Chapter 15
1/85
Earn XP
Description and Tags
Cell Trafficking and Cell Structure
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
86 Terms
citric acid cycle, oxidative phosphorylation
what two metabolic processes take place in the mitochondria
houses genome, DNA replication, RNA synthesis
what are the 3 key functions of the nucleus
metabolic pathways, ribosomes doing translation, signaling pathways, cytoskeleton
what 4 things/processes happen in the cytosol
nuclear envelope
what is the ER continuous with
organelles, plasma membrane, export
where do the proteins made in the rough ER end up
synthesis of membrane lipids, Ca2+ storage
what are the 2 functions of the smooth ER
steroid hormone synthesis
what is unique about smooth ER in endocrine cells
detoxifies alcohol
what is unique about smooth ER in liver cells
sorting, modifying, and transporting lipids and proteins
what is the function of the golgi apparatus
recycled to plasma membrane, exported from cell, hydrolyzed in the lysosome
within the endosome, what are the possible fates of imported materials
endosome
where does every imported material go first in a cell
hydrolytic (allows it to digest materials)
what type of enzymes does a lysosome contain
imported macromolecules, unwanted organelles, unwanted proteins
what things does a lysosome digest
oxidative
what type of enzyme is a peroxisome filled with
produces and removes H2O2, oxidizes and neutralizes toxic molecules, synthesizes phospholipids, beta-oxidation
what are the 4 functiosn of a peroxisome
endoplasmic reticulum, golgi apparatus, peroxisomes, lysosomes, endosomes, ECF
what is part of the endomembrane network and/or connected by vesicular transport
nuclear pores
what connects the cytosol to the nucleus
no
are mitochondria/choloroplasts connected to the endomembrane network
cytosol, nucleus (enters folded via pores), mitochondria/chloroplasts (enters unfolded)
where can a protein go after it is translated by cytosolic ribosomes and how does it enter alternate destinations
signal sequences (a short amino acid sequence within a protein read by sorting proteins)
what directs the sorting of any protein within a cell
ER signal sequence (8 to 15 hydrophobic amino acids)
what dictates where (cytosol or rough ER) a protein is translated by a ribosome and what is this made up of
import signal (at N-terminus, cleavage site nearby, removed during transport), membrane insertion signal (internal location, no cleavage site nearby, forms transmembrane alpha helix)
what two types of ER signal sequences are there and what are their differences
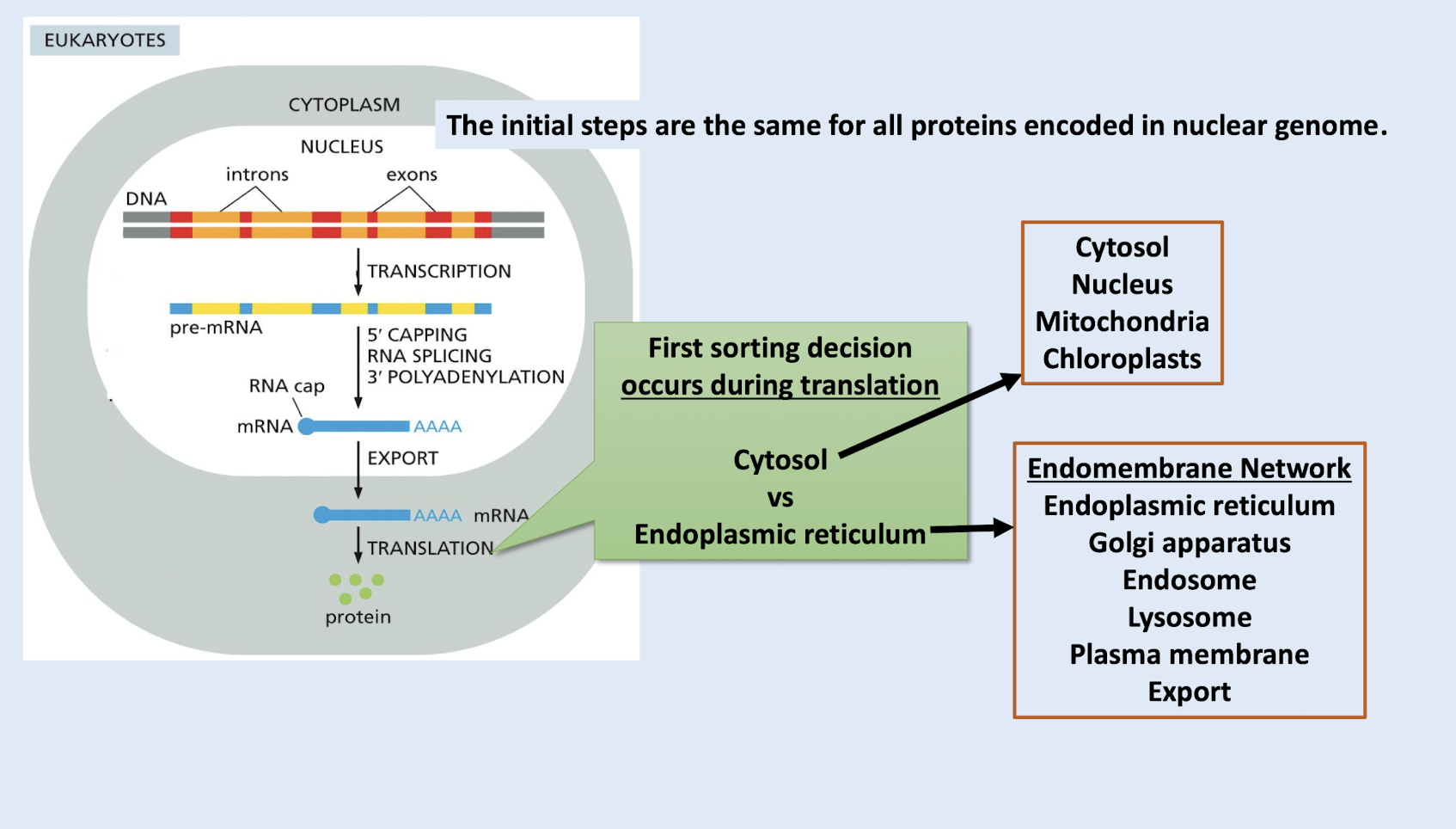
cytosol, nucleus, mitochondria, chloroplasts
where can a protein lacking an ER signal sequence end up
translocator
channel of translocating protein into ER
signal peptidase
this cleaves import signal from precursor protein within rough ER
signal recognition particle
what binds to an ER signal sequence
location of N-terminus and orientation, number of transmembrane regions (single pass or multipass protein)
what about the location of the first ER signal sequence determine the location/function of a protein, what about the total number of insertion signals
within the cytosol SRP binds to ER import signal on growing polypeptide chain and slowing translation, SRP binds to SRP receptor found on rER membrane causing ribosome to bind to protein translocator and ER import signal enters translocator, translation finishes as protein translocator closes, signal peptidase cleaves signal peptide and allows mature protein to fold in ER lumen
how do soluble ER proteins end up in the ER lumen while being translated by a ribosome in the cytosol
once insertion signal is translated on a growign peptide chain, SRP binds to the signal pausing translation and binds to SRP receptor, the protein and signal is pushed into the translocator as translation continues, after translation amino terminus stays in cytosol, there is a transmembrane alpha helix at the insertion singal, and the carboxyl region is in the ER lumen
how do transmembrane ER proteins end up in the ER lumen while being translated by a ribosome in the cytosol
ER soluble protein
for a growing polypeptide with one single ER signal at its N-terminus, will it be an ER soluble protein, single pass protein, or multipass protein
mitochondria, chloroplasts, inside nucleus, cytosol
once a protein is in the endomembrane network, where can it NOT go
there is a diffusion filter lined with disordered proteins, only small solutes and molecues that 1. can interact with the diffusion filter or 2. interact with a protein that can carry you in (nuclear import receptor)
why can’t all proteins enter the nucleus via nuclear pores
a nuclear protein will contain a nuclear localization signal (small amino acid sequence within the polypeptide) recognized by a nuclear import receptor that interacts with cytosolic fibrils and travels down pore proteins via binding to enter the nucleus, dropping its cargo and returning to the cytosol
how does a prospective nuclear protein enter the nucleus if it is unable to enter the nuclear pore by itself
Ran-GTPase
what protein is responsible for making sure imported proteins stay within the nucleus
Ran-GRP binds to a nuclear import receptor once it has entered the nucleus, displacing its protein, the receptor then enters the cytosol triggering Ran-GTPase activating protein (Ran-GAP) to hydrolyze GTP into inactive Ran-GDP which dissasociates from the nuclear import receptor, Ran-GDP enters the nucleus and is converted back to active Ran-GTP via Ran Guanine nucleotide exchange factor (Ran-GEF)
describe the cycle of Ran-GTP to Ran-GDP and how it drives nuclear import
Ran-GTP
which is higher in concentration within the nucleus: Ran-GTP or Ran-GDP
Ran-GTP
which can bind to a nuclear import receptor: Ran-GTP or Ran-GDP
a nuclear export receptor (exportin) bound to cargo cannot leave the nucleus without Ran-GTP also binding to it (Ran-GTP follows the same regeneration cycle as seen in nuclear import)
how does Ran-GTP drive nuclear export (don’t include regeneration cycle)
after mitosis (proteins need to be resorted into nucleus), transcription factor regulation (protein in nucleus = transcription on; protein out of nucleus = transcription off)
how is nuclear import and export crucial to a cell - 2 reasons
peroxisome
what is the only organelle that receives proteins from both the ER (endomembrane network) AND cytosol
folded proteins transport through pores in peroxisome, membrane proteins and lipids integrate via vesicular transport
how do peroxiomes take in proteins from cytosol, how do they from the ER
some are encoded in mitochondrai DNA and translated in mitochondria; most are encoded in nuclear genome, translated in cytosol, then imported unfolded after translation by import receptor protein recognizing its N-terminal signal sequence, after translocation the signal peptide is cleaved
how do mitochondrial (and chloroplasts) proteins end up in the mitochondria - 2 ways
outer membrane, inner membrane, intermembrane space, matrix
after being imported to the mitochondria, what are the 4 possible specific destinations a protein could end up based off of other signals and mechanisms
lipid-carrying proteins transfer them from ER
mitochondria have their own membrane and need lipids, how do lipids enter the mitochondria (hint they cant enter via vesicles)
proteins have specific signals and vesicles have specific surface targeting proteins
how is vesicle transport controlled, aka how does each protein end up in the right vesicle and location
clathrin
this is an outer coat protein which drives the budding of vesicle and helps to capture the correct cargo; is multidirectional
golgi (buds outwards to endomembrane network), plasma membrane (buds inwards during endocytosis to endosome)
what two locations can clathrin (a coat protein) be in and how does this determine the vesicles direction
adaptin
what confers cargo specificity to clathrin vesicles
different types of these result in different destinations of the vesicle
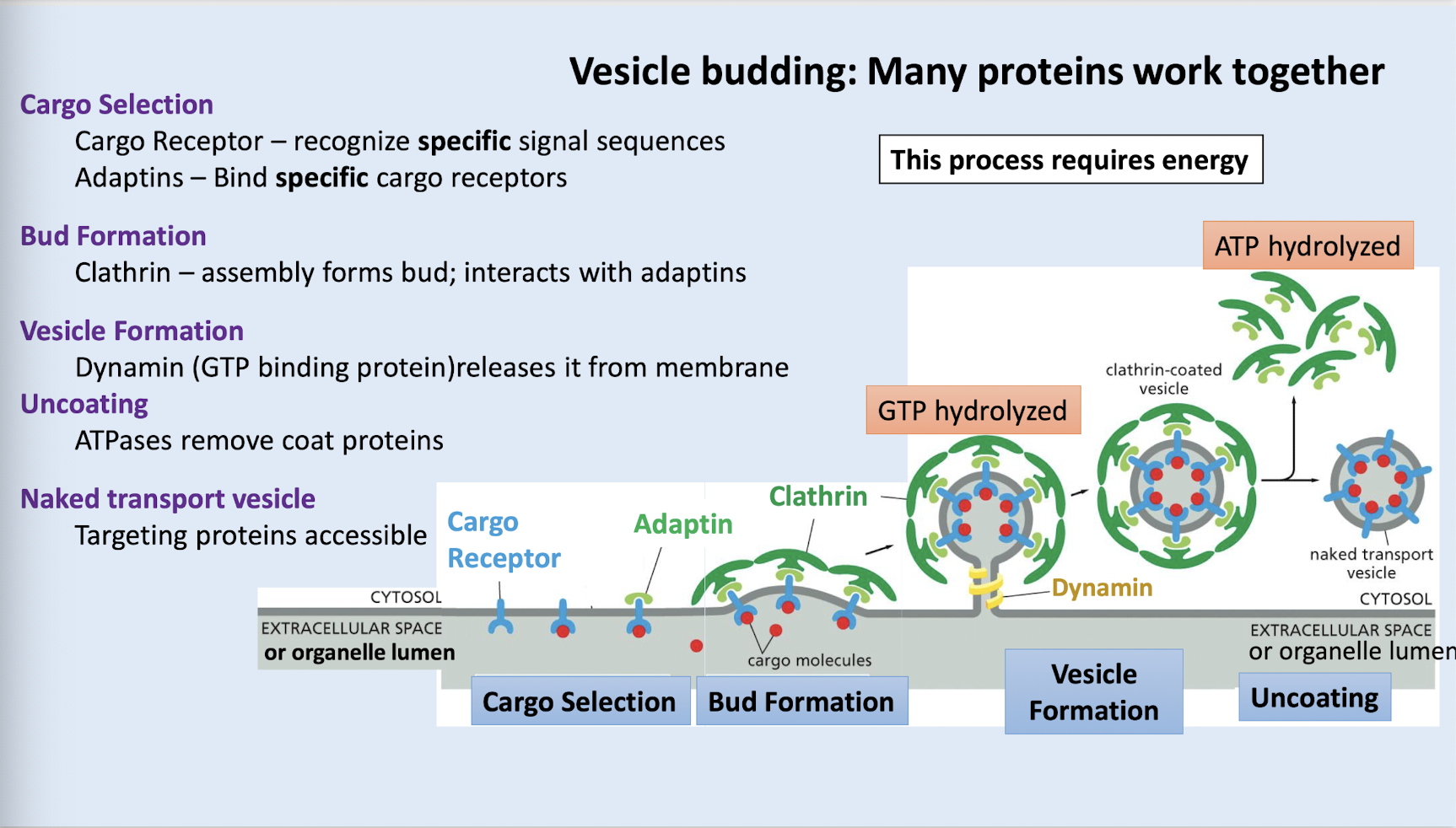
cargo selection (cargo receptor recognizes signal sequences and binds to cargo; adaptins bind to the cargo receptors)
bud formation (clathrin binds to adaptins to form bud)
vesicle formation (dynamin a GTP binding protein releases the bud from the membrane)
uncoating (ATPases remove coat proteins - adaptin and clathrin)
naked transport vesicle is revealed (now targeting proteins can access vesicle)
what are the steps of vesicle budding and what proteins played a role
motor proteins, microtubules, vesicle specific
vesicles are often transported by ___ ___, usually along ____; motor proteins and direction are ____ specific
membrane
vesicle docking is ____ specific
tethering (specific Rab-GTPase on vesicle binds to a thethering protein on target membrane)
docking (vesicle-SNARE and target-SNARE interact)
fusion of membranes
describe the process of vesicle docking and include what proteins make this process membrane specific
the v and t-SNAREs interact to form a bundle of a-helices that wind together to squeeze out water, allowing membranes to get close enough to fuse together
how are v and t-SNARE proteins crucial to vesicle fusion
its toxin cleaves SNAREs prevent all docking and fusion of vesicles, resulting in weakness and paralysis, inabilty to breath (specifically from acetylcholine being unable to be released as a nt)
what affect does clostridium bolutlinum have on a cellular level and what affect does this have on the body
this temperature sensitive mutation misfunctions at higher temperatues and causes endocytosis to be blocked after bud formation of vesicles, paralysis results because neurotransmitters can’t be recycled
what affect does a dynamin mutation have on the body
glycosylation
what process creating a certain type of membrane and secreted proteins begins in the ER (hint: is N-linked, occurs during translation via oligosaccharyl transferase)
oligosaccharyl transferase
what enzyme moves a standard oligosaccharide to a growing peptide chain within the ER lumen, beginning glycosylation and creating an immature glycoprotein
disulfide bonds, glycosylation
what 2 modifications can occur to a protein within the ER
ECF (stabilize protein’s folded structure allowing it to remain functional longer)
where would a protein with disulfide bonds (created in the ER) likely end up: cytosol or ECF
are exported to cytosol as garbage and degraded in proteasome
if the ER only exports good proteins, what happens to proteins which failed folding
sudden increase in protein expression or a mutant protein accumulating, reduces demand on ER or increases ER capacity
within the ER, why would too many unfolded proteins accumulate, how is cell behavior altered to address this problem
excessive unfolded proteins bind to sensors on ER membrane, activated sensors inhibit protein syntehsis and slow the cell cycle, the expression of ER proteins is increases, including chaperones; apoptosis
within the ER, when too many unfolded proteins accumulate, what is inhibited and what is increased via the binding of what to what; lastly if this cycle continues for too long what happens to the cell as a whole
ER (requires an ER retention signal), golgi apparatus via vesicles
what two locations can a protein go after being folded and modified in the ER
endosome to lysosome, plasma membrane, ECF
where can proteins travel to after the golgi apparatus
constitutive secretion
this is the constant secretion of plasma membrane components and extracellular proteins (signals, enzymes, etc.) with no signal sequence required
specialized secretory
regulated secretion occurs in ____ ____ vesicles only and its exports are unique to the type of cell (ex: hormones, mucus, digestive enzymes, neurotransmitters)
selective aggregation, regulated vesicle fusion
what are the two types of regulated secretion a cell can do
selective aggresion
a type of regulated secretion in which secretory proteins aggregate in the golgi and are packed into a secretory vesicle at high concentrations and taken to ECF when the vesicle is full
regulated vesicle fusion
this type of regulated secretion is when secretory vesicles accumulate and wait at their final location for the signal required for fusion and release
hormones, neurotransmitters, action potential
what types of signals can trigger regulated vesicle fusion (a type of regulated secretion)
antitrypsin
this is a protease inhibitor secreted by the liver into the bloodstream, moderates the activity of secreted proteases
protects lung tissue via inhibiting neutrophil elastase
lung disease
chronic obstructive pulmonary disease in younger pateients without a history of smoking
liver disease
cirrhosis and carcinoma without a viral hepatitis or alcohol abuse
decreased serum levels of antitrypsin
what is alpha-1 antitrypsin deficiency diagnosed
encodes a functional protease inhibitor but it cannot escape the ER due to misfolding (aka antitrypsin is not present in the blood)
what does the mutant gene do to antitrypsin
liver disease due to failed secretion of antitrypsin, lung disease due to overactive proteases
what are the effects and why of alpha-1 antitrypsin deficiency (an example of failed exocytosis)
endocytosis
when membrane components return to organelles and fluid is returned into the cell
two types: pinocytosis and phagocytosis
pinocytosis
this type of endocytosis is when small vesicles form at plasma membrane and extracellular fluid and molecues are captured
two types: non-selective and selective; both occur in the same vesicles
balances fluid and membrane loss from constitutive secretion
what is the purpose of non-selective pinocytosis
recycling to plasma membrane, transcytosis of recptor/cargo, degradation in lysosome
after endocytosis, all molecules go to the endosome; after that what 3 paths can a molecule go
ATP-driven H+ pumps acidify endosome and some receptors release cargo at low pH
what happens within the endosome
vesicle transport delivers hydrolytic enzymes, lysosomal proteins arrive from golgi, ATP powered H+ acidify the organelle as it moves away from the plasma membrane until the pH is 4.7-5.0
an endosome, with a pH of 5-6, eventually turns into a more acidic lysosome, how does this happen and what is the final pH
lysosome enzymes require a highly acidic environment to work, glycosylated membrane proteins facing the lumen
what prevents the lysosome from digesting the rest of the cell
phagocytic cell’s receptors recognize target, psuedopods engulf target, phagosome fuses with lysosome to be digested
describe the process of phagocytosis within an animal immune system
autophagy
when a cell eats parts of itself
extreme starvation, certain fate changes (ex: RBCs lose mitochondria), remove damaged organelles
in what examples would a cell do autophagy