Topic 5: Homologous Recombination
1/97
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
98 Terms
What is homologous recombination:
Physical exchange of DNA segments between two DNAs of identical or very similar sequences
What does any exchange of DNA segments involve?
Must involve breaking the DNA and resealing the breaks
→ Can be viewed as a DNA repair mechanism
What are three applications for homologous recombination?
1) Creates genetic diversity in gametogenesis
exchange between non-sister chromatids of homologous chromosomes can involve 100s of alleles
2) DNA repair of nicks and DSBs and spontaneous induced mutations during DNA replication
3) Biotechnology: targeted gene knockouts
4) Cellular processes such as mating type switching in yeast and intronic transposition
How many pairs of sister chromatids is involved in HR? How many strands does each chromatid contain? How many strands total are in involved in HR?
→ 2 pairs of sister chromatids
→ 4 chromatids
→ 2 strands/chromatids = 8 strands total
What is the first step in repairing DSBs by HR?
DSB in sister chromatid is processed where the 5’ ends of both strands are degraded by exonucleases and unwinded by helicase (5’ DNA resection) which forms ssDNA 3’ overhangs coated by SSB
What happens following the formation of 3’ overhangs in DSB repair (HR)?
The SSBs are displaced from ssDNA and replaced by recombinase RecA/Rad51 to form a filament that searches and invades homologous sequence
What occurs following the coating of RecA/Rad51?
One of the 3’ overhangs invades a homologous donor (first strand invasion) forming a D-(displacement) loop and heteroduplex DNA
Then second strand invasion occurs with other 3’ overhang
What occurs following both strand invasions in DSB repair (HR)?
DNA polymerase extends the two invading strands from the 3’ OH using the homologous chromosome as template to extend and restore the lost information at the DSB
Two DNA crossovers are formed which are then resolved in several ways to complete the HR repair process
What are the two different pathways to complete the HR repair process?
Synthesis-Dependent Strand Annealing (SDSA) pathway
Double-strand Break Repair (DSBR) pathway
What are the steps in the SDSA pathway?
Invading strands dissociate by SRS2 helicase and anneal back to each other, followed by DNA replication and ligation to complete the process
SRS2 disrupts D-loop
No heteroduplex DNA
No crossover formation
The main DNA repair mechanism in somatic cells
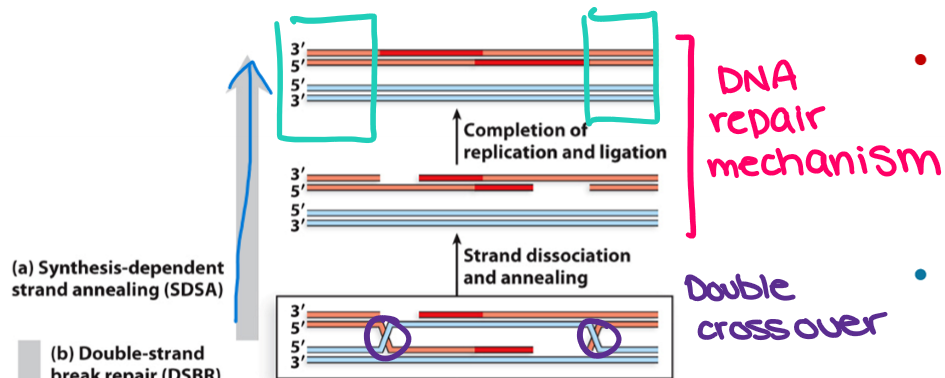
What are the steps in DSBR pathway?
Invading strands do not dissociate during DNA replication and cause four-way helical DNA junctions or cross-overs (Holliday junctions) that undergo two symmetrical nicks by resolvases and then ligated
D-loop stays intact, and movement of Holliday junctions (branch migration) increases the length of heteroduplex DNA
Depending on how the Holliday junction is resolved, crossover products can be generated, whereby allelic combinations flanking the DSBs no longer resemble the parental chromosomes
Predominant in meiosis, not mitosis
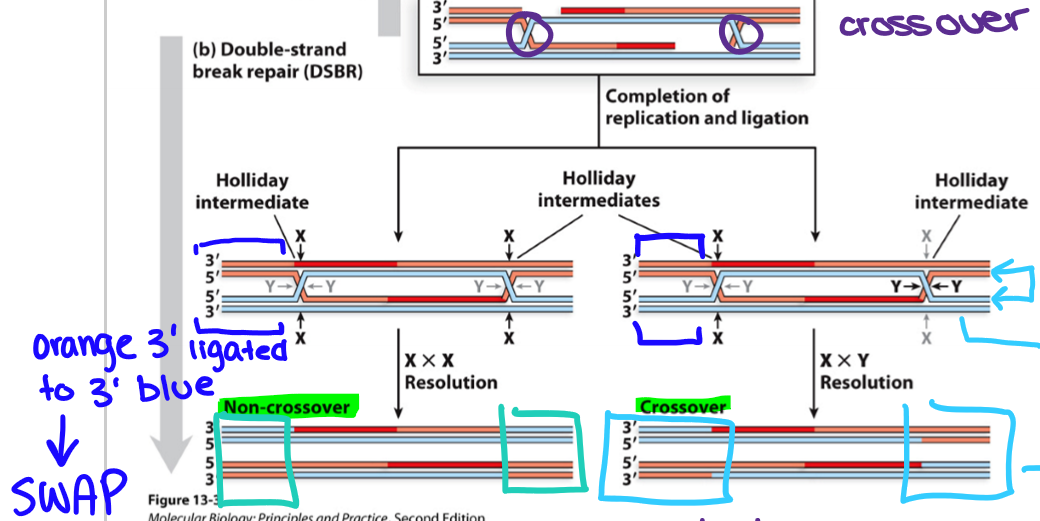
How are Holliday junctions resolved (one crossover)?
Need to cut two of the four strands (of inner two chromatids) with the same sequence and polarity at the site of the cross (two cleavage sites possible) and then ligate the nicked strands
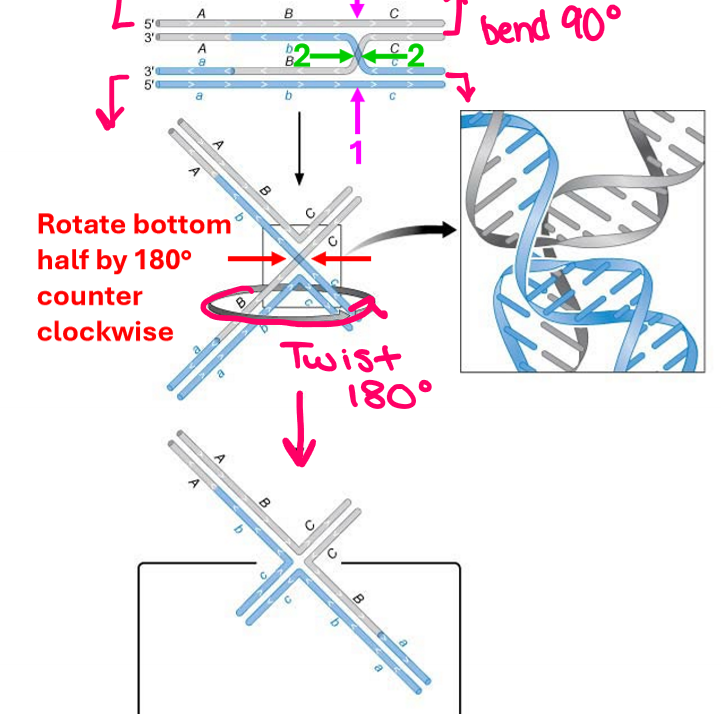
How do you draw a crossover site to better visualize it without strands crossing?
Draw as an open cruciform structure by bending ends at 90 degrees and rotate bottom half by 180 degrees
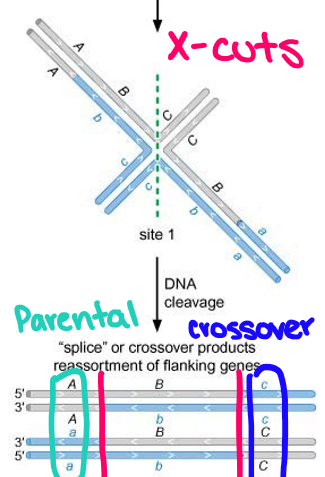
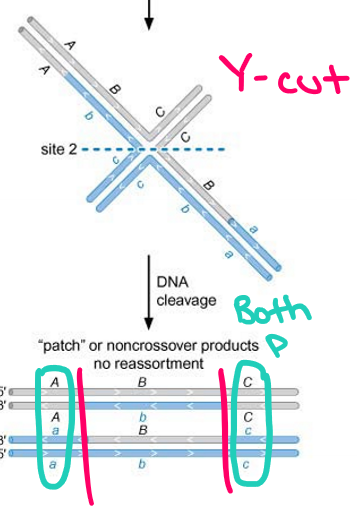
What does site 1 refer to? What kind of products does it create?
Vertical mode of resolution or X-cuts: nick occurs in the two strands composed entirely of the parental DNA resulting in the “spliced” product after ligation of the nicks
Spliced products: crossover occurs between genes flanking the DSB whereby alleles of genes flanking the DSB are no longer parental (A-c and a-C)
What does site 2 refer to? What kind of products does it create?
Horizontal mode of resolution (Y-cuts): nicks occur in the two strands that have a mixture of regions from both parental and the DNA strand
Patched products: non-crossover whereby alleles of genes flanking the DSB remain parental (A-C and a-c)
How many possibilities of resolving Holliday junctions with ‘x’ and ‘y’ are there with two crossovers?
4 possibilities: 1-1, 1-2, 2-2, 2-1
Where do horizontal and vertical cleavage sites occur on DNA strands?
Occur on DNA strands with the same orientation
What happens if two crossovers are cleaved with the same site? (e.g. 2-2 or 1-1)
Noncrossover products
What happens if two crossovers are cleaved with different sites? (e.g. 1-2 or 2-1)
Crossover products
How do crossovers/Holliday junctions move along DNA?
By repeated melting and formation of base pairs (hydrogen bonds)
What occurs each time the Holliday junction moves?
Base pairs are broken in the parental DNA molecules (green circles) while identical base pairs are formed in the recombination molecule (red circles)
What does heteroduplex DNA contain that was not in the parental DNA?
Mismatched bases
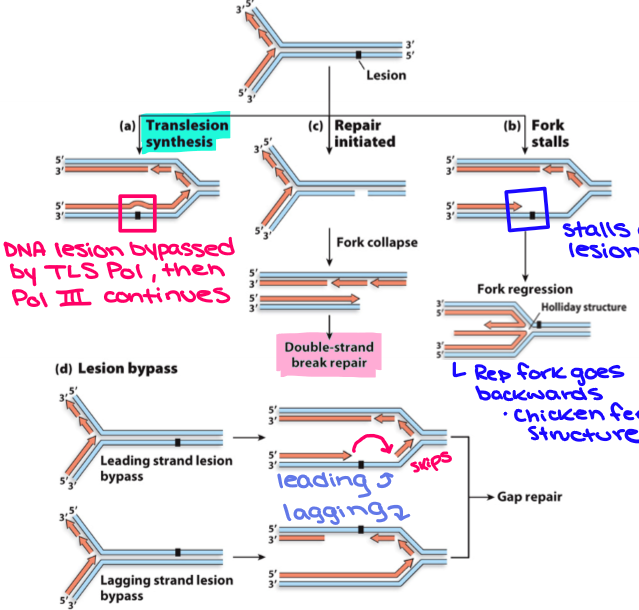
A lesion on the template strand is not repaired in time before collision with replication fork, what can all be done in response to it?
TLS synthesizes DNA across the legion
Fork stalls when DNA pol III reaches lesion, but sugar phosphate backbone is intact. Fork is moved backwards past the lesion whereby leading and lagging strands are unwound from template and rewound to form a duplex (also known as the chicken foot intermediate)
Fork encounters nick in template → fork collapses → DSB
Replisome blocked by lesion but then proceeds to restart replication downstream from lesion leaving a gap in daughter strand (more common in lagging)
2-4 can be fixed by recombinational DNA repair
How does a collapsed fork lead to DSB?
Nick in template strand stops DNA replication as replisome disassembles and fork collapses creating a DSB
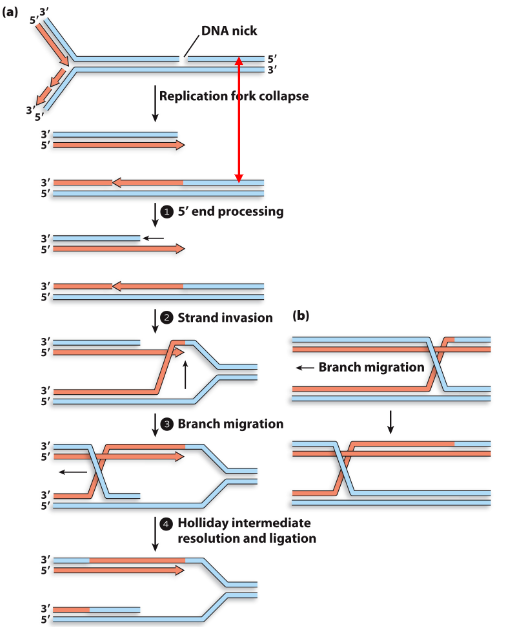
When HR repairs a collapsed fork, what serves as a template to repair the DSB and reconstruct the fork?
Other duplex DNA (sister chromatids) is intact, thereby serving as a template to repair the DSB and reconstruct the fork
How is a three-stranded junction formed in HR repair and reconstruction of a collapsed fork?
1) 5’ end of the strand at DSB is degraded/processed, and strand invasion occurs with 3’ extension
2) 3’ extension after strand invasion is not extended, forming a three-strand junction
What happens following the formation of a three-stranded junction in HR repair and reconstruction of a collapsed fork?
3) Branch migration moves the three-stranded junction backwards to form a stable-four strand Holliday junction
What does resolving and ligating of the Holliday model produce in HR repair and reconstruction of a collapsed fork?
4) Resolving and ligating of Holliday model produces two intact DNA duplexes encompassing the original DSB
How is the fork reconstructed in HR repair and reconstruction of a collapsed fork?
5) Fork is reconstructed by reloading the replisome to restart DNA replication
What acts as a barrier for DNA replication causing the fork to stall?
A lesion on the DNA template strand
What first occurs to deal with a lesion on the template strand (HR repair)?
Fork regression: reversal of fork from the lesion by RecG helicase whereby the leading and lagging strands detached from the template strands and bind to each other while the lesion strand is annealed to the other undamaged template strand
What is created by fork regression? (HR repair of stalled fork)
A Holliday structure whereby four DNA strands are connected at a central point; a chicken foot intermediate
This buys time for the repair pathway to fix the lesion
What occurs following the formation of the chicken foot intermediate if the lesion IS repaired?
If lesion is repaired, the leading and lagging strands of the regressed fork is degraded by exonucleases until the base of the fork (resolving the Holliday junction) allowing the fork to restart after replisome loading

What occurs following the formation of the chicken foot intermediate if the lesion is NOT repaired?
If the lesion is not repaired, the short DNA arm is replicated, followed by branch migration away from the lesion until a fork structure is reconstructed at the lesion
What generates a gap in a daughter strand?
When DNA pol skips over a lesion that has not been repaired and starts DNA synthesis after the lesion
What is the first step in gap repair after fork bypass (HR)?
1) Recombinase RecA binds the DNA segment of the template strand (gap) and promotes a strand invasion of the undamaged sister chromatids (there is no 3’ extension here)
What is the next step following strand invasion in gap repair after fork bypass (HR)?
2) branch migration created two Holliday junctions
3) displaced strand serves as undamaged template DNA to fill in the gap by DNA polymerase (high fidelity unlike TLS)
What happens following resolution of Holliday structures in gap repair after fork bypass (HR)?
Holliday junctions are resolved which fills in the gap with an undamaged DNA segment and the lesion can then be repaired by NER/BER pathways
What does RecA do?`
Enzyme in Bacterial Recombination:
Recombinase binds to ssDNA 3’ extension to form a nucleoprotein filament for strand invasion of a homologous region
looks for homology for strand invasion
What does RecBCD do?
Enzymatic complex in Bacterial Recombination:
generates 3’ extensions for recombination repair of DSBs
What does RecFOR do?
Loads RecA on ssDNA during gap recombination repair
What does RuvABC do?
Enzymatic complex in Bacterial Recombination:
recognizes and binds Holliday junctions and promotes branch migration
also creates two nicks to resolve it
How does RecBCD generate 3’ extensions?
RecBCD complex binds to both sides of a DSB and RecB (3’-5’) and RecD (5’-3’) helicases begin unwinding the duplex (ATP hydrolysis)
3 attached components
What activity does RecB have that RecD does not? How fast does it travel?
RecB has endonuclease activity that will degrade both strands during unwinding (with ATP hydrolysis)
RecB travels slower than RecD so there is a ssDNA loop that forms ahead of RecB
What alters RecBCD nuclease activity?
When complex runs into an 8-mer Chi site which is bound tightly to RecC
What does tight binding of RecC to Chi prevent?
Prevents further degradation of 3’ end, but degradation of 5’ end is increased → producing a 3’ extension for strand invasion
What does RecBCD through RecB nuclease domain interact with to promote its assembly on 3’ extension?
Interacts with RecA recombinase and promotes its assembly on 3’ extension ssDNA prior to SSB binding
What has higher affinity for ssDNA, SSB or RecA?
SSB, so they need to be displaced for RecA to bind ssDNA section of gap repair
What recruits RecFOR to the 5’ end of the gap?
RecF
What does RecO and RecR do?
Bond ssDNA and SSB proteins and causes a conformation change displacing the SSB proteins
What direction does RecFOR load RecA in on ssDNA?
RecFOR facilitates RecA loading and filament nucleation in a 5’-3’ direction on the ssDNA
What are 5 characteristics of RecA?
1) Ubiquitous DNA strand-exchange protein
2) Binds ssDNA and assembles cooperatively to form a nucleoprotein filament up to several thousand subunits long
3) Assembly involves nucleation 5’-3’ on ssDNA and disassembly occurs at non-growing end (ATP hydrolysis)
4) Filament searches for homologous sequences and aligns the ssDNA with the duplex DNA for strand invasion (synapsis)
5) strand invasion initiates intertwining of ssDNA and duplex DNA
What kinds of DNA can RecA promote strand exchange between?
ds linear and ss linear DNA
ds linear with ss circular
ds linear and ds linear
ds linear with ss gap
Upon strand invasion of RecA-coated ssDNA filament what happens?
RecA-mediated branch migration occurs by ATP hydrolysis
What determines the length of heteroduplex DNA?
Branch migration or movement of Holliday junction in 5’-3’ direction relative to RecA-coated ssDNA
What does extreme case of branch migration cuase?
Heteroduplex length = entire DNA molecule
How is branch migration detected?
By mixing circular ssDNA and linear homologous dsDNA, RecA and ATP followed by gel electrophoresis
What is RuvA’s role in RuvABC?
Two tetramers that recognize and bind to the four helices of the Holliday junction
What do the two RuvB hexameric translocases bind to?
Bind to RuvA tetramer and two helices
Undergoes ATP hydrolysis to catalyze the rapid exchange of base pairs for branch migration
Where is DNA propelled from in RuvB’s?
Propelled outward through the ‘donut hole’ of each RuvB moving the Holliday junction by thousands of bp/s along homologous chromosomes
What happens once Holliday junction is away from the lesion?
RuvC resolvase dimer is recruited to RuvAB and nicks two of the homologous strands with the same 5’3’ polarity
Then ligase seals the nicks and the recombination products can be “spliced” (crossover) or “patched” (noncrossover) depending on cleavage pattern by RuvC
How many stages are there in homologous recombination in eukaryotic meiosis?
Four:
Leptotene
Zygotene
Pachytene
Diplotene diffuse stage:
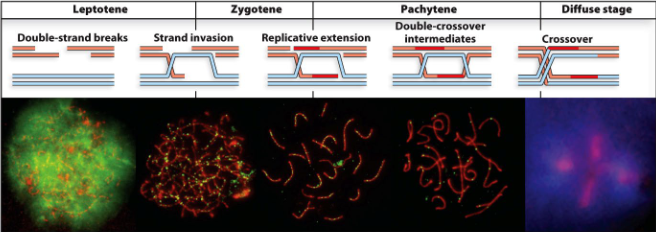
What is the leptotene stage?
Chromosome condensation starts, homologous chromosomes start to find each other, DSBs occur
What is the Zygotene stage?
Homologous chromosomes pair up (synapsis) and held together by the synaptonemal complex containing Rec8 meiotic cohesin; strand invasion starts
What is the Pachytene stage?
Crossing over (chiasma) and branch migration
What is the diplotene diffuse stage?
Synaptonemal complex dissolves, homologous chromosomes start coming apart and decondenses
What does Spo11 do?
Spo11 introduces DSBs in chromosomal DNA to initiate meiotic recombination
Acts as a dimer where each subunit makes a nick in one of the DNA strands
What does Spo11 have in its active site that allows it to create DSBs?
Tyrosine attacks phosphodiester backbone and a 5’ phosphotyrosyl linkage is formed
Spo11 remains covalently attached to the 5’ DNA ends of the DSB are are initial sites for processing and formation of 3’ extensions
What binds to each Spo11 complex?
Mre11-Rad50-Xrs2 (MRX) complex binds to each Spo11 complex and cleaves the DNA on the 3’ end of Spo11 liberating a short DNA fragment with Spo11 attached to 5’ end
What does Sae endonuclease do?
Degrades DNA strands a bit more from 5’ end
Which nucleases further degrade 5’ ends to create long 3’ ssDNA extensions?
Sgs1 helicase and Dna2 and Exo1 nucleases
in eukaryotes Exo1 is 5’-3’ unlike prokaryotes
What coats the ssDNA?
RPA
_____ promotes the assembly of two RecA-class recombinases ____ and ____, as well as displaces ___ to form nucleoprotein filaments
Rad52 (like RecBCD), Dmc1, Rad51, RPA
Once 3’ extensions have been formed at Spo11 DSB sites, what is the site ready for?
Strand invasion and recombination
What are the two outcomes for DSBs in Meiosis?
Non-crossover (like SDSA): invading strand dissociates and pairs with its complement on the other side of the break followed by further strand extension and ligation
Crossover: Two Holliday junctions are formed and cleaved by resolvases to generate recombinant meiotic products
What is gene conversion?
When an allele of a gene is lost and replaced by an alternate allele (can occur in crossover and non-crossover recombination)
Considering Mandelian genetics and AA/aa parents, what is the genotypes of the 4 gametes?
Should have 2 A gametes and 2 a gametes
With AA/aa parents and non-mendelian genetics (gene conversion) what would the genotypes of the 4 gametes be? What is it due to?
A, a, a, a OR A, A, A, a due to mismatch repair of mispairing of two different alleles
mismatch repair enzymes will randomly choose a strand to repair and depending which one will cause gene conversion to occur
What does budding yeast serve as?
A Eukaryote model system, with advances genetics, molecular biology, biochemistry, and functional genomics methodologies allowing for faster progress in characterizing genes
many cellular processes are conserved
What are the two mating types of budding yeast and how do cells differ under varying conditions?
Two mating types (alpha and a) determined by alpha and a alleles of MAT locus
Exists as alpha and a haploid or alpha/a diploid cells under nutrient-rich conditions
Under poor conditions, alpha and a cells will mate, undergo meiosis, and form spores
How many times can haploid cells switch mating types?
Once every generation
What do the MATa/MATα genes encode?
Encode transcription factors that activate and repress genes that define the mating type
→ the mating type gene expressed in the MAT locus determines the mating type of the haploid cell
Where is the genetic information for MATα stored but not expressed?
In the adjacent gene HMLα (left)
Where is the genetic information for MATa stored but not expressed?
In the adjacent HMRa (right)
What makes a DSB in the MAT locus?
HO endonuclease that recognizes MAT locus sequence
What does intrachromosomal recombination refer to?
DSB is repaired by gene conversion whereby a DNA sequence is copied from a donor template (HMLα/HMRa) and inserted into the broken site (MAT locus) on the same chromosome
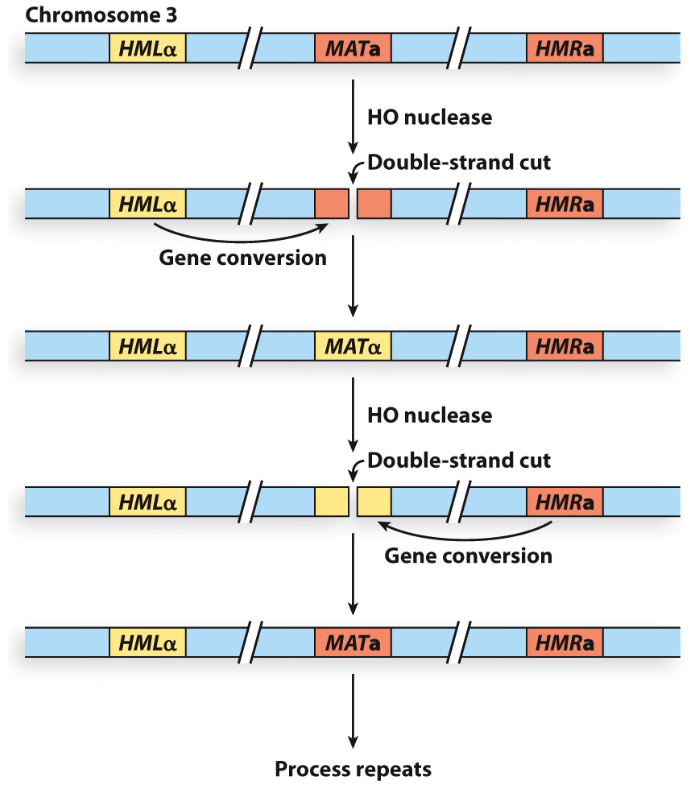
What pathway repairs the HO-induced DSB?
SDSA pathway (non-crossover)
Describe the first step in repairing HO-induced DSB by SDSA pathway
1) Free DNA ends undergo 5’ resection to generate 3’ ssDNA overhangs
What binds the 3’ overhangs?
Rad51 recombinase binds 3’ overhangs and initiates strand invasion at the homologous portion of the HMLα cassette
What does the invading 3’ end serve as?
A primer for DNA polymerase to copy HMLα cassette
What does the nuclease then remove?
The second old strand at the MAT locus
Once the invading strand is long enough, what happens?
It dissociates and pairs with a homologous gene segment in the original MAT locus
What is the last step?
Remaining gap filled by DNA pol by second strand synthesis and then is ligated
→ Now completed gene conversion from MATa to MATα in the MAT locus without any crossing over
How are DSBs in somatic cells repaired?
Mitotic recombination during S and G2 (presence of sister chromatids) (homologous recombination)
What delayes the cell cycle until recombination repair is complete?
DNA damage checkpoints
What is the pathway like in Mitotic DSBs? Is it similar to meiosis?
Pathways of mitotic recombination is like meiosis (SDSA and DSBR)
1) 5’ resection by nucleases
2) RPA coats ssDNA 3’ overhangs
3) Rad52 and BRCA2 displace RPA and load Rad51 mitotic recombinase to initiate strand invasion and resolved by SDSA or DSBR
What does mitotic recombination of a heterozygote between non-sister chromatids result in?
Homozygosity and mosaicism
What is gene targeting using homologous recombination used for?
Precision modifications such as gene deletions and fusion of epitope tags
linear targeting DNA construct with flanking homology arms to guide an insertion or alteration of chromosomal locus
gene targeting is stable and heritable
Elucidate function of genes by determining their spatial and temporal gene expression, phenotypic analysis and protein purification