BIOL 2044 - bacterial stress response
1/17
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
18 Terms
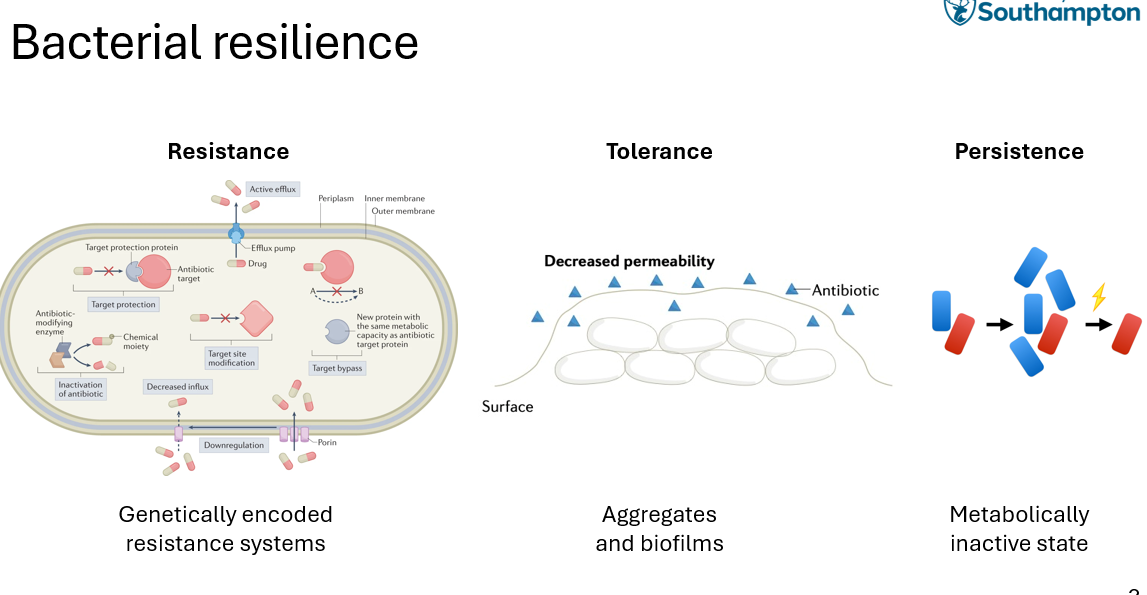
bacterial resilience - defining resistance, tolerance, persistance
RESISTANCE
implies genetically encodes systems
includes mutation, modification
E.g: active efflux —> mechanism to ensure that AB doesn’t see target, bacteria produces a target for the AB so that it doesn’t bind to it
TOLERANCE
bacteria grows in biofilm to produce a physcial barrier so AB cant penetrate
PERSISTANCE
bateria can eneter a dormant state so that they can survive stressful conditions
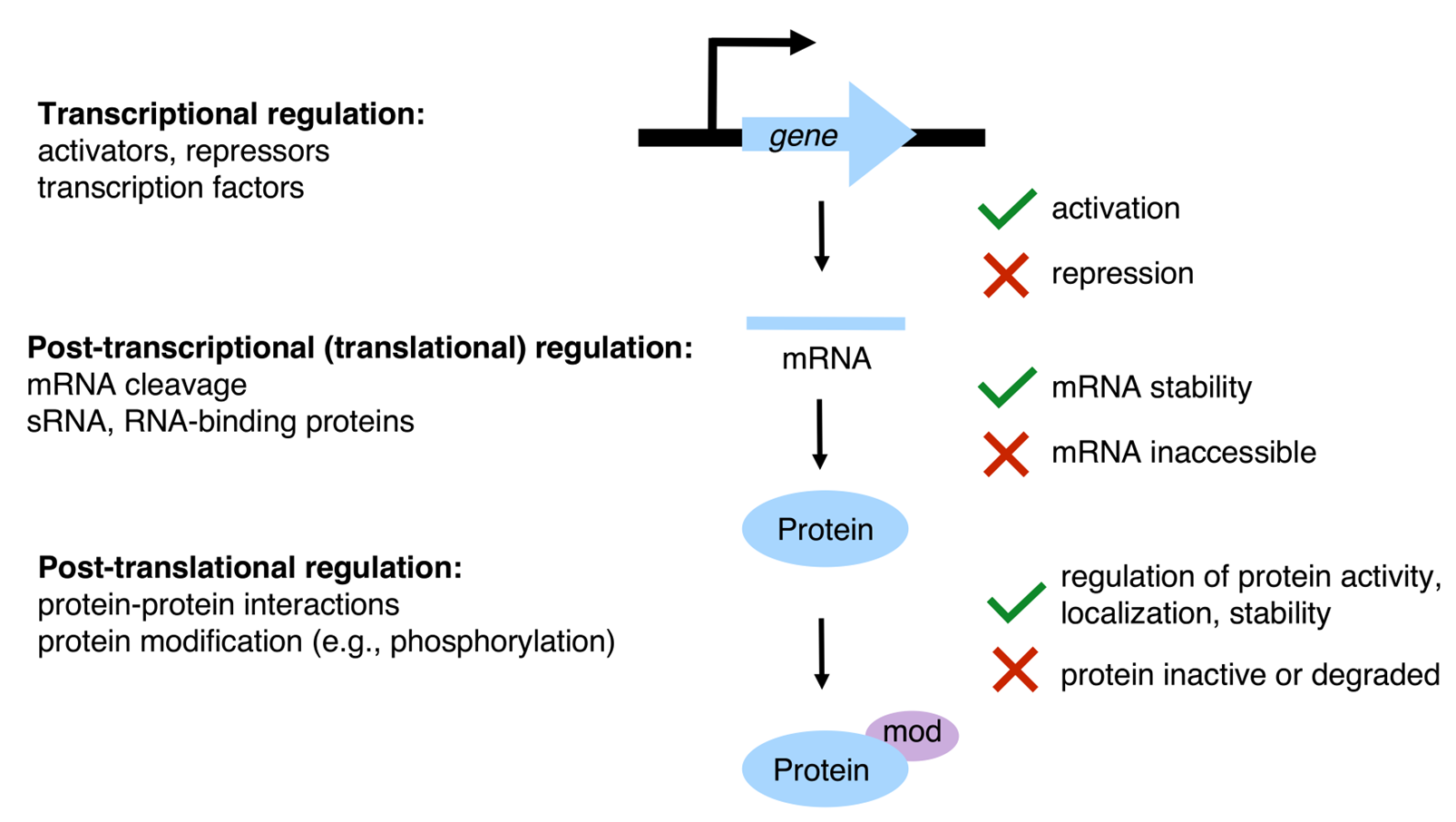
gene regulation in bacteria
occurs at 3 levels:
transcriptional regulation
expression of activators, repressors, TFs
post transcriptional regulation
at the mRNA level
mRNA cleavage, sRNA, RNA binding proteins
can downregulate protein translation by changing RNA stability
post translational regulation
at the protein level
protein-protein interaction, protein modification
can target proteins for degradation
ensure that bacteria can respond quickly at the appropriate level
E.g: if bacteria changes environments and requires a diff set of proteins, then will respond at the transcriptional level
E.g: if toxins are present then intervention at the proetin level best to modify the protein-protein interactions
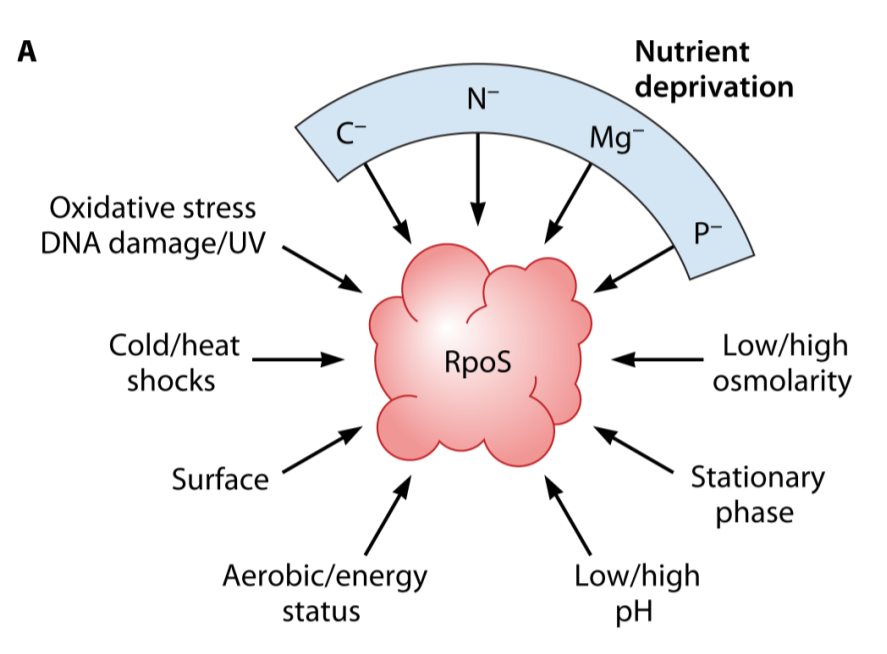
sigma factors
sigma factors not all the same
housekeeping sigma
other stress sigmas which can downregulate certain programmes and upregulate others
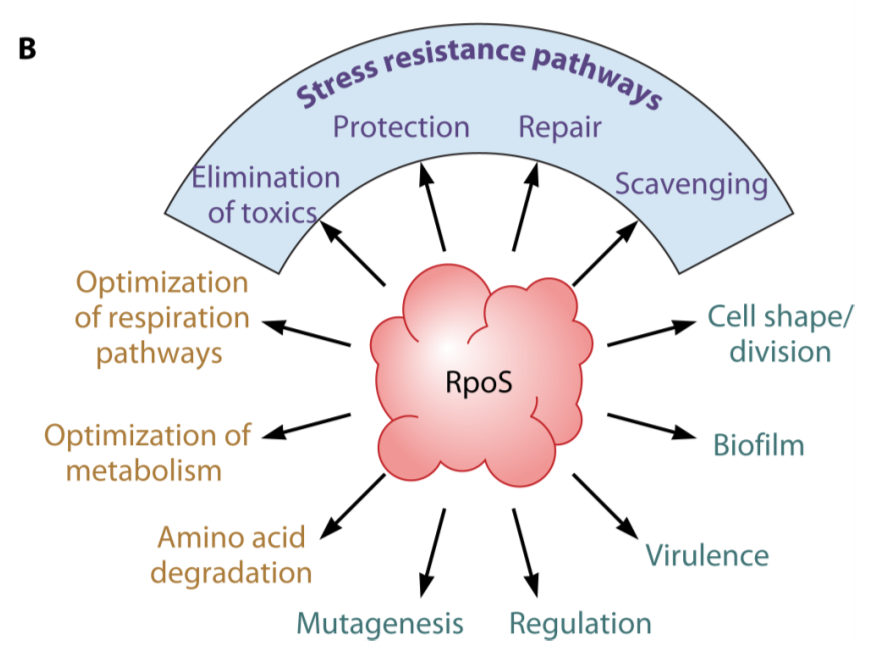
RpoS - general stress response sigmas
nutrient depletion/starvation
co-ordinated many stress responses
elimination of toxins
protection
repair
scavenging
activated at gene transcription, mRNA transcription and protein level by:
N-
Mg-
P-
C-
difference in pH, temp, oxidative stress, osmolarity
genes controlled by the RpoS regulon containing over 400 genes
stringent response
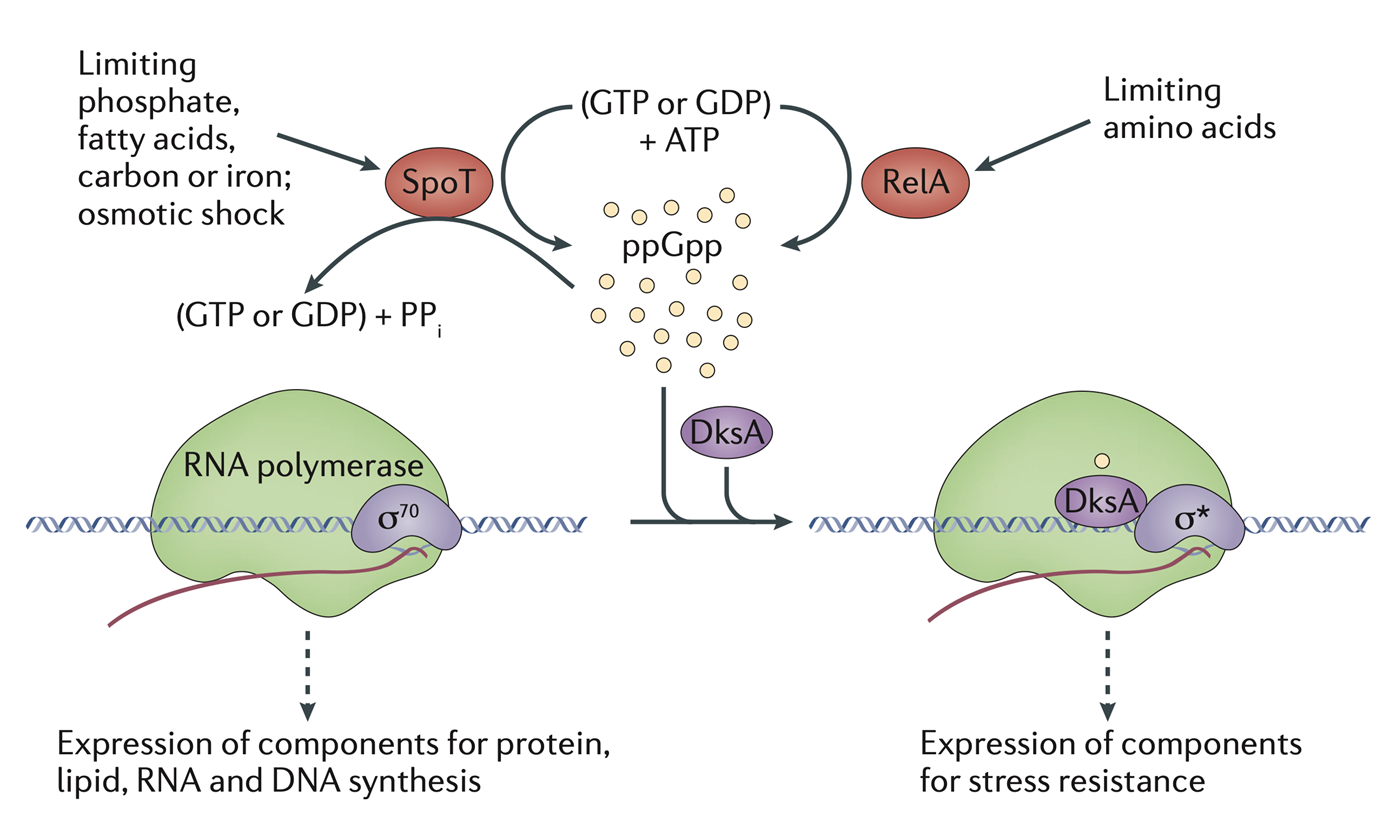
mechanism of RpoS regulation
when uncharged tRNAs (tRNAs with no amino acids attached) theyre detected by RelA which becomes active
Rel A synthesises ppGpp and pppGpp from ATP and GTP
SpoT helps to regulate the (p)ppGpp levels
(p)ppGpp then binds DskA TF which binds RpoS sigma which sequesters core RNAP to transcribe stress resistance genes
nutreint depletion causes sigma 70—>RpoS
SpoT and RelA kinases become active in response to low carbon, iron, phosphate, fatty acids
accessory genes
sigma factors general/specific stress response are core genes - all E. coli have them
some mechanism in accessory genes
located in plasmids, transposons, genomic islands
transferred by horizontal gene transfer
can encode:
AB resistance
virulence factor
metabolic adaptations
stress response and survival mechanisms
phage defence
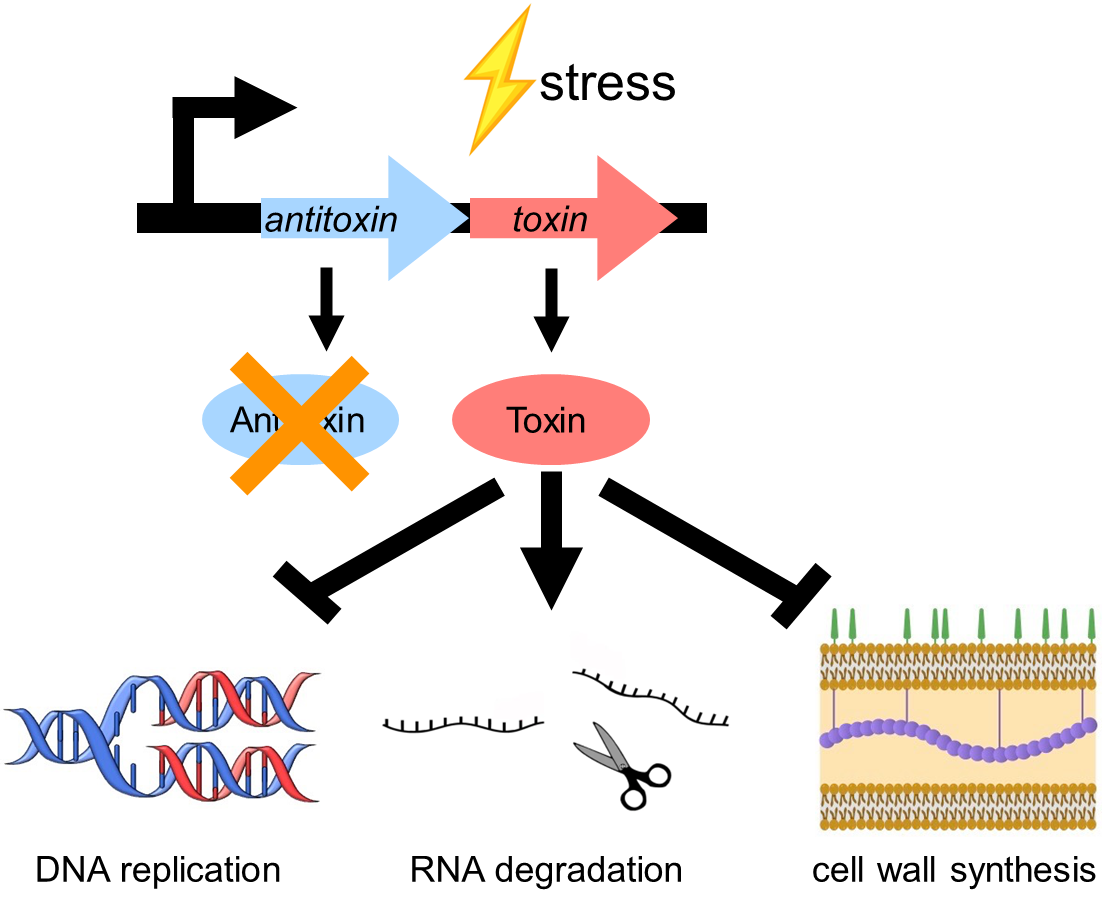
toxin/antitoxin systems - genetic elements involved in carrying these functional traits
roles of toxin/antitoxin systems
dormancy/lysis
antiphage defence
stress tolerance
persister cell function
some require this for infection
important for understanding AB resistance
each bacterial species has their own toxin/antitoxin sequences in chromosomes
varies among bacterial strains belonging to the same species —> can be problematic as when studied in the lab the genes wont be the same as in the patient
mainly found on chromosomes but also found in plasmids having a structural role
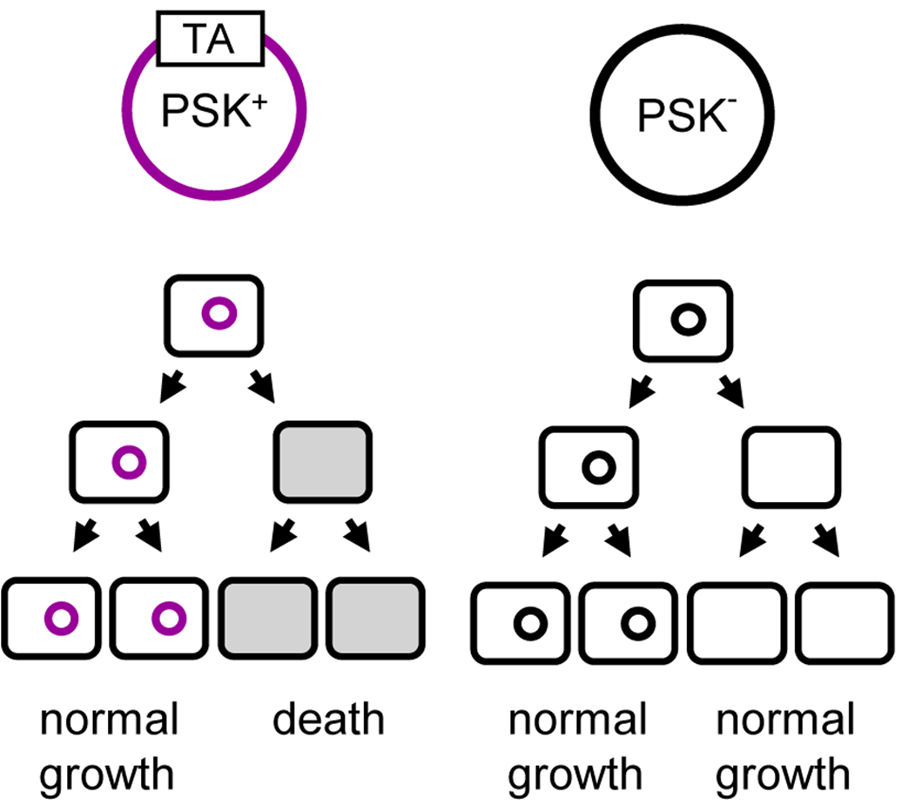
antitoxin is usually less atble —> if antitoxin degraded the toxin may kill the cell
if T/A plasmid in the cell is not present and not actively producing T/A the antitoxin remaining in the cell will degrade faster leaving the toxin to do its job and kill the cell (has a structural role)
if bacteria has a plasmid without T/A genes then progeny can survive with or without but this ensures that only bacteria with T/A plasmid survive if their parents also have this system
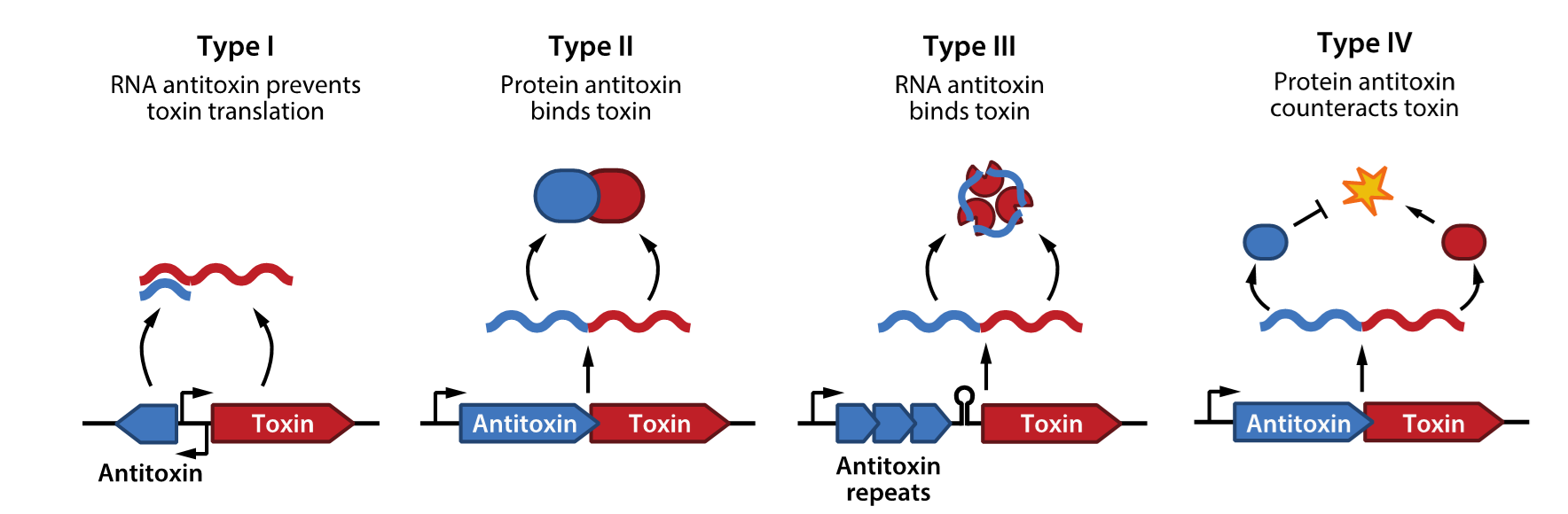
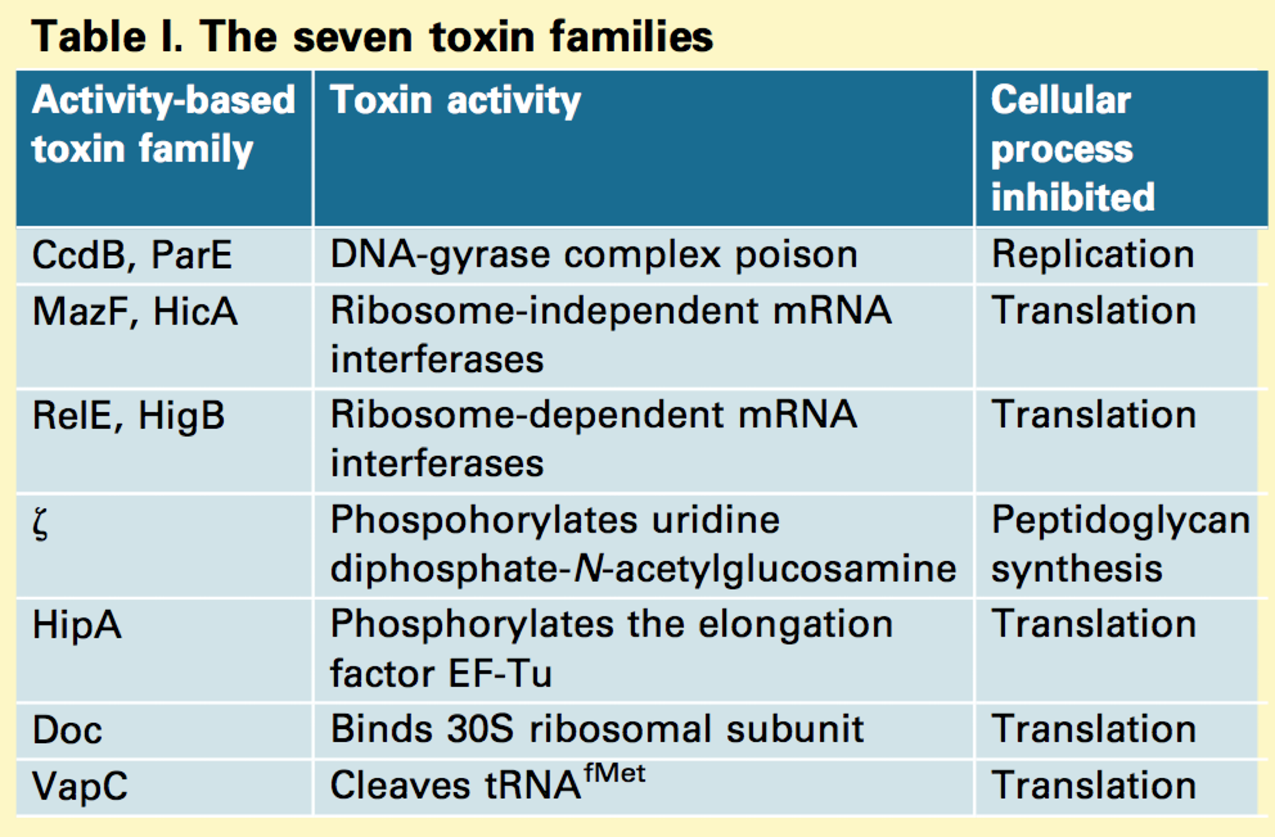
classification
8 types
3 party antitoxin/toxin systems
based on nature and mode of action
TYPE 1
antitoxin mRNA
antitoxin prevents toxin translation
TYPE 2
antitoxin protein that binds toxin
TYPE 3
RNA antitoxin
binds toxin
TYPE 4
protein antitoxin that counteracts toxin
intermediate players
Type 2 example
antitoxin/toxin in operon, transcribed at once
stress upregulates proteases which degrade antitoxin —> leaves toxin to do its role
an interfere with DNA replication, impede cell wall synthesis, degrade RNA
when theres no stress antitoxin binds to toxin which acts as a repressor and downregulates transcription of T/A
many diff toxin failies
complex network
dont undergo complex regulation
undergo crosstalk with one another and regulate the expression of stress response gene
E.g: all have diff affinities for tRNA so the network is very complex
no bacterial genome that doesnt have toxin/antitoxin system (some have more than 1) —> even found in plants and higher organisms
degrading mRNAs not needed in diff environment, can be good for cells to adapt to new conditions quickly
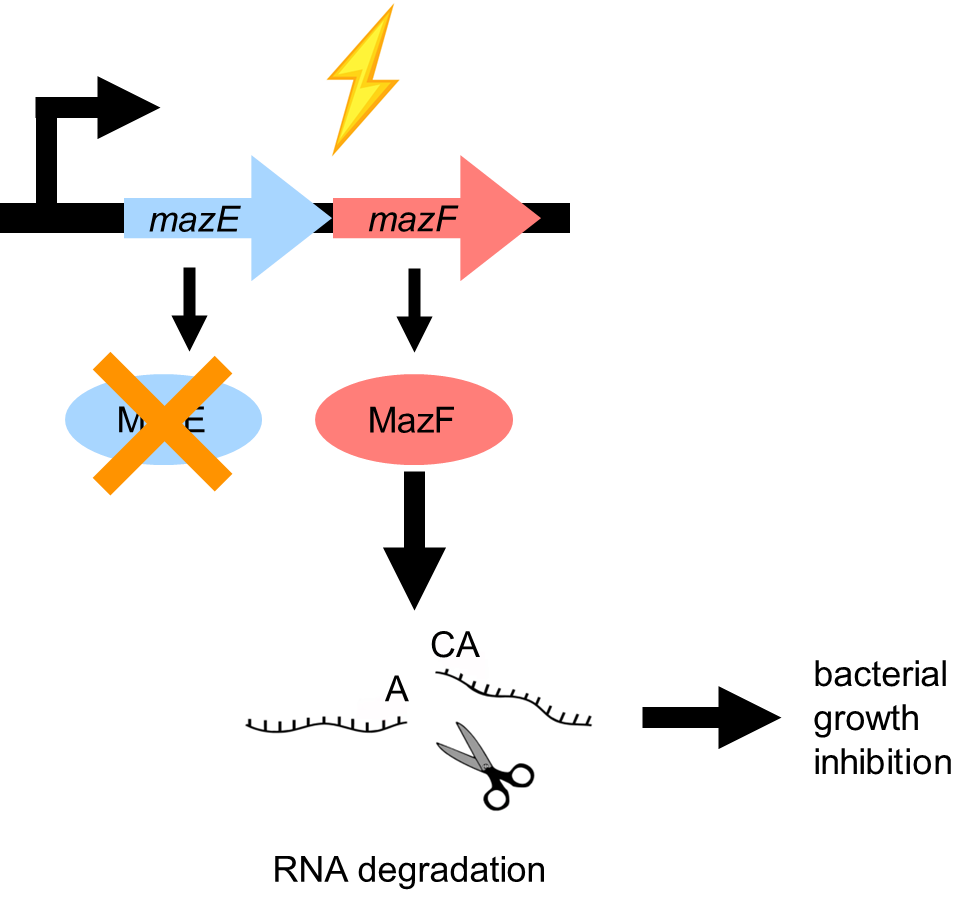
type 2 A/T system —> Maz E/F system in E. coli
MazF= toxin, MazE= antitoxin
when stress is present antitoxin is degraded
toxin then degrades RNA t the ACA sites —> doesnt sirectly induce lysis
bacteriostatic not bacteriocidal
in no stress MazE binds MazF —> no downstream effects and acts as repressor of transcriptional unit
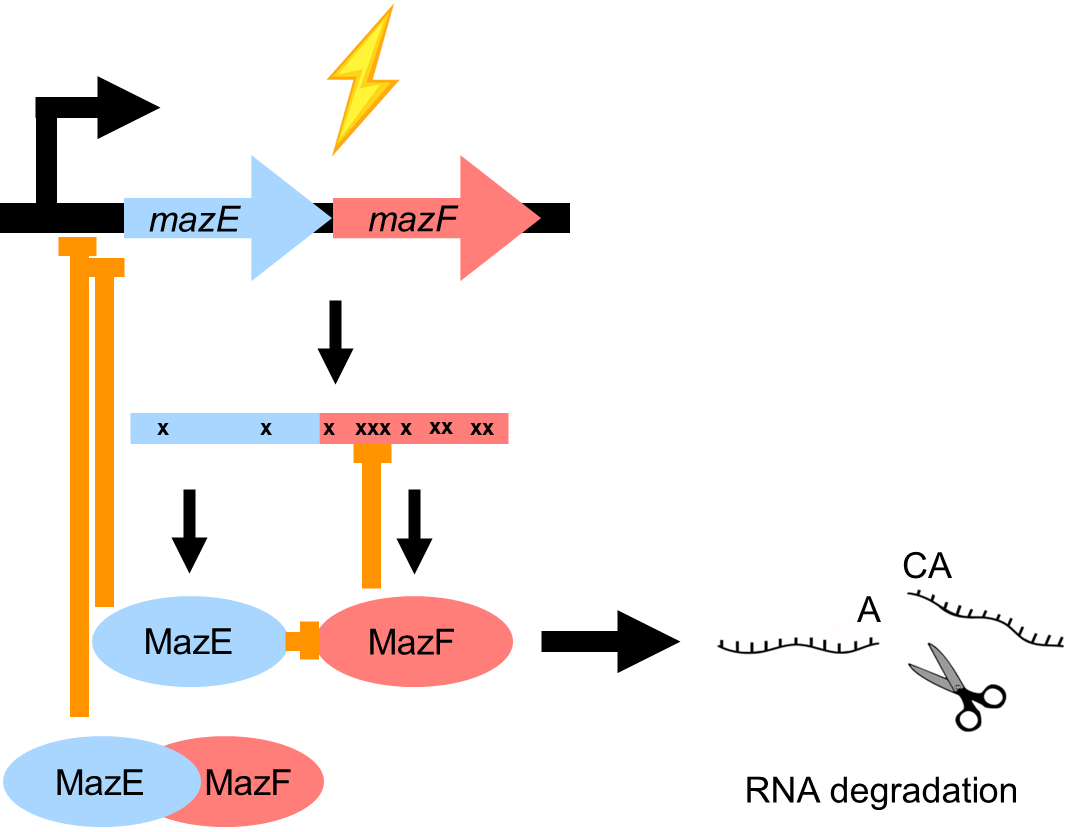
ACA motif not rare —> MazF cleaves its own RNA —> causes dynamic regulation of bacteril cellular processes
causes oscillating MazF levels during period of stress
fast regulatiom of MazEF underlies growth heterogeneity (due to RNA degradation)
beneficial for bacteria as at any point a fraction of the stressed population in the ready to exit state (bet hedging)
role in phage defence
endonucleases such as Ma2F are involved in phage defence
cleave mRNA of DNA phage genomes and RNA phage genomes
abortive infection (toxin induced cell lysis/metabolic arrest)
with no Abi can produce progeny phage particles
with Abi interupts the spread of infection - phage adsorbs to cell and is taken in, interrupts replication cycle
cell either lysed or enters metabolic arrest so tht the phage cant spread
only works when phage DNA/RNA already there although there are other mechanisms to prevent phage entry
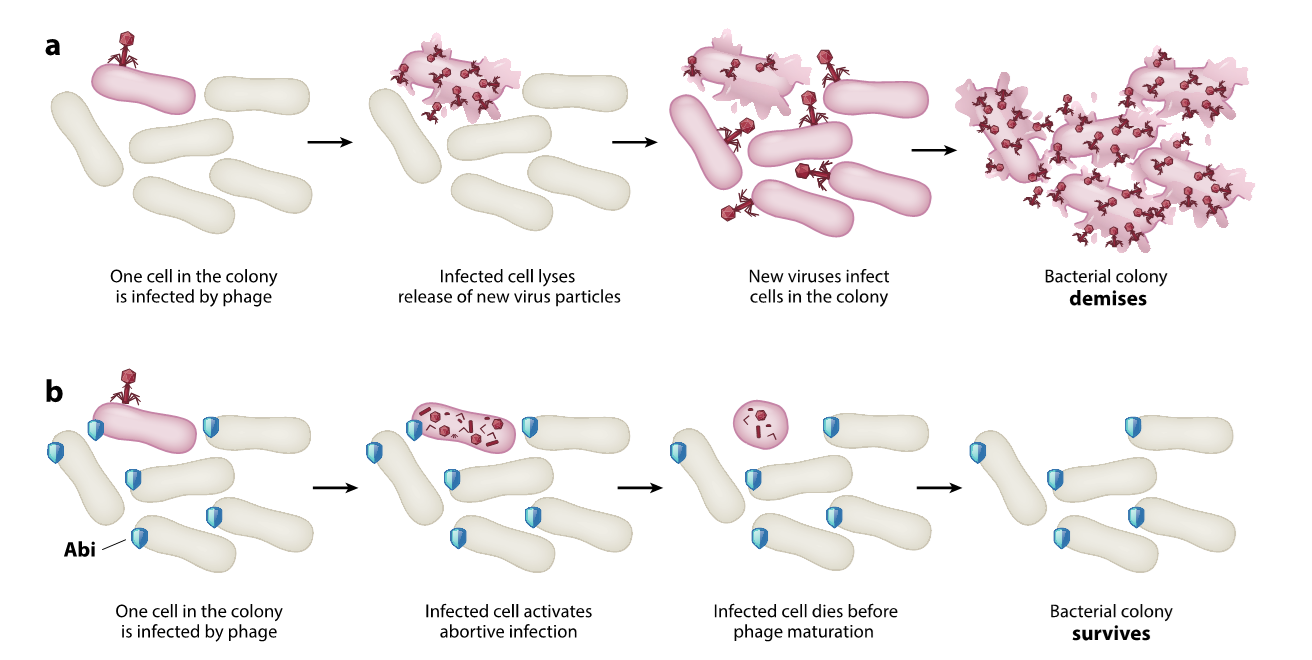
mechanisms of defence
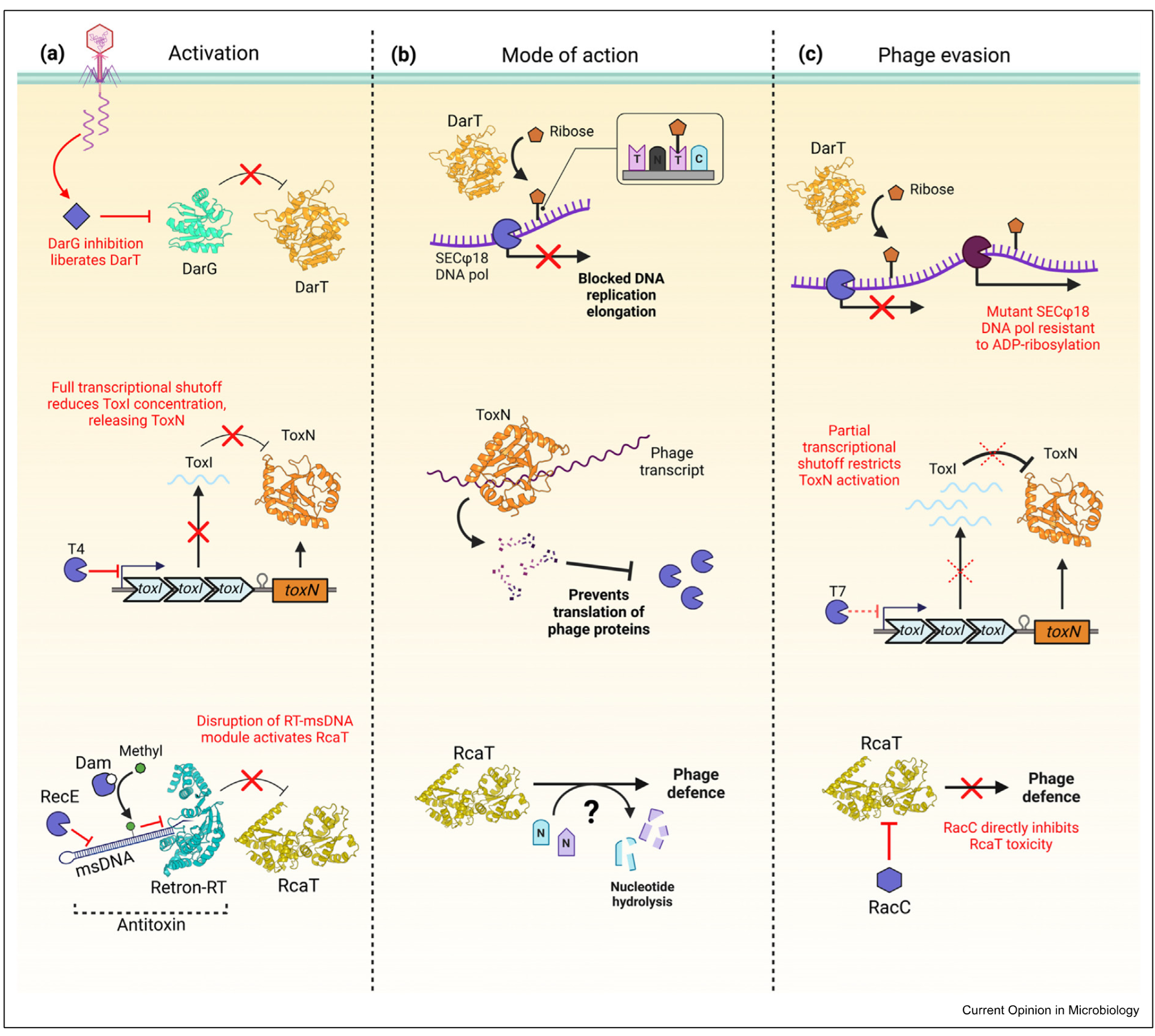
DarTG
ToxIN
Retron-sen2
DarTG
type 2 system - toxin and antitoxin proteins
DarG antitoxin, DarT toxin
DarT is an enzyme which decorates phage DNA with ribose —> pol stalls and phages cant replicate
phage evasion: mutates polymerase so it can replicate DNA when ribose still there
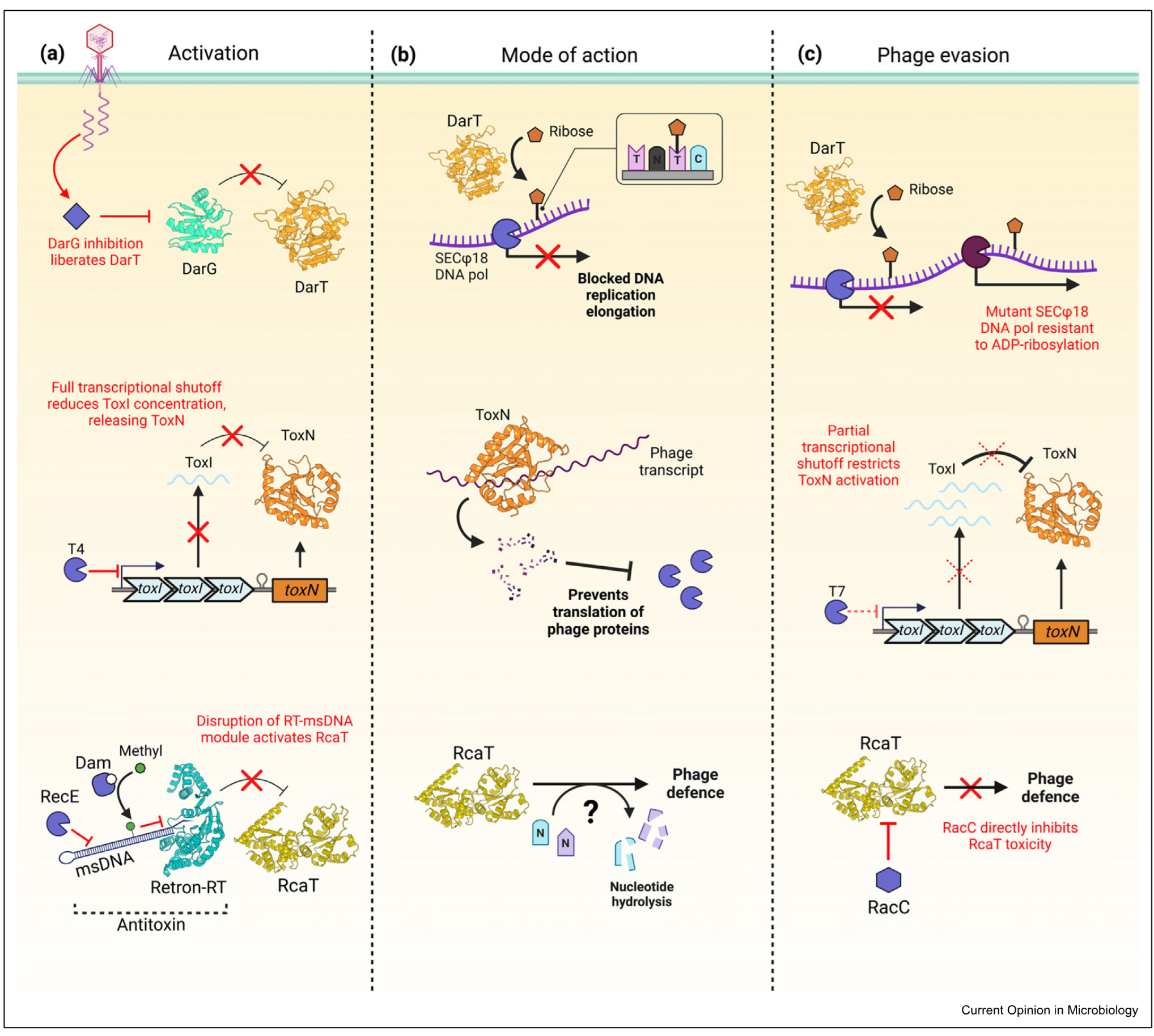
ToxIN
type 3 system
toxin is a protein, antitoxin is a repeat of small RNAs
toxN is toxin
bacteria shuts down trancription - prvenets phage propogation and bacterial propogation
when transcription shut down antitoxin degraded and no more is made —> activates toxin
ToxN degrades phage trancript
phage counteracts this only allowing partial transcription
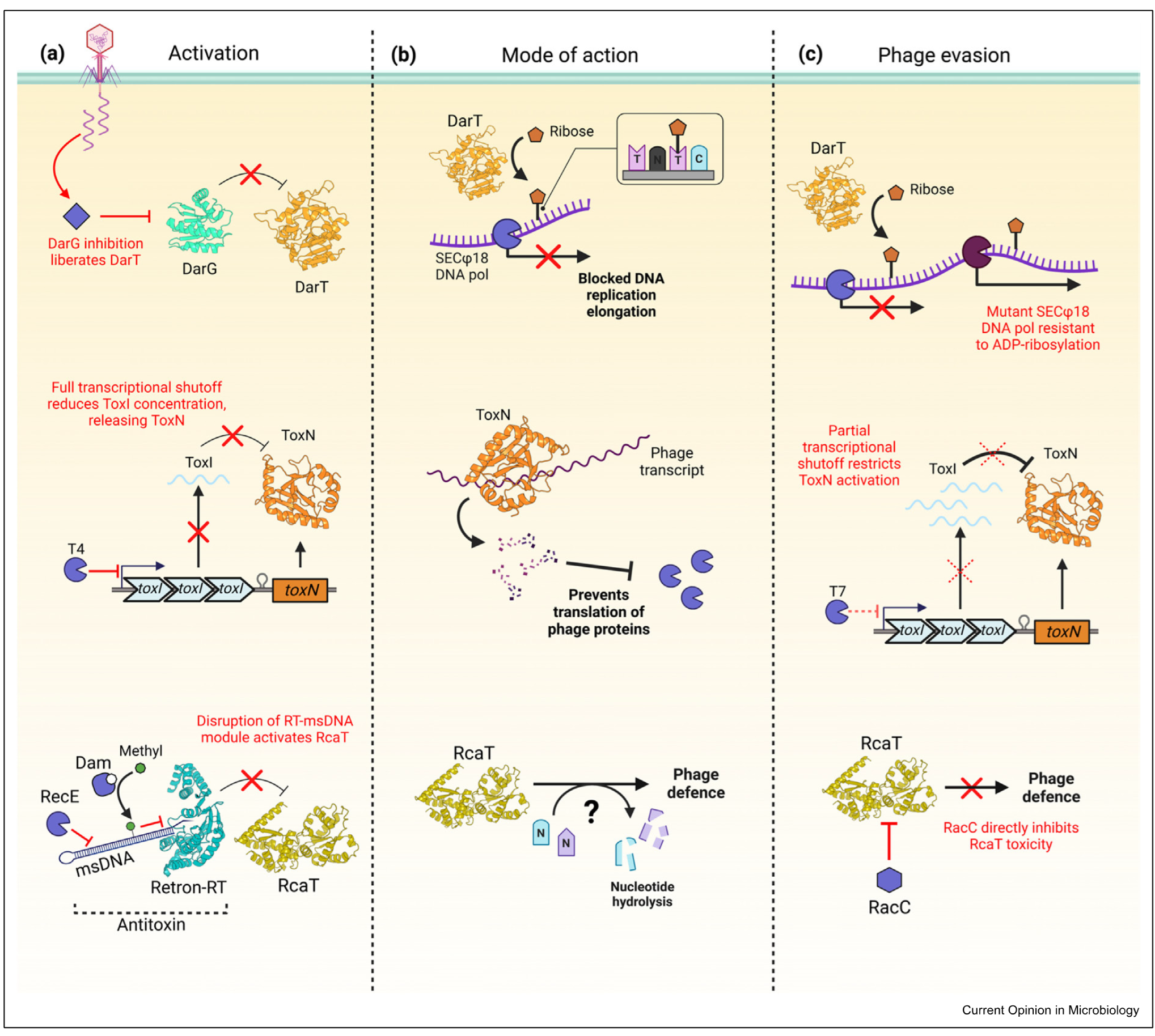
Retron Sen 2
3 player system
toxin is RcAT, antitoxin is reverse trancriptase that produces multicopy starnd ssDNA made up of Retron RT and msDNA
onlly stable in one form - when msDNA bound to retron RT
during pahge infection diff degradation mechanisms take place which effect stability of antitoxin
msDNA degraded so that it cant neutralise the toxin
during phage infection diff degradation mechanisms take place which affect stability of antitoxin
msDNA can also be methylated to have the same effect
toxin doesnt degrade RNA but nuclease hydrolase interferes with DNA and RNA so cant produce progeny
as a countermeasure pahge has antitoxin neutralise protein (RaCC) which binds the toxin and renders it inactive
phages vs antibiotics
PHAGES
can be very specific (can have broad host range though)
replicate at the site of infection
have no side effects
development of resistance
our body has response to phage but not immune response
finding new pahges is fast
ANTIBIOTICS
non specific
not concentrated at the site of infection
multiple side effects
developing new ABs is slow
best therapies are now thought to be combination therapies of pahges and antibiotics
phage therapy
if patient infected with AB resistant bacteria —> isolate and identify strain
in vitro screening of specific lytic phages isolated from various environmental and clinical samples or sources from pahge banks
safety/efficay trials
inhaled
applied to wounds
bladder irrigation
orally
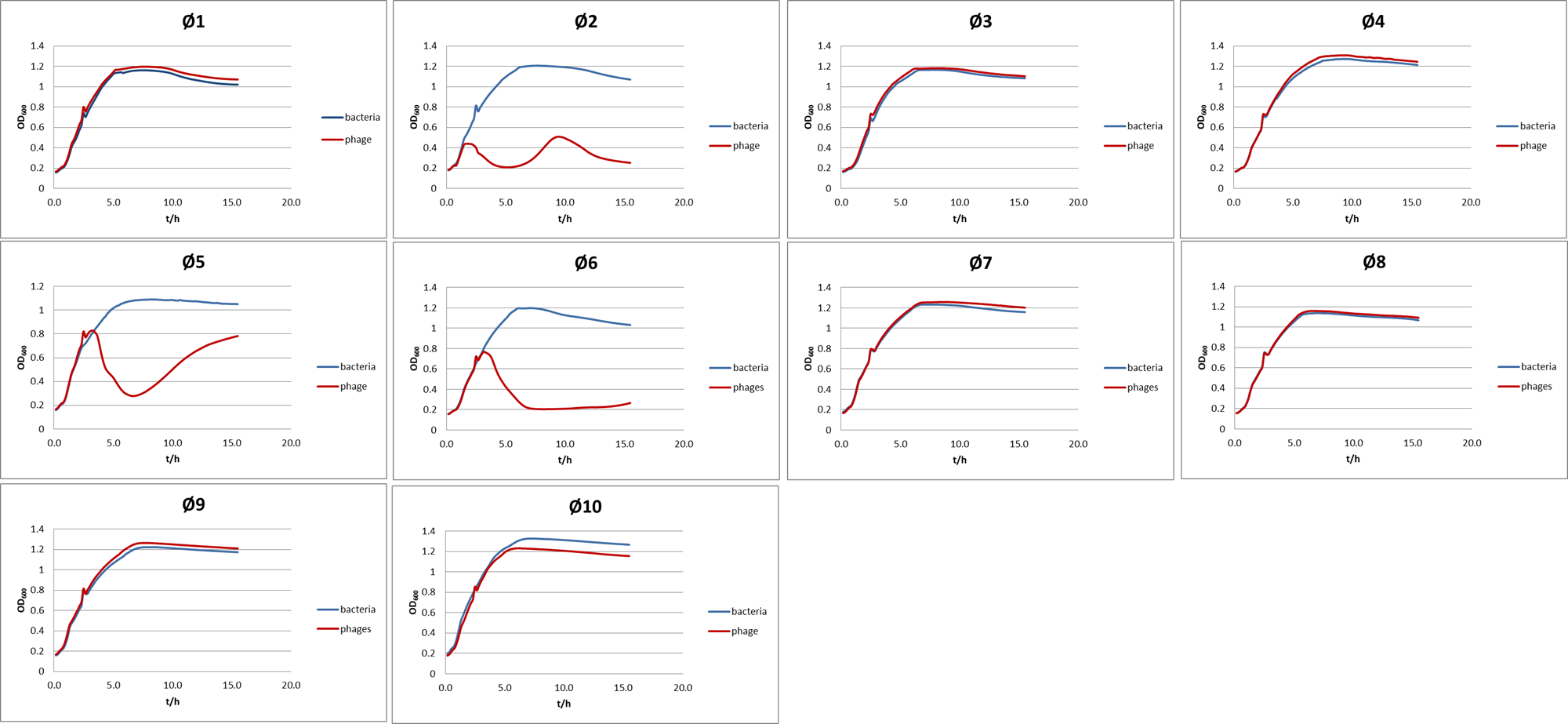
how do we screen phage collection
add 180ul LB to 10ul of bacteria
add 10ul of pahge for each (and the control)
measure absorbance at 600nm every 10 min for 12h (allow bacteria to grow 2-3 hours before adding phage
if theres a decrease in the phage line OD600 with bacteria and phage then its able to supress bacterial growth
sometimes when challenged with phages different modes of action ensures that the bacteria cannot develop a counter defence mechanism —> killed before then
also want to ensure phage only kills strain of interest
can be deemed costly especially due to the personalised treatment for phage