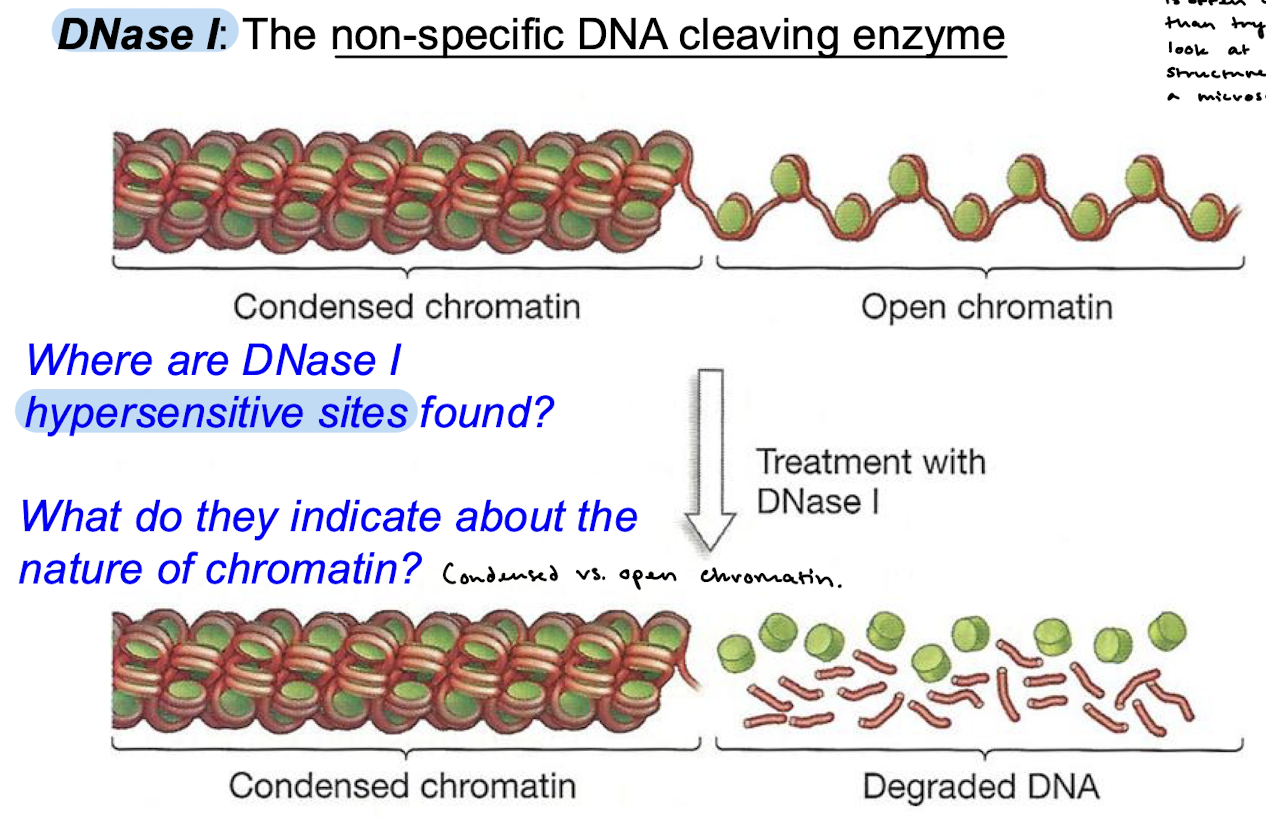
What is the purpose of using DNase I in lab? Why DNase and not a microscope?
To measure chromatin accessibility. DNase I is a non-specific DNA cleaving enzyme and will digest open chromatin into degraded DNA. Treatment with DNase provides us with hypersensitive sites that indicate where open chromatin is found. Using DNase I is often cheaper than trying to look at chromatin structure under a microscope.
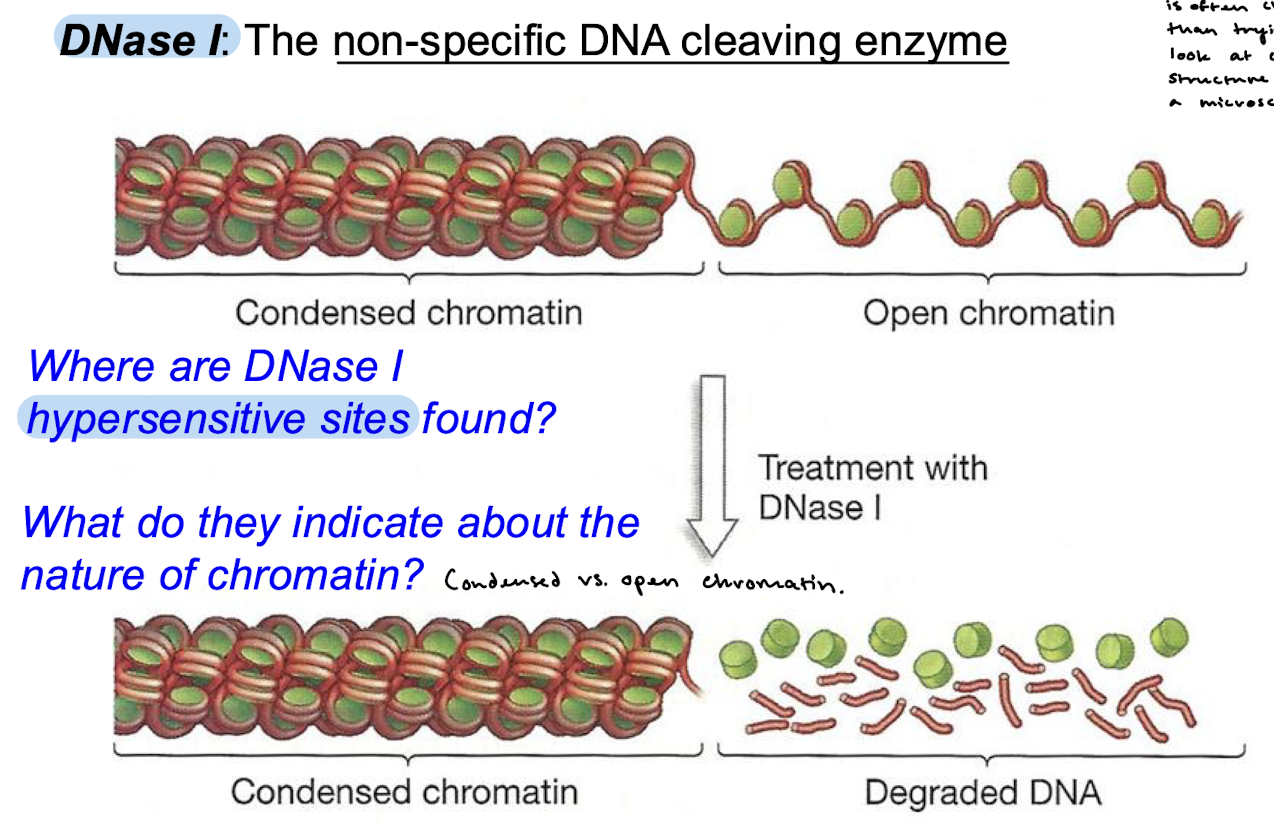
Regions that have high sensitivity to digestion of DNase I are called…
DNase I hypersensitive sites
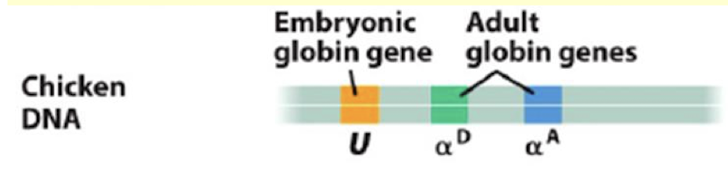
DNase I sensitivity was tested on different tissues and at different times in development. Based on the DNA, recall where the hypersensitive sites are for each stage of development and explain the importance of this: Erythroblasts in the first 24 hours, at 5 days, and at 14 days. What about brain cells throughout development?
This provides information on what genes are open chromatin and are actively being expressed at a given stage of development.
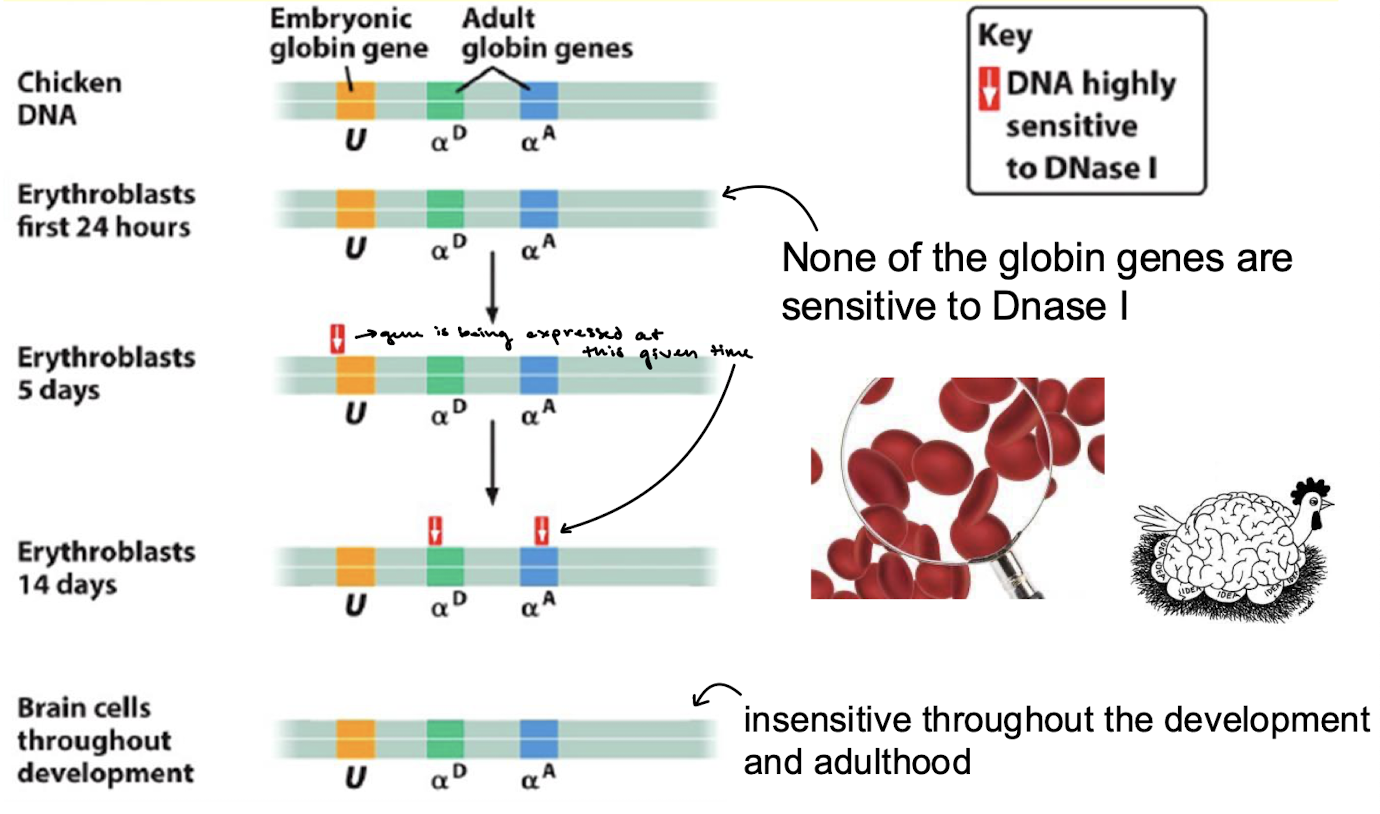
What is the function of yeast TF GAL4? Where does it bind?
It activates genes that are involved in galactose metabolism. Gal4 recognizes 5’-CGG(N)11CGG-3’ sequence which are found approximately at 4,000 sites (only binding the sites of these 4,000 that are open chromatin).
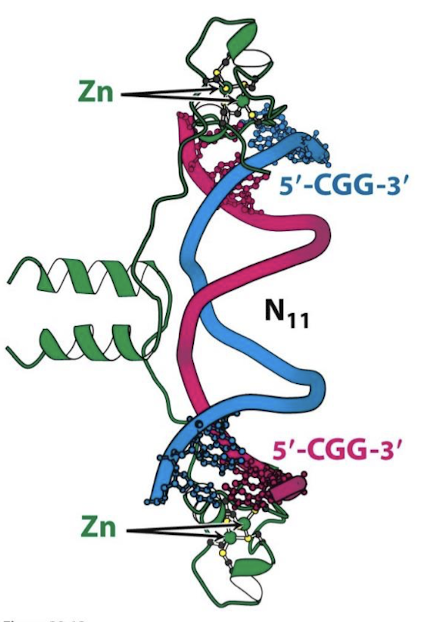
What is ChIP (chromatin immunoprecipitation) and why is it done?
ChIP is used to identify DNA-binding sites of a specific protein and the locations of modified histone proteins.
What did ChIP (chromatin immunoprecipitation) say about GAL4 and its 4,000 binding sites? What does this imply about eukaryotic transcription?
ChIP showed that Gal4 binds only to 10 sites when the cells are growing in galactose. Thus, in eukaryotes, binding sties for a transcription factor are not always available like in prokaryotes.
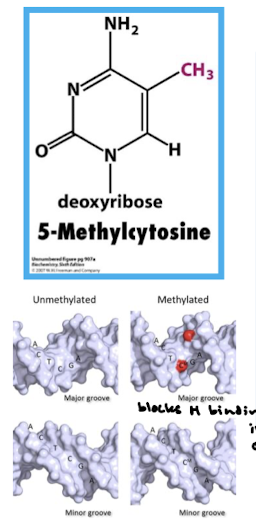
In what organisms is DNA methylation found? On what bases and where in the sequence? Where on the gene? What is the effect?
DNA methylation is found in vertebrates and takes place on those cytosines that are followed by guanines, e.g. CpG. Methylation takes place in the promoter DNA. Methylation is a mechanism to keep genes transcriptionally silent.
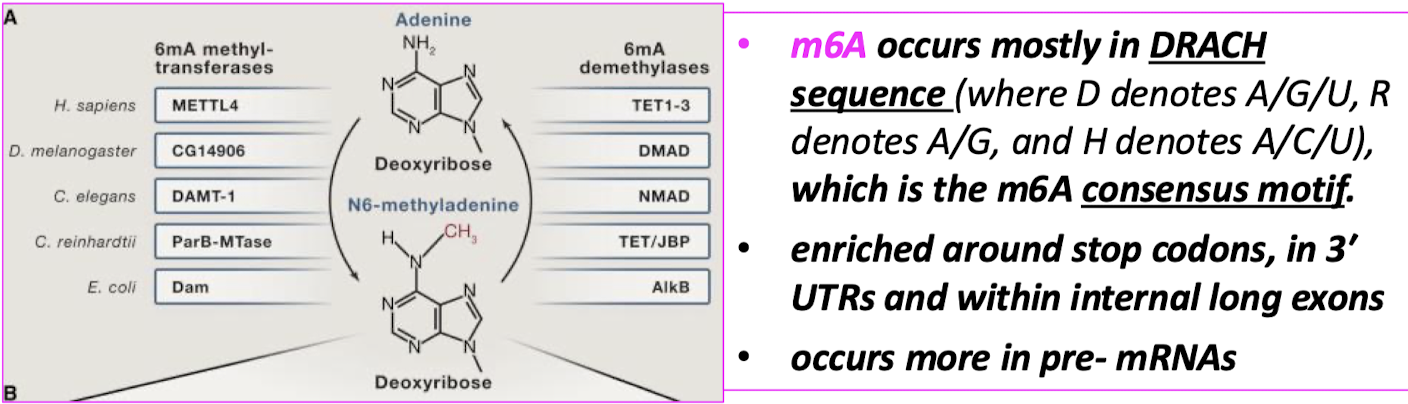
Where does m6A occur (what sequences)? Where are these sequences enriched? In what molecules does this occur more in?
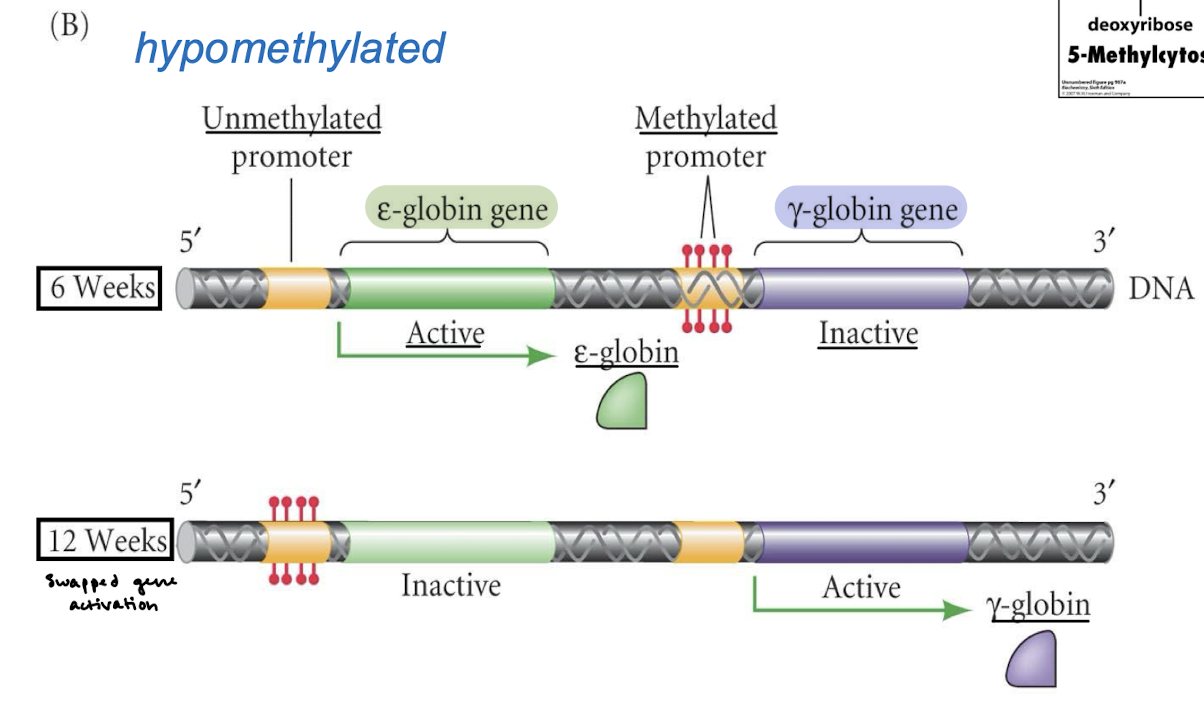
Methylation of globin genes in human embryonic blood cells: Which promoter is methylated at 6 weeks and which at 12 weeks?
The gamma-globin gene is methylated at 6 weeks and the epsilon-globin gene is methylated at 12 weeks.
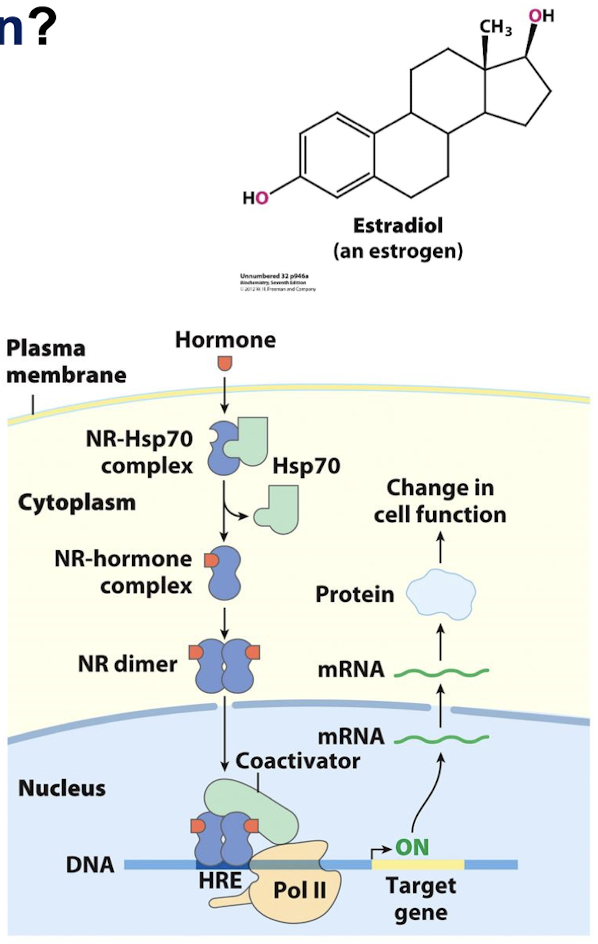
Describe how estrogens and other hormones are detected and brought into the cell/nucleus for regulation of transcription.
Hydrophobic molecules like estradiol and other hormones easily cross cell membranes and bind to the estrogen receptor or nuclear hormone receptor. The receptor is then located to the ERE/HRE (estrogen response element/hormone response element) with the consensus sequence: 5’-AGGTCANNNTGACCT-3’.
What are the two highly conserved domains of the nuclear hormone (NH) receptor? What structures within these domains interact with other molecules?
DNA-binding domain: Zn-based domains bind specific DNA sequences, alpha helix contacts the major groove
Ligand-binding domain: alpha helices arranged in three layers.
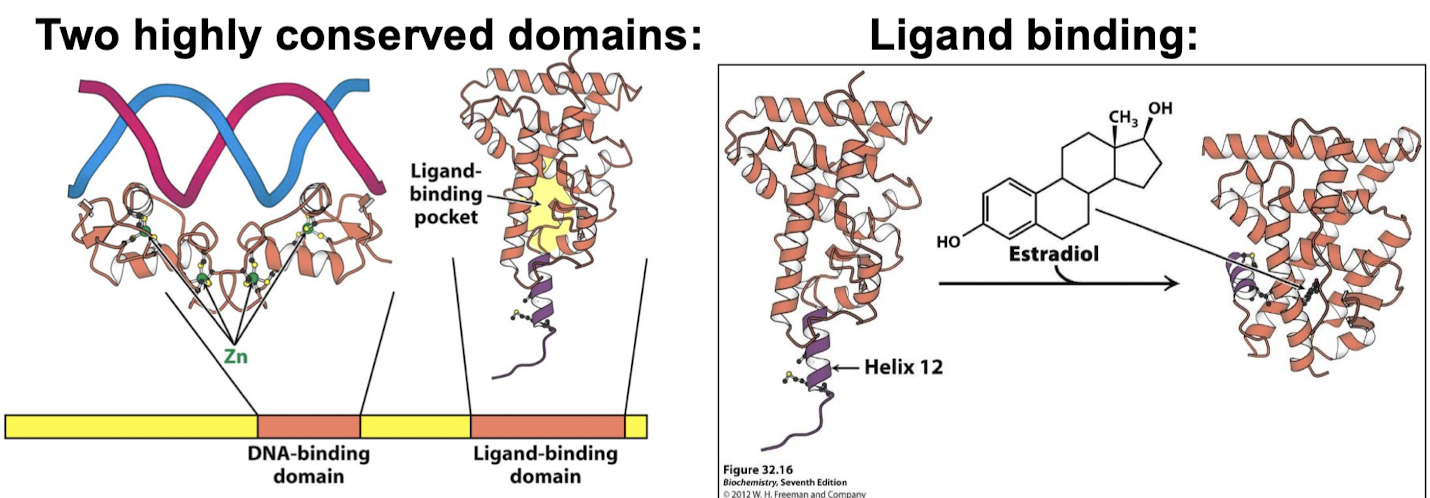
What change is made when the ligand binds the ligand-binding domain of the nuclear hormone receptor?
Binding changes the conformation of the receptor when it binds its ligand but this does not alter the ability of NH receptors to bind DNA. The binding of the ligand changes the conformation of the receptor dimer and allows coactivator binding.
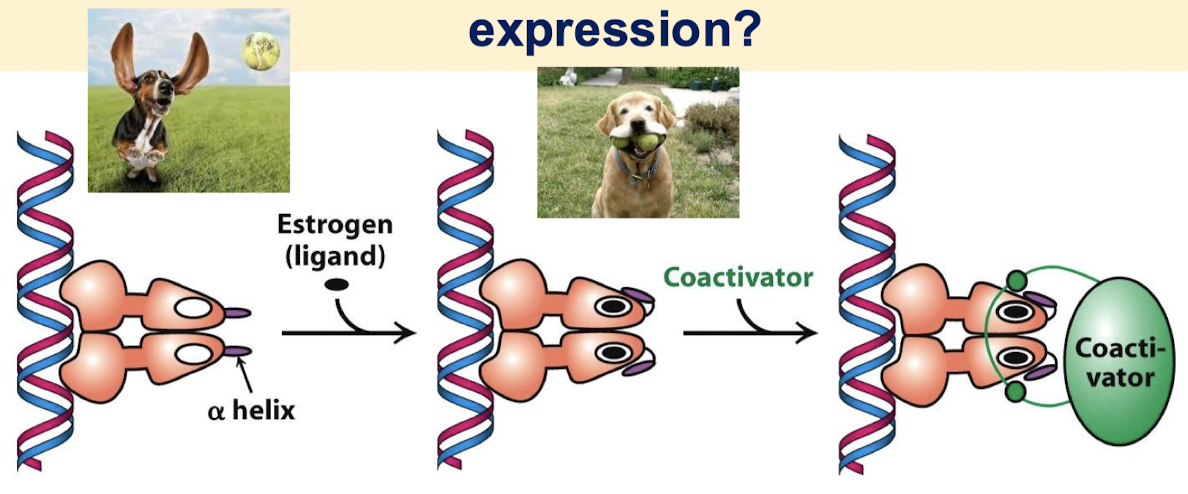
What are some examples of hormone receptors that follow the same mechanism as the estrogen receptor?
p160 family:
SRC-1 (steroid receptor activator-1)
GRIP-1 (glucocorticoid receptor interacting protein-1)
NcoA-1 (nuclear hormone receptor coactivator-1)
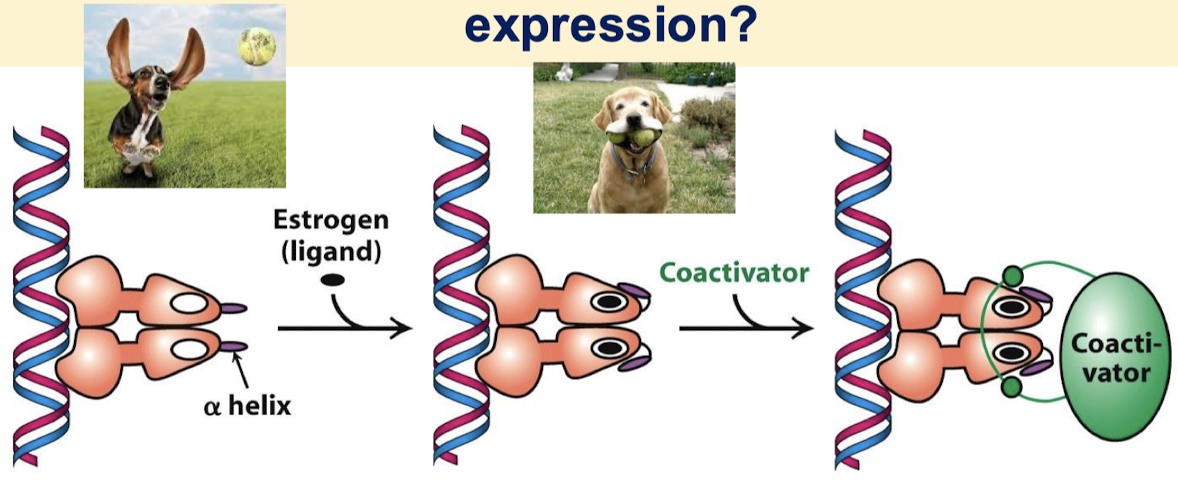
How do coactivators modulate transcriptional activity?
Coactivators have the ability to covalently modify N-terminal tails of histones and other regions on other proteins.
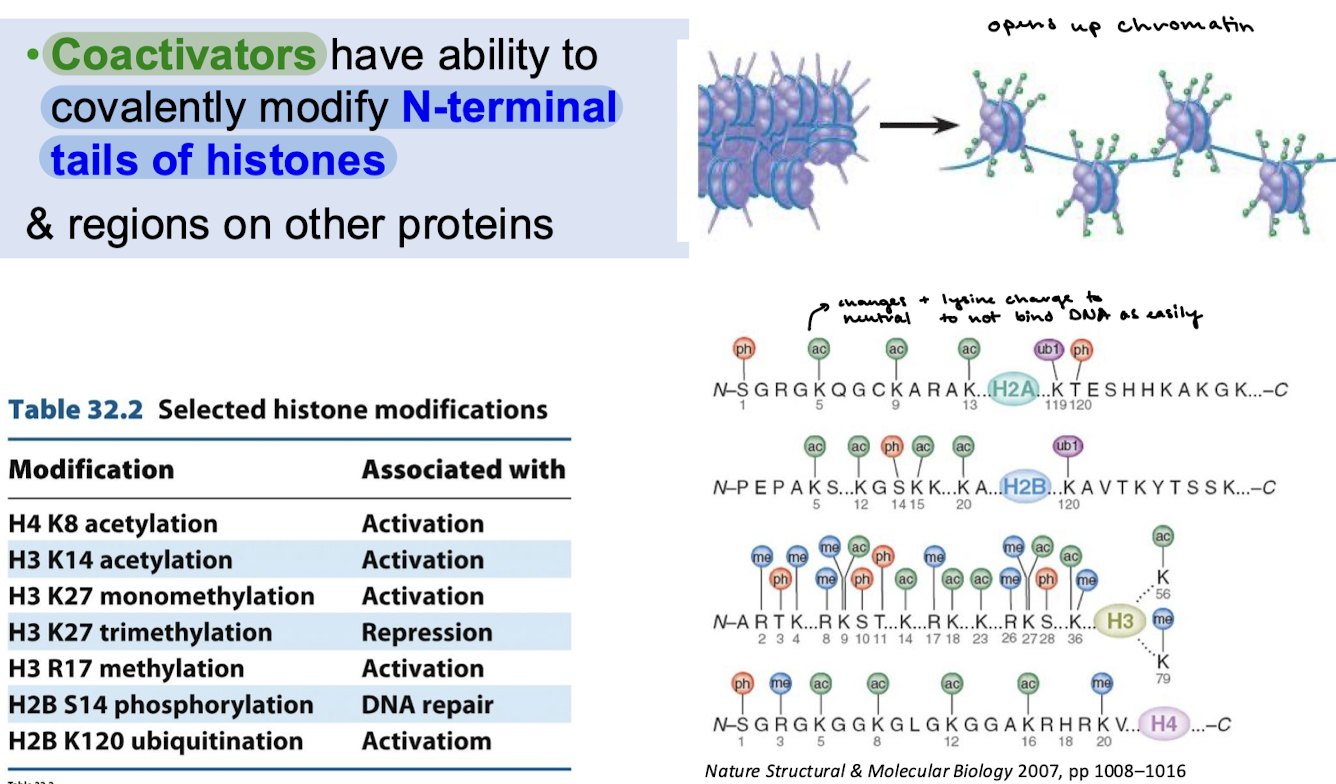
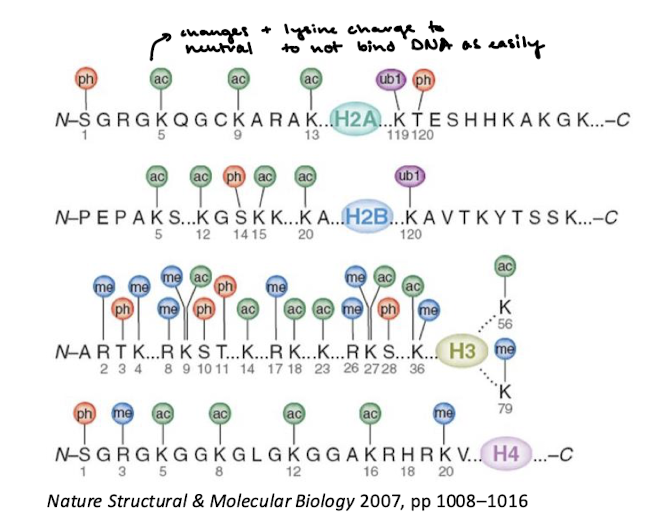
What is the effect of acetylation of N-terminal tails of histones?
This changes the positive charge of lysine to a neutral charge so that the tails cannot interact with DNA as easily.
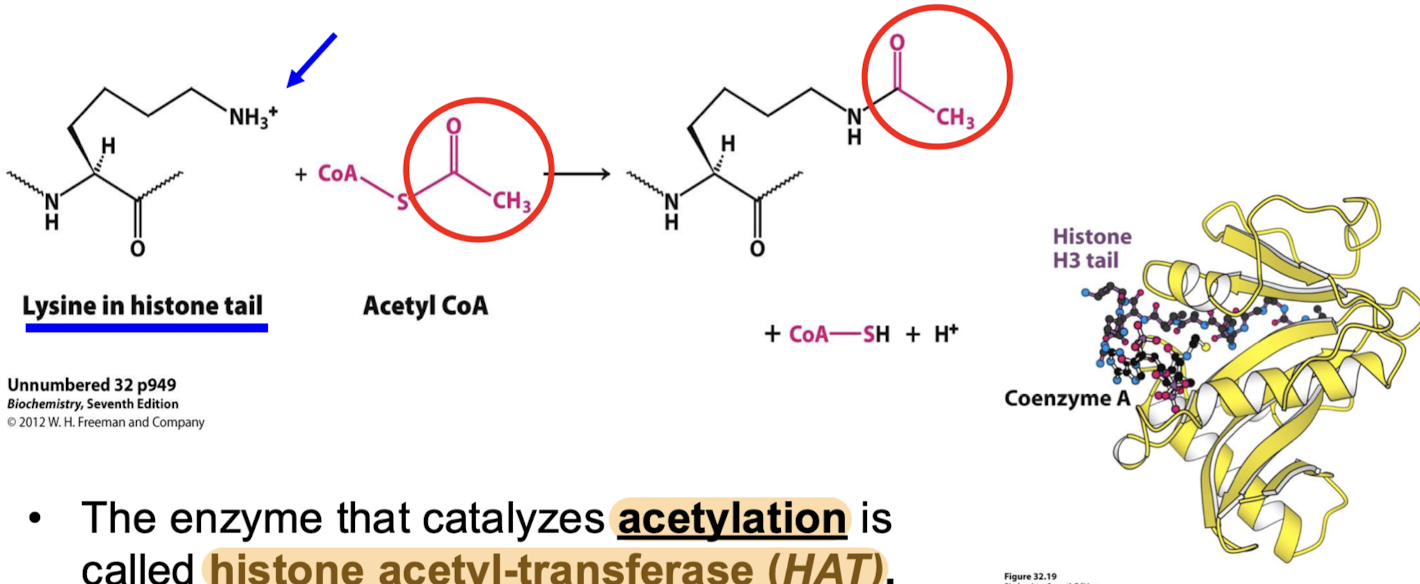
Some coactivator proteins recruit proteins that transfer acetyl groups. What are these proteins, and where does the acetyl group come from? Where is it moved to?
The enzyme that catalyzes acetylation is called histone acetyltransferase (HAT). They transfer the acetyl group from acetyl CoA to the specific lysine residues of the histone tails.
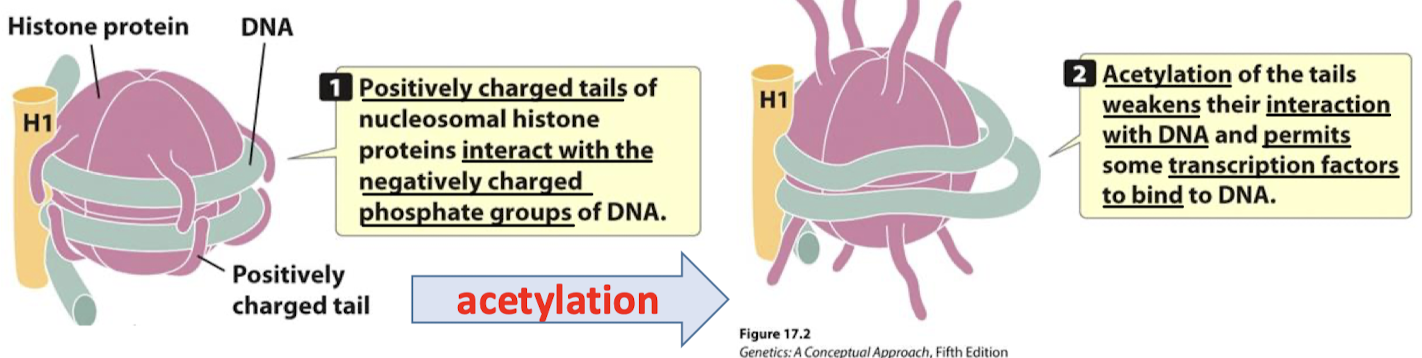
What are the consequences of histone acetylation?
Positively charged tails of nucleosomal histone proteins interact with the negatively charged phosphate groups of DNA. Acetylation of the tails weakens their interaction with DNA (DNA affinity for histones decreases) and permits some transcription factors to bind to DNA (loosening allows/exposes other regions of the DNA to the other factors).
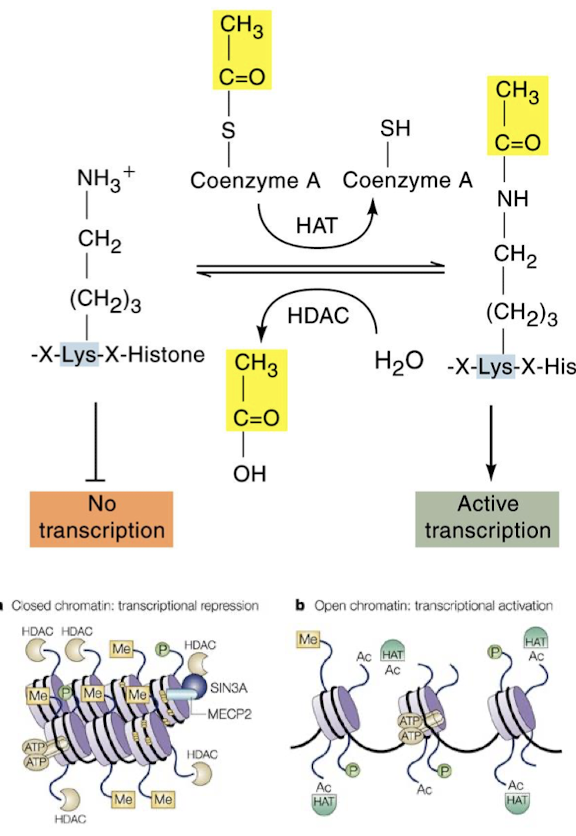
When is HAT bound? What does it result in? How is this undone?
HAT is bound to transcriptionally active chromatin. It acetylates the histone tails and allows transcriptional activation. Histone deacetylase (HDAC) removes the acetyl group from the histone tail and represses transcription.
What do acetylated lysines on the histone tails interact with? What are these called?
They may interact with the acetyllysine-binding domain of another group of factors that activate transcription. This domain is called the bromodomain (BRD) and the factors are called bromodomain proteins.
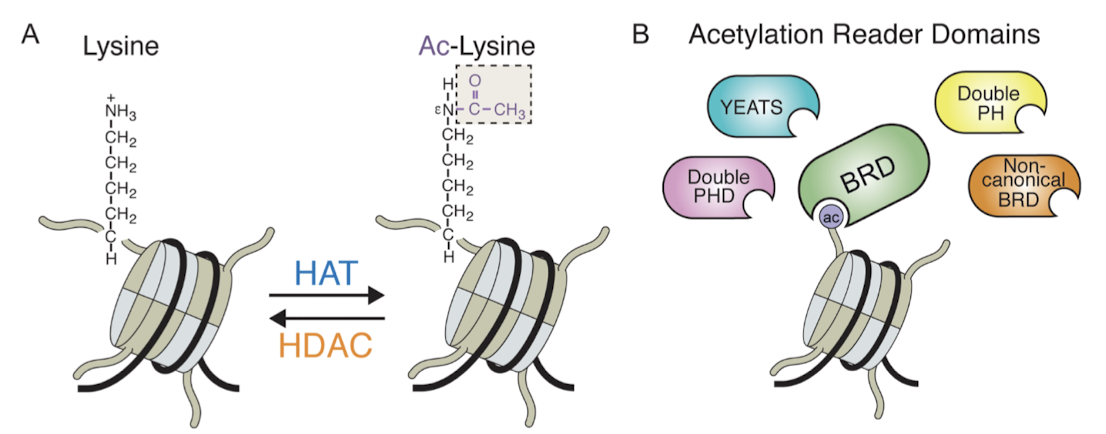
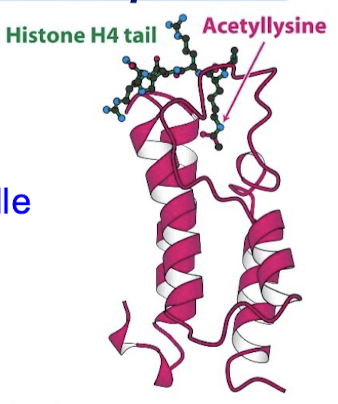
What is the structure of a bromodomain?
Comprises about 110 aa that form a four-helix bundle containing a peptide site at one end.
What complex do bromodomains form? What kind of activities is this complex capable of? What is the function of the complex?
Bromodomain proteins form a large chromatin remodeling complex that has ATPase and helicase activities. The complex alters nucleosome conformation and structure.