Genetics Biology 2C03 - Evans Post Midterm 1
1/247
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
248 Terms
What are the features of bacteria that are useful to geneticists?
Genome simplicity
Haploid genomes (mostly), mutations can be observed directly
Short generation times
Large progeny numbers
Easily and inexpensively maintained and propogated
Numerous heritable differences
What are the characteristics of bacterial genomes?
Small (fewer genes)
DNA associated with histone proteins and supercoiled
Circular (mostly)
No introns (mostly
Often only one chromosome (not always)
No mitochondria
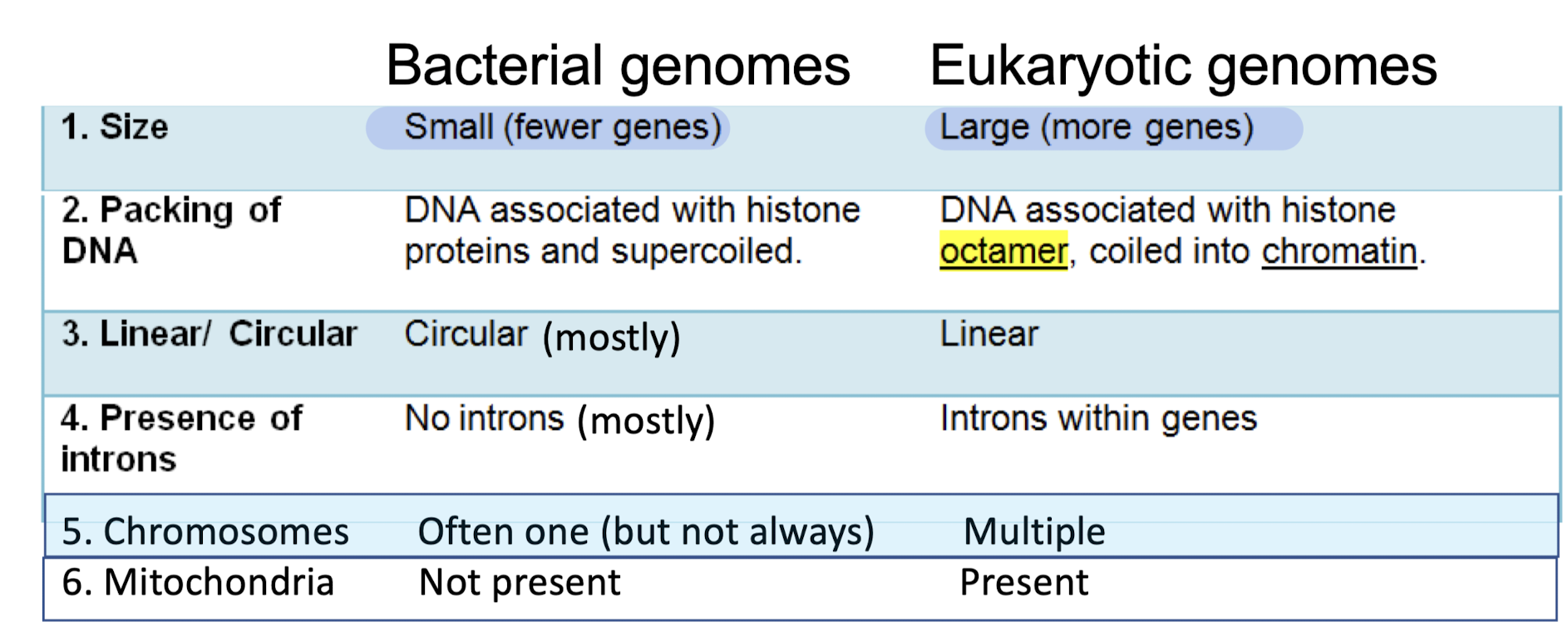
What are the characteristics of eukaryotic genomes?
Large (more genes)
DNA associated with histone octamer, coiled into chromatin
Linear, multiple chromosomes
Introns within genes
Mitochondira present
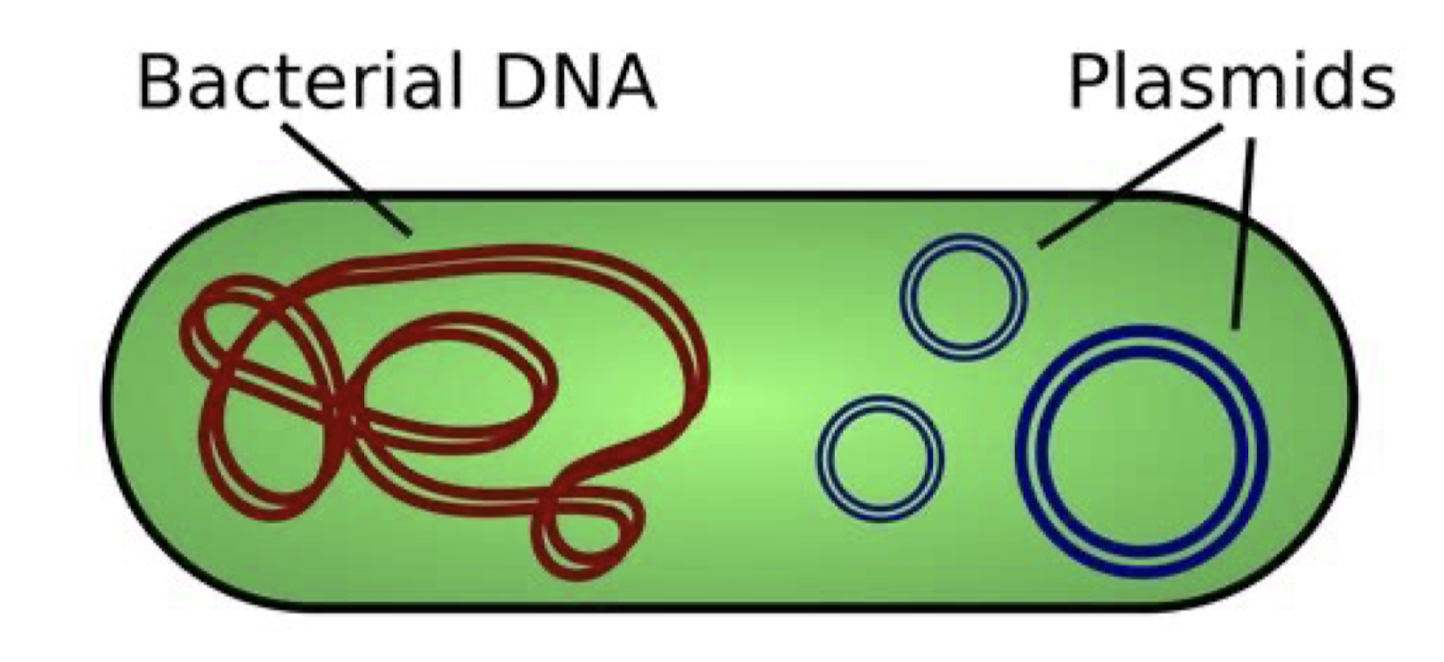
What are plasmids?
Small double-stranded circular DNA molecules containing nonessential genes that ure used infrequently
“extrachromosomal DNA”
sometimes plasmid DNA is incorporated into the bacterial genome
What are F (fertility) plasmids?
Contains genes that assist with transfer of that plasmid to another host bacterial cell
What are R (resistance) plasmids?
Contains genes that confer antibiotic resistance that can be transferred from donors to recipients
a strain resistant to a antibiotic is carrying a R plasmid
a strain susceptible to a antibiotic is carrying no R plasmid
What are the 3 types of lateral gene transfer between bacteria?
Conjugation, transformation and transduction
succesful gene transfer requires that the donor DNA is incorporated into the genome of the recipient
What is conjugation?
The transfer of replicated DNA from a donor cell to a recipient cell through tempoary contact
requires physical contact, establish bridge beweten 2 cells, initiate DNA transfer across the bridge
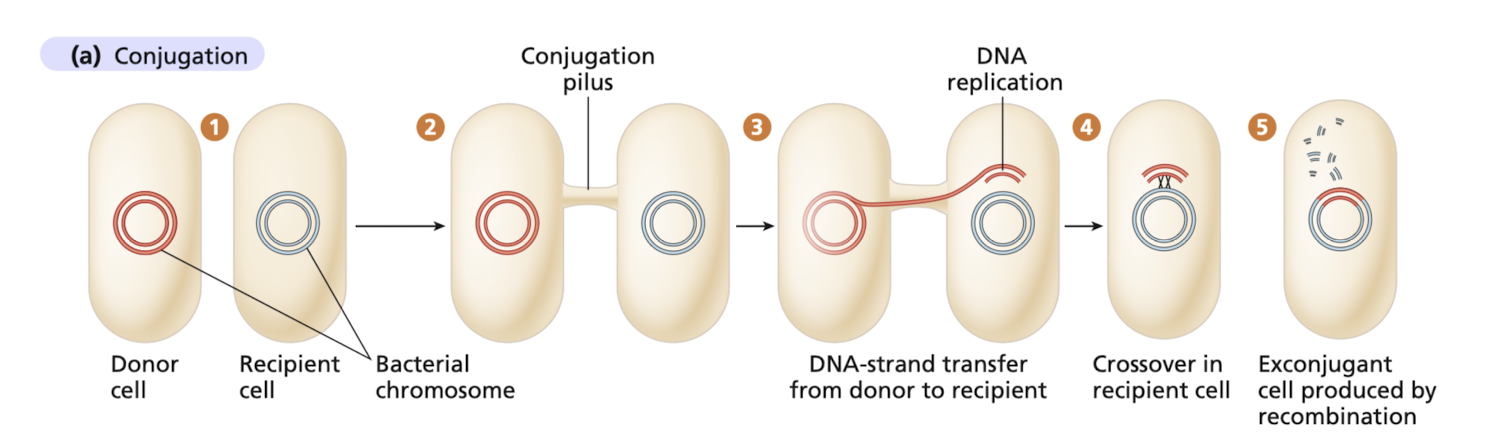
What is transformation?
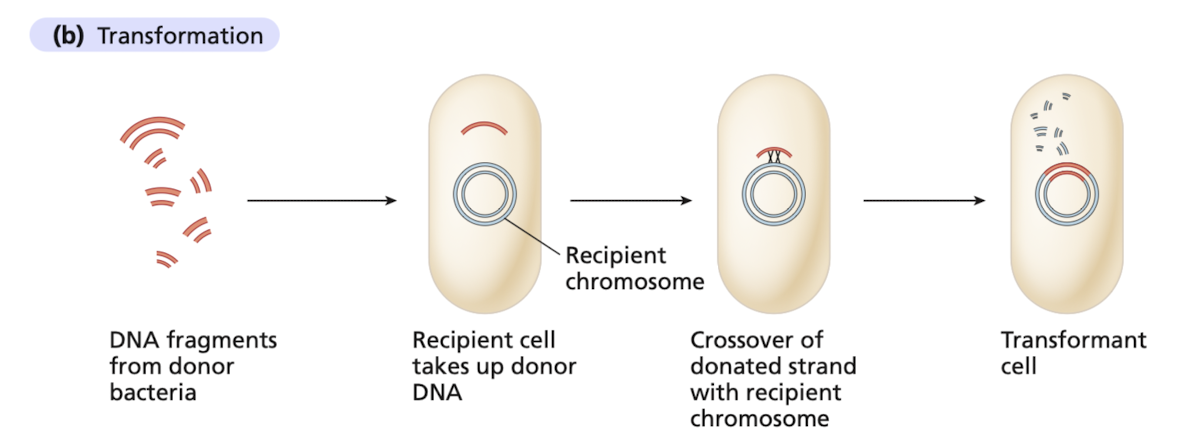
The uptake of DNA from the environment
derived from a donor cell, from the growth medium of the recipient
What is transduction?
Transfer of DNA from donor bacterium to recipient bacterium by a viral vector (bacteriophage)
can happen if the virus accidentally incorportates donor bacterial DNA
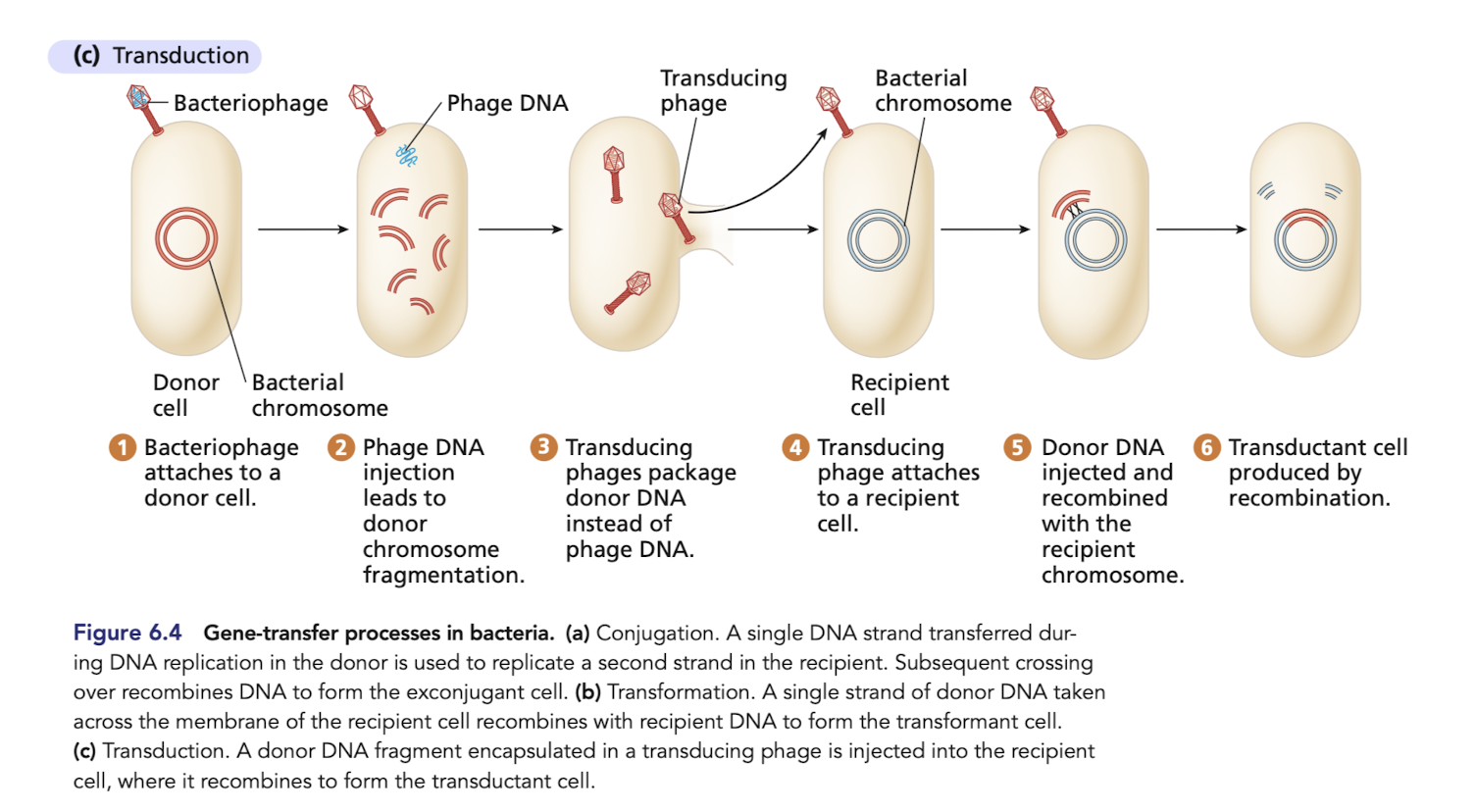
What is lateral gene transfer?
The transfer of genetic material between individual bacteria/archea between organisms that are in the same generation
pervasive: ~12% of genes in a average bacterial genome
biased: involves genes related to pathogenicity and antibiotic resistance
What is the fertility factor?
The ability to act as a donor, determined by the “F factor”
Donors: F+
only donors initiate conjugation
hereditary
Recipients: F-
In some types of conjugation, the donor carries a F plasmid and the recipient does not
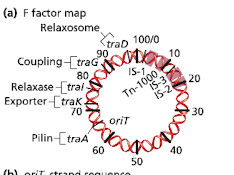
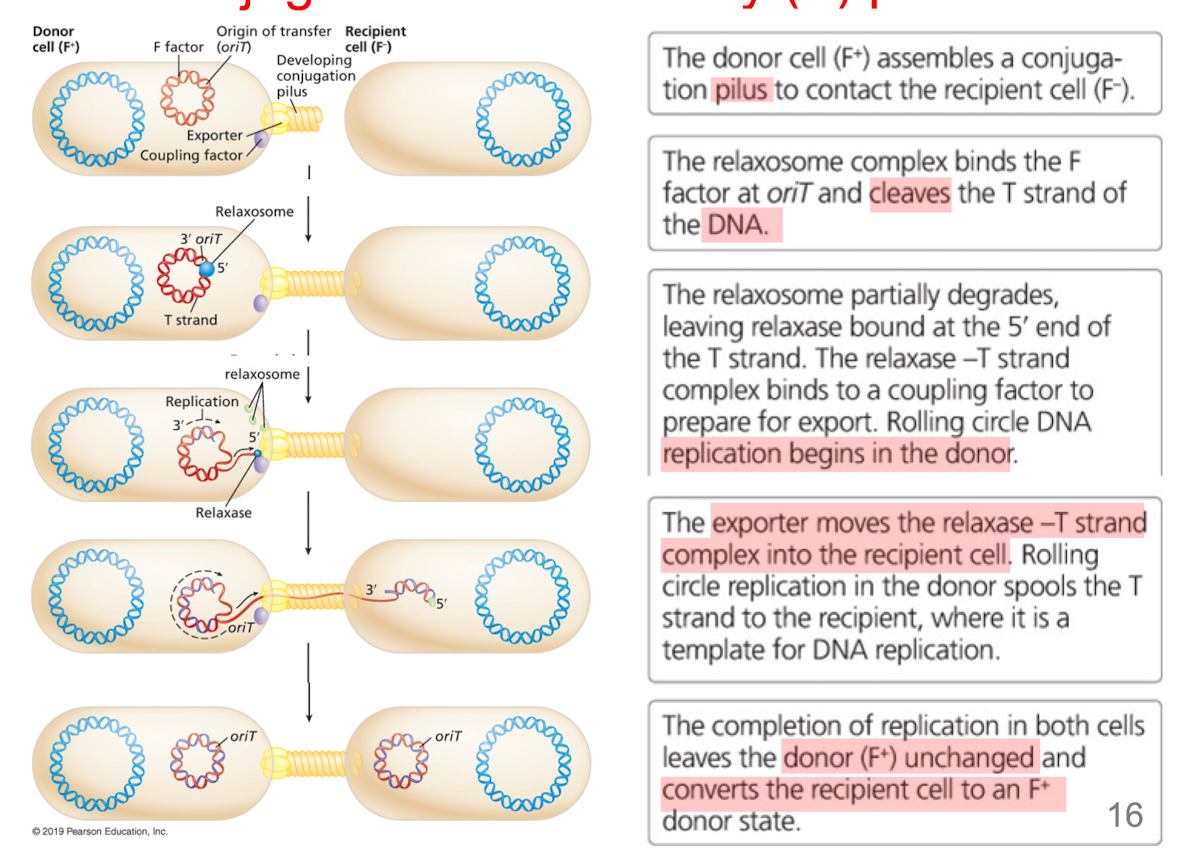
How does conguation with a F (fertility) plasmid work?
DNA transfer begins at F factor sequence “oriT”
conjugation pilus allows passage of single DNA strand
relaxosome binds oriT, cleaves T strand
replication begins in donor, T-strand is template
replication in both cells leaves donor (F+) unchanged and converts the recipient to a F+ donor state
Conjugation with Hfr strains
Conjugation with a donor F+ strain does not transfer the bacterial genome - it only transfers the F plasmid
F plasmid can become incorporated into the bacterial genome
converts bacteria into a high frequency recombination (HFR) strain
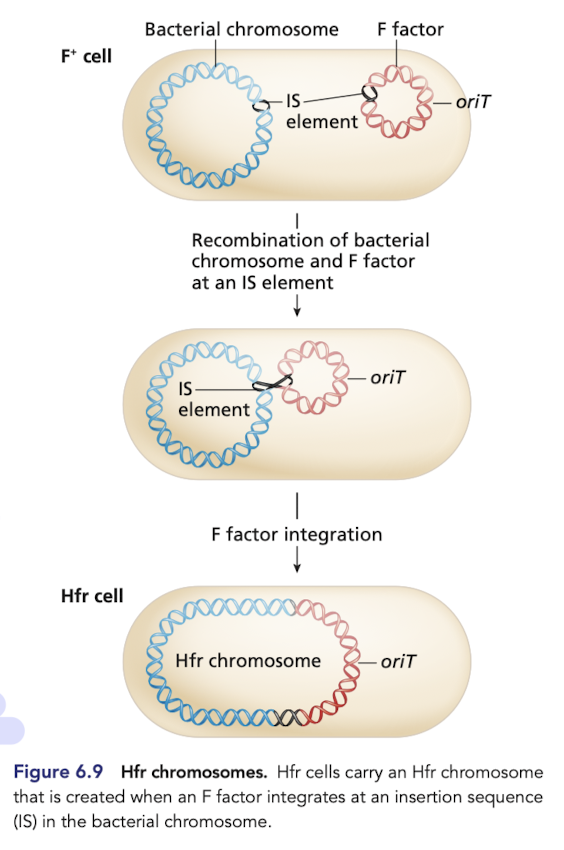
Conversion of a F+ to a Hfr strain
Very rare, integration event takes place at IS elements that are shared by plasmids and bacterial chromosomes
recombination of bacterial chromosome and F factor at an IS element produces the HFR chromosome
Conversion with a Hfr strain
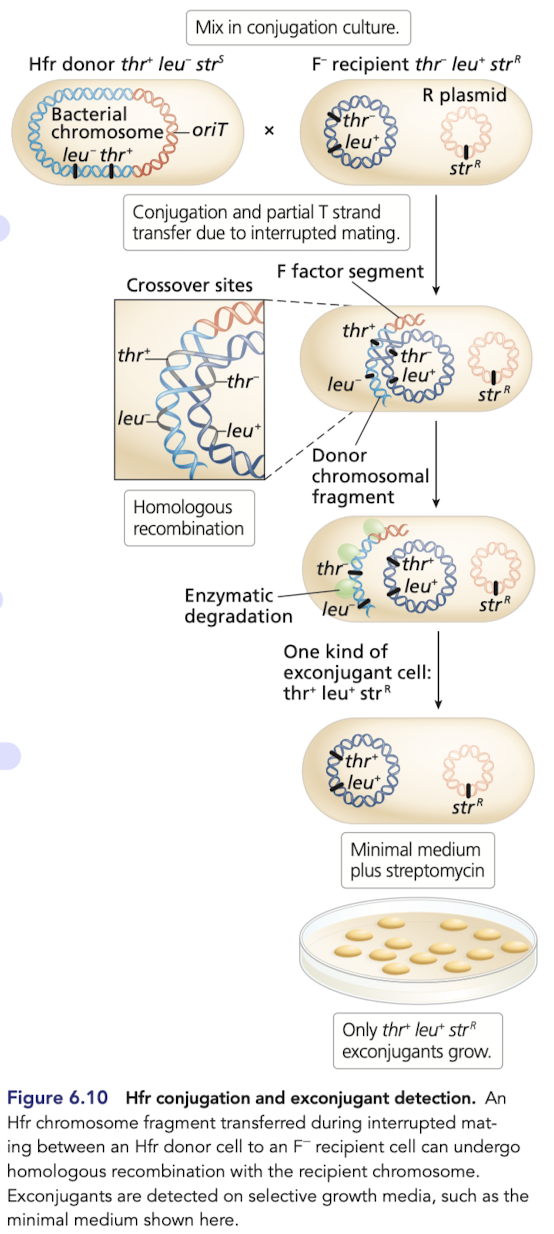
Gene transfer from Hfr to F- is similar to F+ x F- conjugation
donor bacterial chromosomal genes can be tranferred to the recipient
homologous recombination occurs between the transferred linear DNA and the circular chromosome of the recipient, generating recombinants
What is a exconjugant cell
A recipient cell that has had its genetic content modified by receiving DNA from a donor cell
What is interrupted mating?
The cessation of conjugation caused by breakage of the conjugation pilus
stops conjugation before the Hfr chromosome can be completly transferred from the donor to the recipient
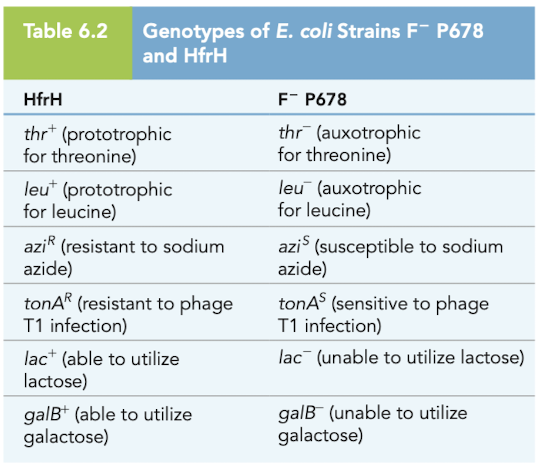
Time of entry mapping experiment part 1
Cells of HfrH and P678 mixed
samples plated on selective media to evaluate phenotypes of exconjugants
plated on leu-, thr-, str+ plates
donor HfrH is str-sensitive, so it dies
only exconjugants that got thr+ and leu+ from the donor can grow
Time of entry mapping experiment part 2
Known: thr and leu are closer to the origin of transfer in HfrH
utilize the leu+ thr+ str^r exconjugants on a 2nd plate to determine which other donor alleles have undergone recombination
azi - 8 minutes
tonA - 10 minutes
lac - 16 minutes
galB - 25 minutes
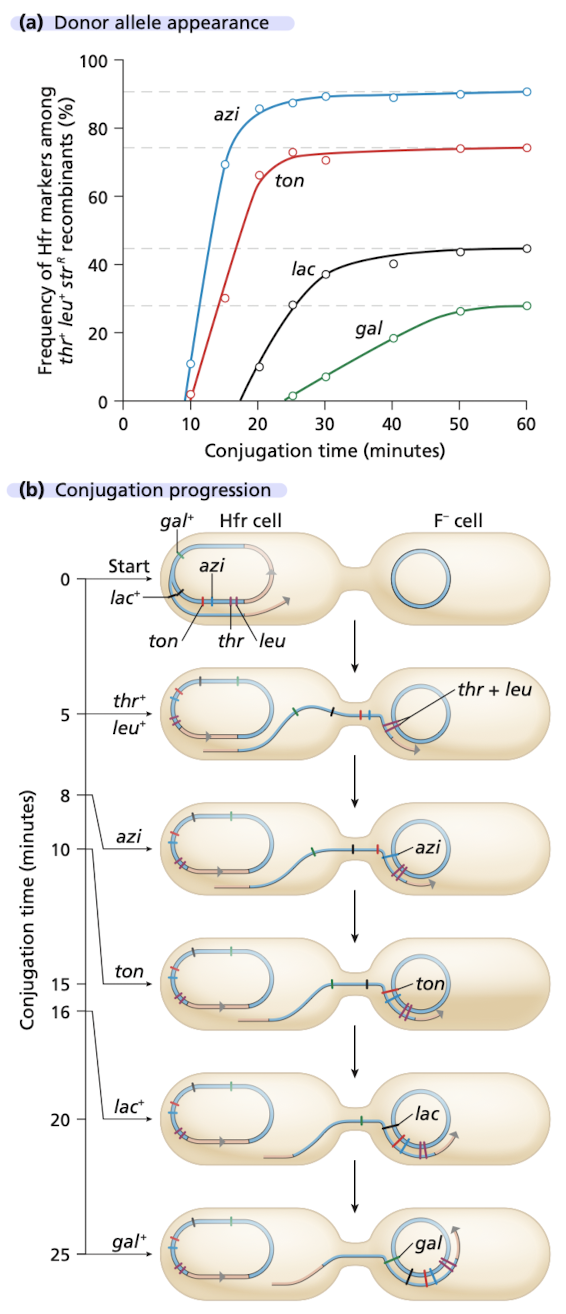
Time of entry mapping experiment part 3
Mapping information for a single Hfr strain is limited b/c the conjugation pair usually breaks apart before the entire genome is transferred
liklihood of gene transfer drops of quickly with distance from oriT
one can use multiple Hfr strains to map all of the genes in a species
F factor may integrate into different locations on a bacterial chromosome
F factor can be integrated into a Hfr chromosome in either of 2 orientations
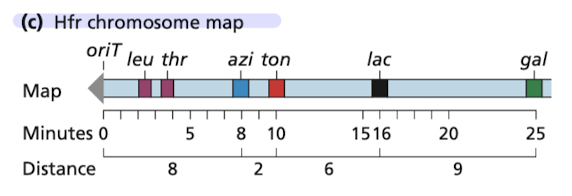
Overlapping linear maps from different Hfr strains
Individual Hfr maps laid out so that the genes in different maps align
mins of conjugation between a given pair of genes will be the same in each Hfr strain transferring the gene pair, no matter their oritentation in the Hfr chromosome
map indicates location of each integrated fragment on the circular chromosome, its orientation and the gene order/distance in minutes
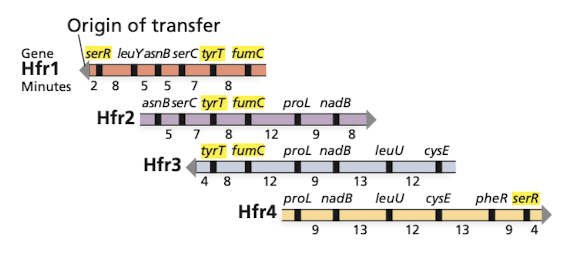
F factor excision from Hfr strains
F’ donor - contains functional but altered F factor dervied from imperfect excision of the F factor out of the Hfr chromosome
includes genes from the bacterial chromosome in the F plasmid
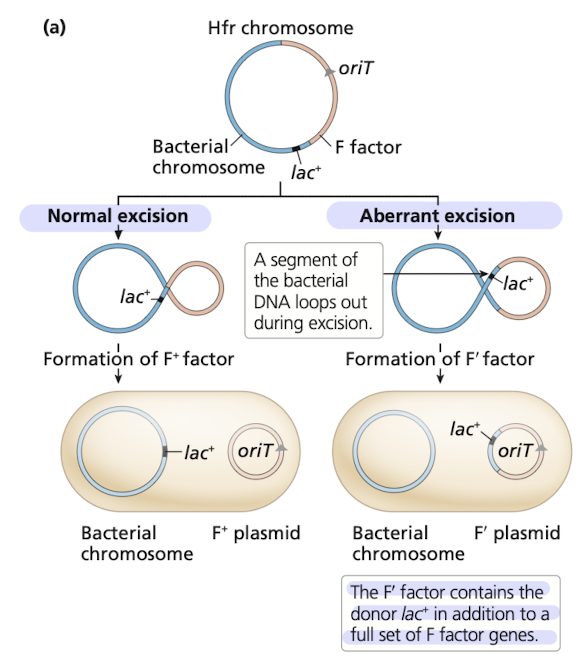
How do F’ plasmids make “partial diploids”?
F’ plasmid has a gene from a donor cell that is transferred to a recipient cell
recipient cell will contain two copies of a gene
one from the donor, and one frmo the original gene in the recipient
exconjugant can act as a F’ donor
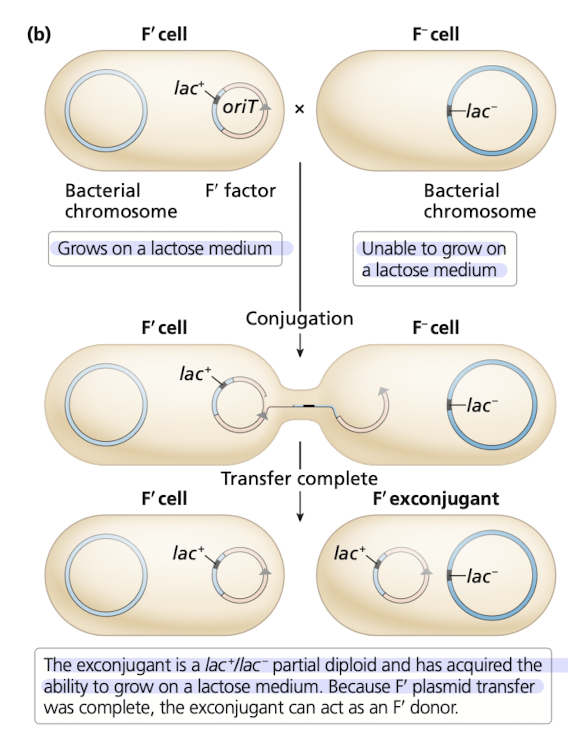
What are the outcomes of different types of conjugation?
Note 2 cases where exconjugant is converted ot donor state
2 cases where bacterial genes are transferred to exconjugant
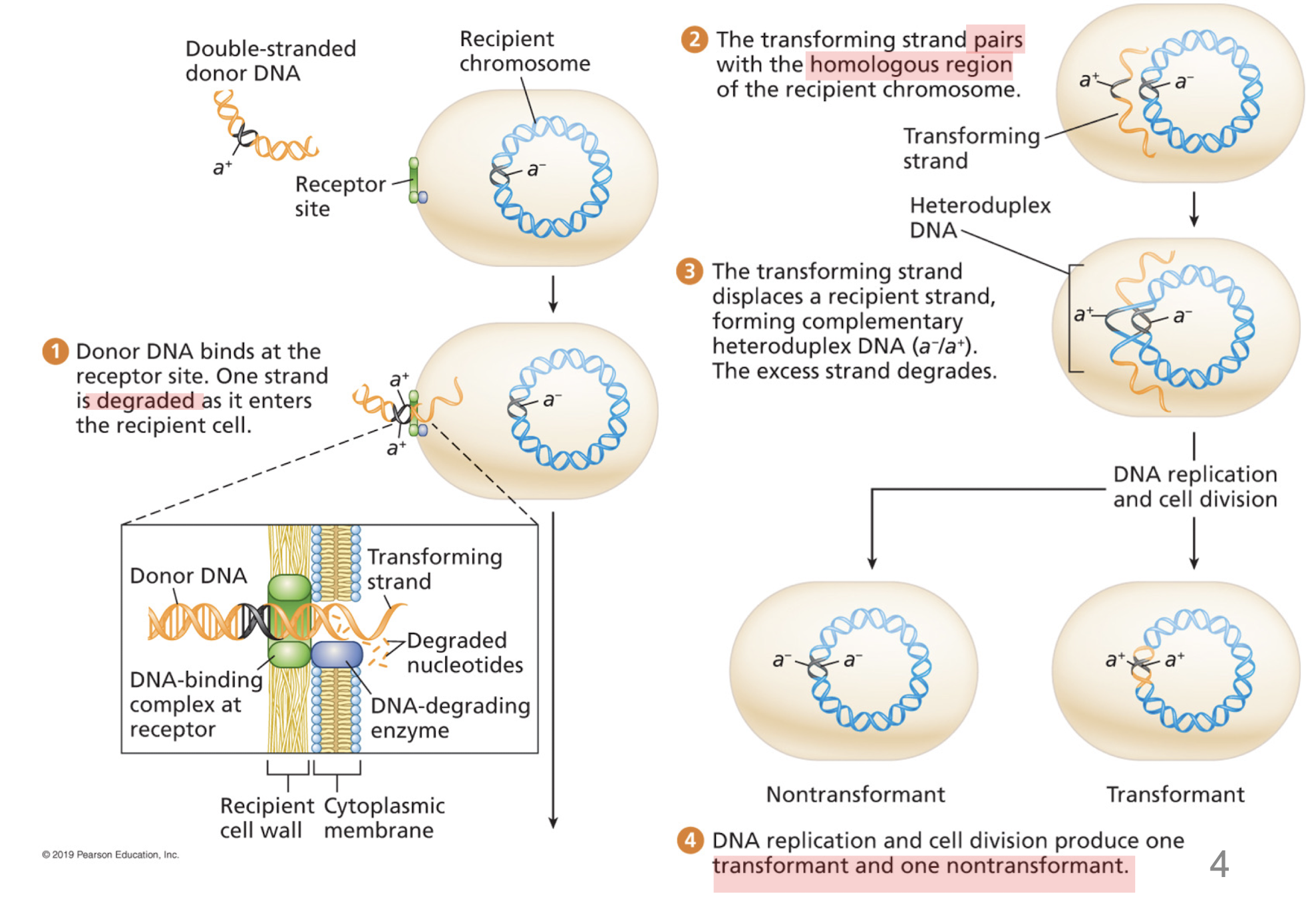
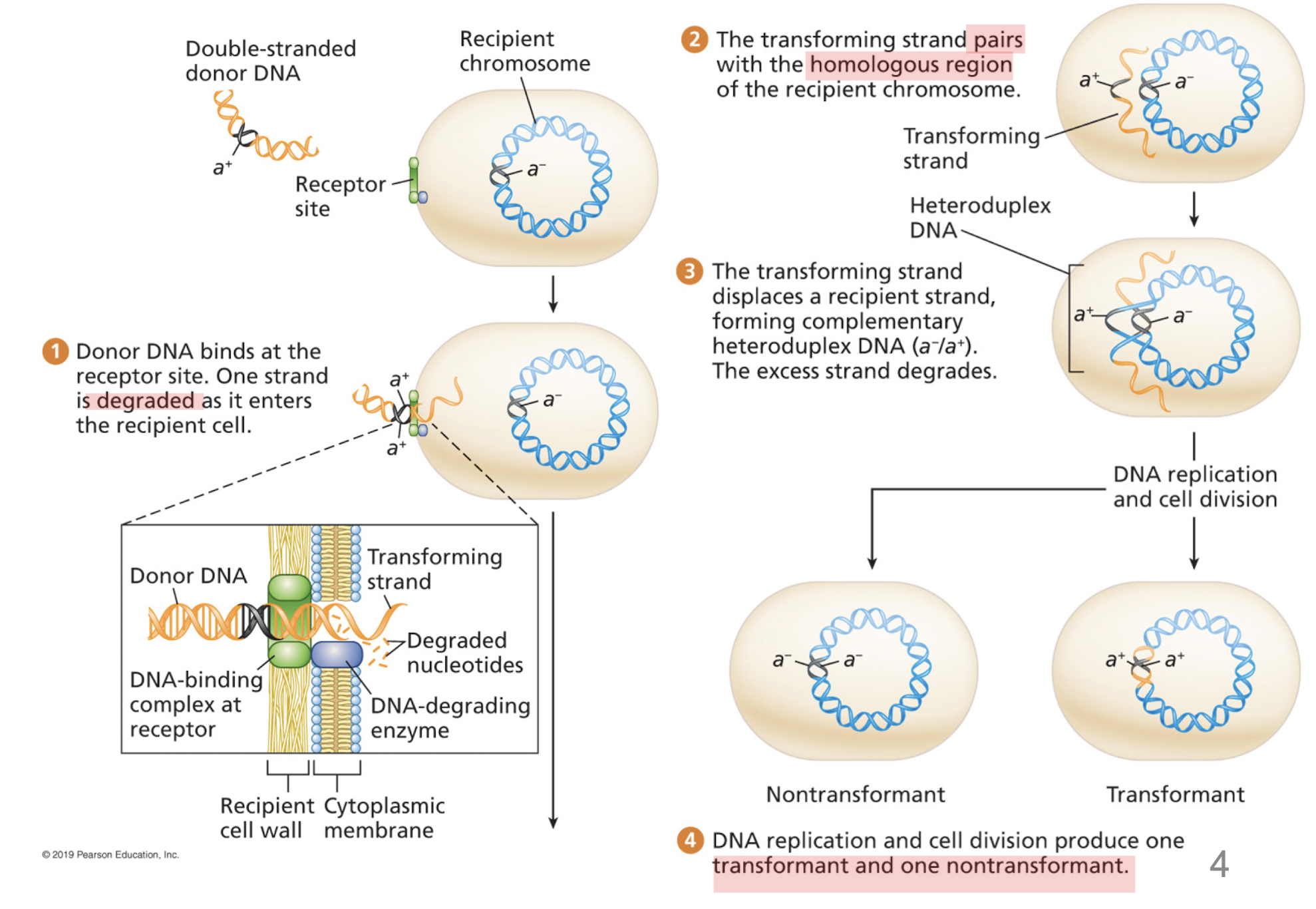
How does transformation work?
degradation by one of the strands
transforming strand invades a complementary region of the recipient chromosome
heteroduplex DNA
one strand from recipient cell, complementary transforming strand derived from the bacterial donor
transformant cell (carries transforming strand/newly synthesized complementary strand) and nontransformant (retains recipient chromosome)
What is a “compotent cell”?
A cell capable of being transformed
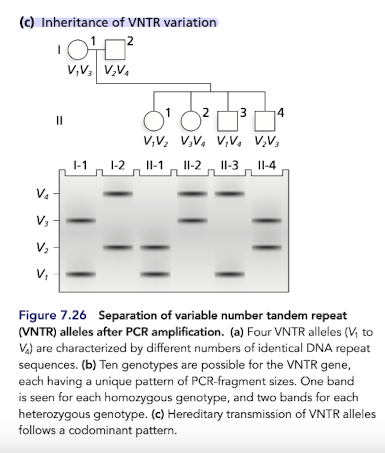
What does co-transformation of multiple genes indicate?
Indicates that they are closely linked (provides info on the genetic map of the donor and recipient)
What are the two types of life cycles in bacteriophages?
The lytic and lysogenic cycles
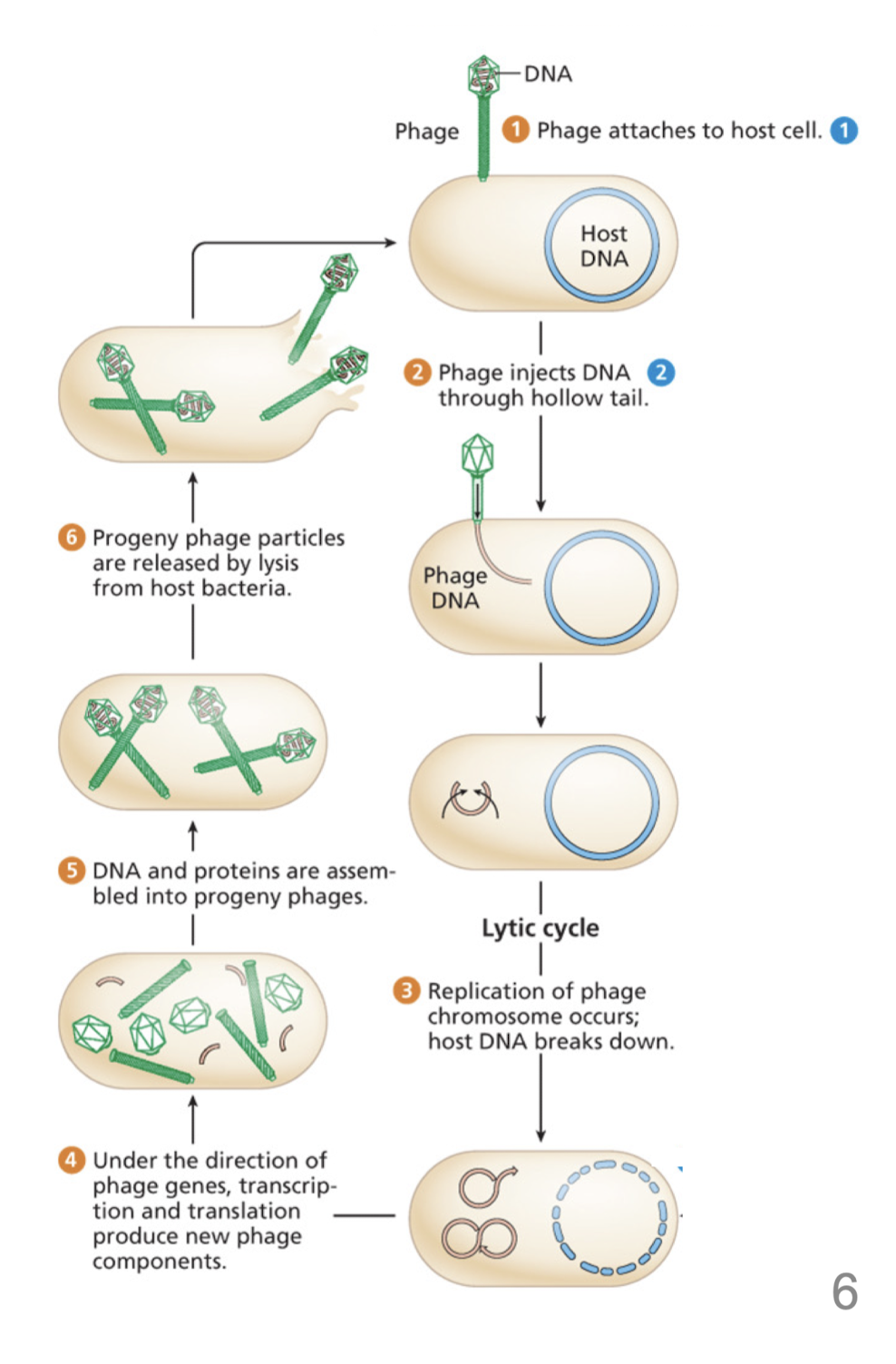
Describe the lytic cycle
Phage genome is not incorporated into the bacterial genome
occasionally, donor genes are incorporated in the phage
DNA can be inserted into a recipient cell
produces transductant when recombination integrates the DNA into the recipient genome
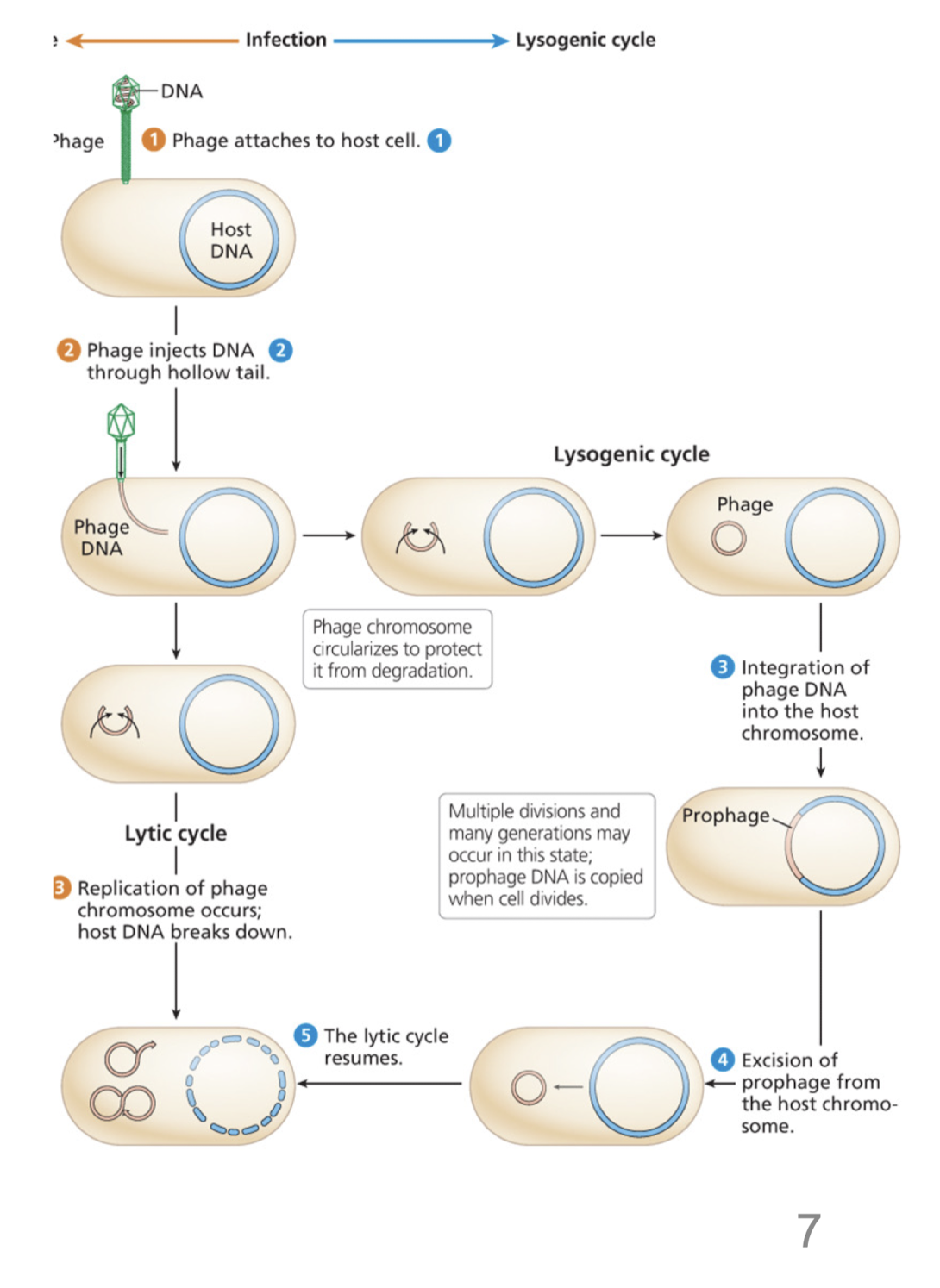
Describe the lysogenic cycle
Phage genome is incorporated into the bacterial genome
resulting prophage undegoes multiple cell divisions before the phage genome is excised and the lytic cycle continues
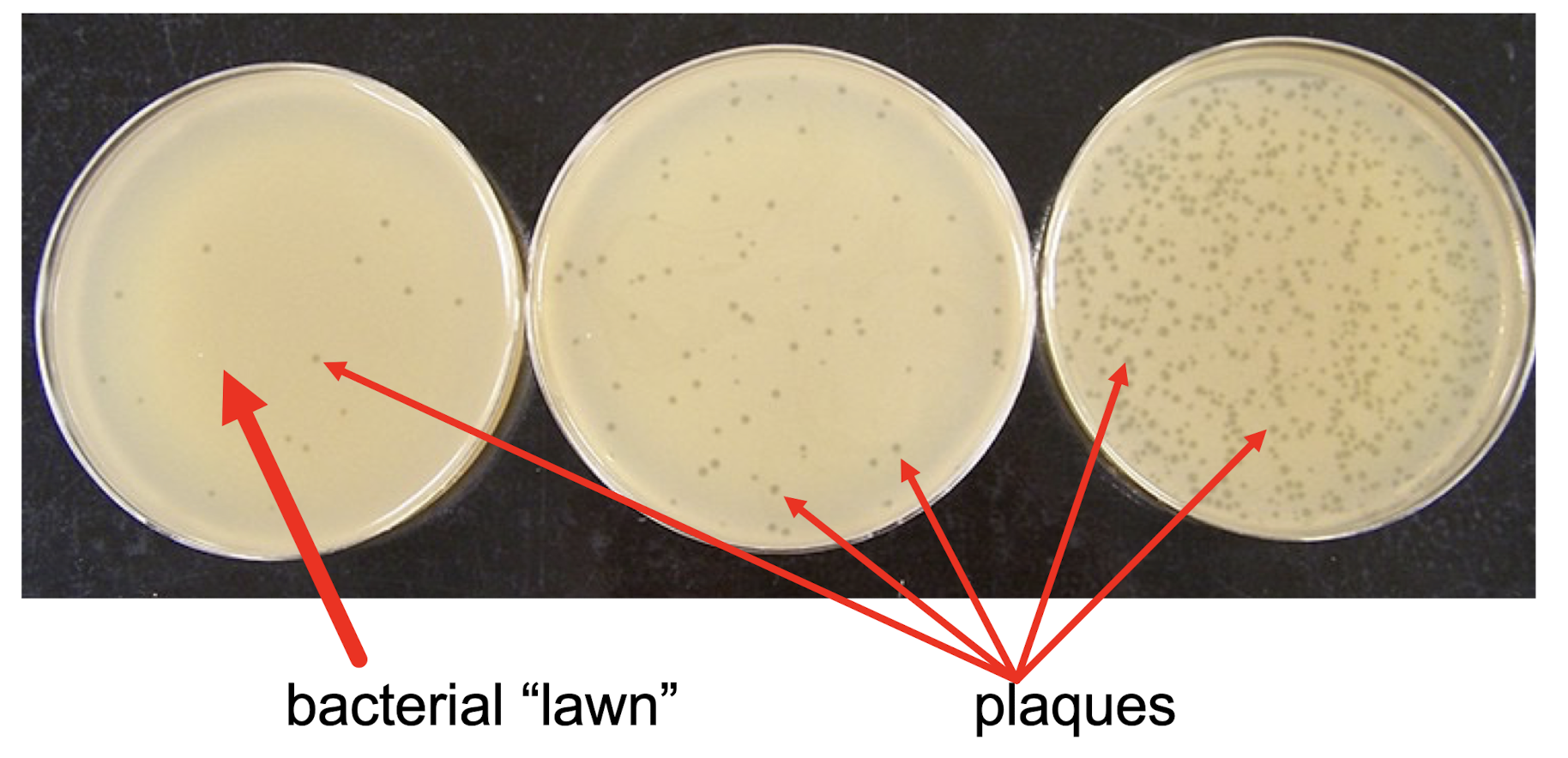
What is plaque?
When bacteriophages infect bacteria, plaque forms - dead bacteria
if genes in the bacteriophage are defective, plagues either don’t form, or don’t look like wildtype plaques
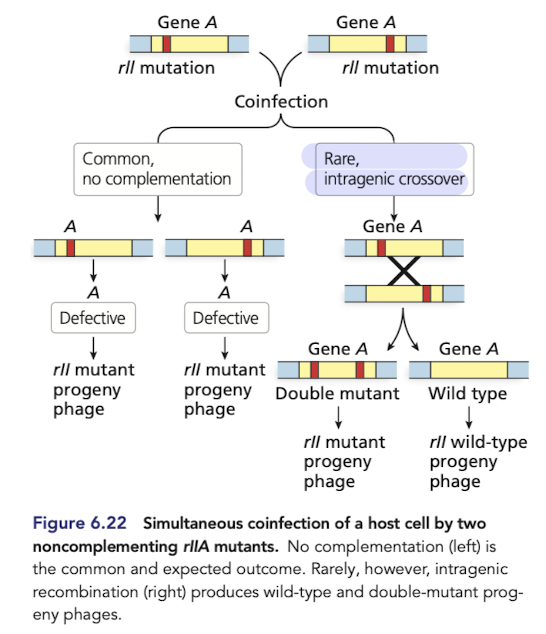
How does fine-structure mapping using lamda phage reveal recombination within genes? part 1
Bacteria can be coinfected by phage strain with different mutations that prevent plaque development
strains with different mutaitons in the same gene fail to complement (plaques don’t form or look weird)
but, recombination can occur within defective genes to generate a functional wildtype allele (normal plaques form)
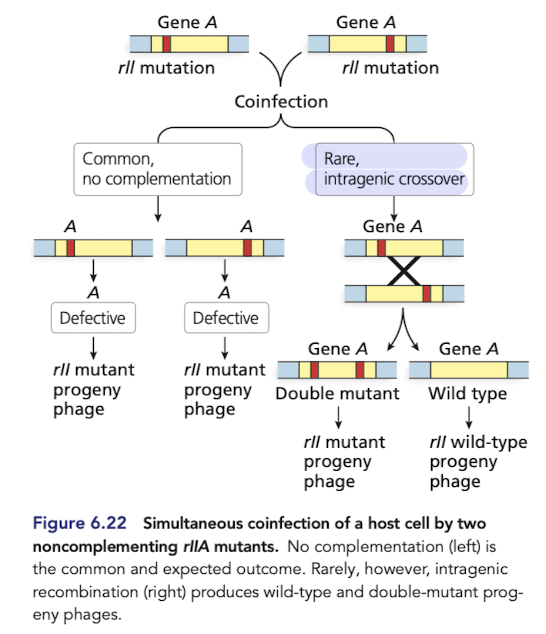
How does fine-structure mapping using lamda phage reveal recombination within genes? part 2
These experiments (Seymour Benzer), demonstrated that agenes are composed of smaller, individually mutable elements (later discovered to be nucleotides)
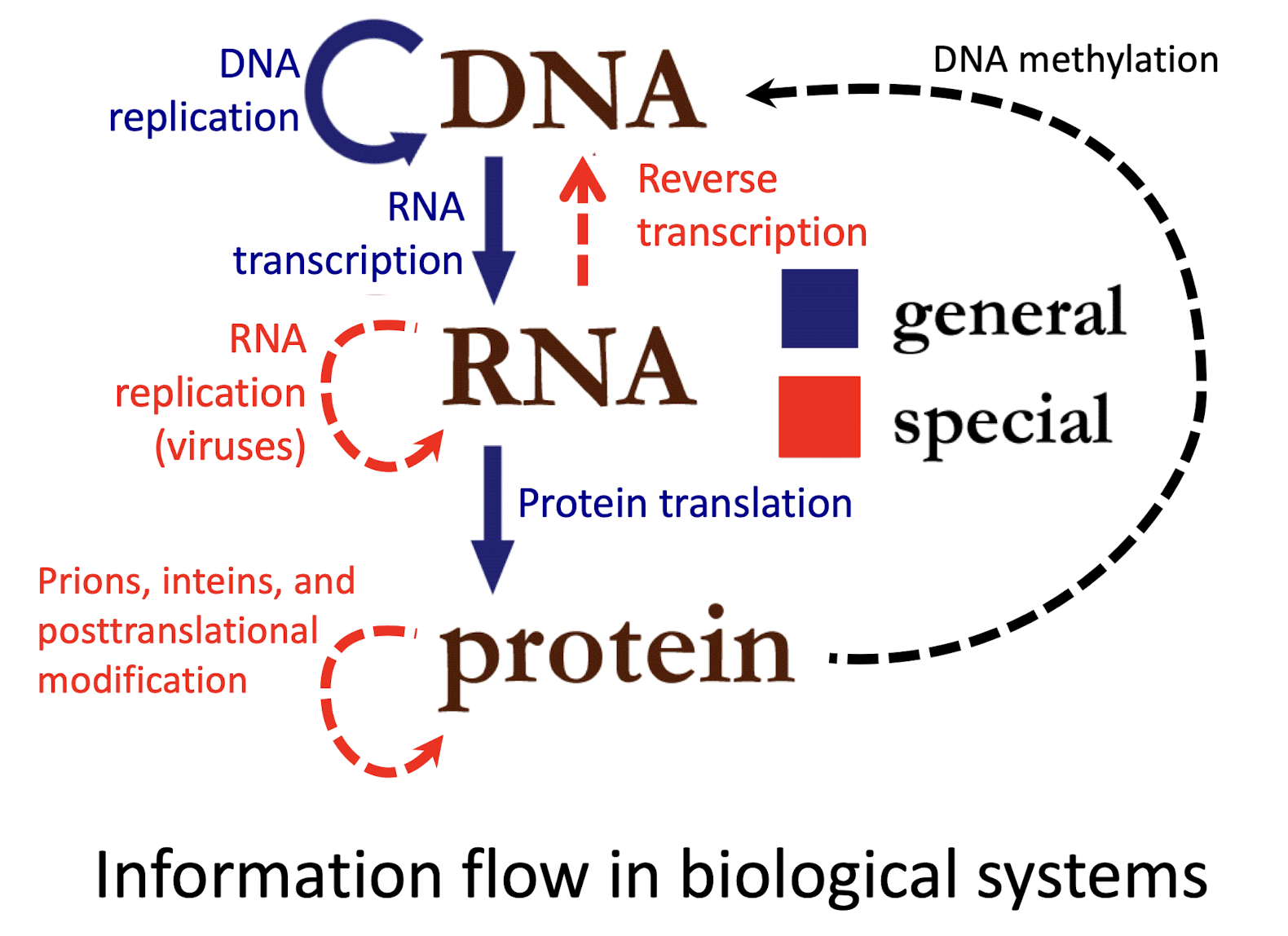
What is the central dogma of molecular biology?
Identifies DNA as the repository of genomic information and describes is key role in the production of RNA transcripts of genes leading to the production of polypeptides
How do we know that DNA and not protine is the heredity material?
Protein contains sulfur and almost no phosphorus
DNA contains phosphorus but no sulfur
bacteriophages attach to the outside of bacterial cells and inject DNA
emtpy “ghost” phages, no longer have DNA but are made up of protein
separated from bacteria throguh centrifugation
P labelled is in the pellet contained in the infected bacteria - confrims DNA is the hereditary molecule passed by the infecting phage into the host cell
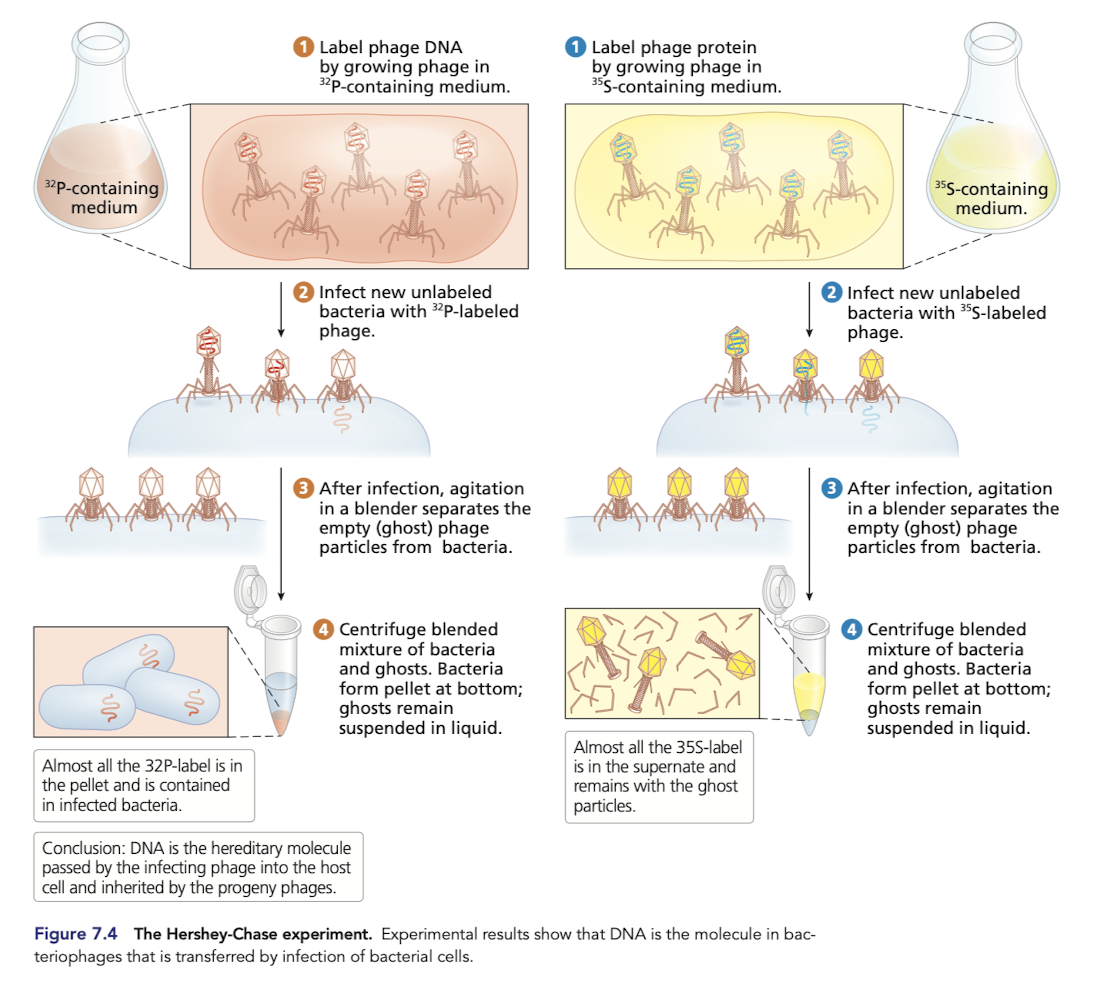
In the first part of the experiment, where only the protein was labeled, how did the researchers know that something (DNA) was transferred to the bacteria after shaking off the phage protein coat?
When the bacteria were plated, plaques formed indicating transforming has occured
In the second part of the experiment, where only the DNA was labeled, how did the researchers know that DNA was actually transferred to the bacteria, as opposed to some other information-containing substance?
They put plates on film and the radiation made dots on the film
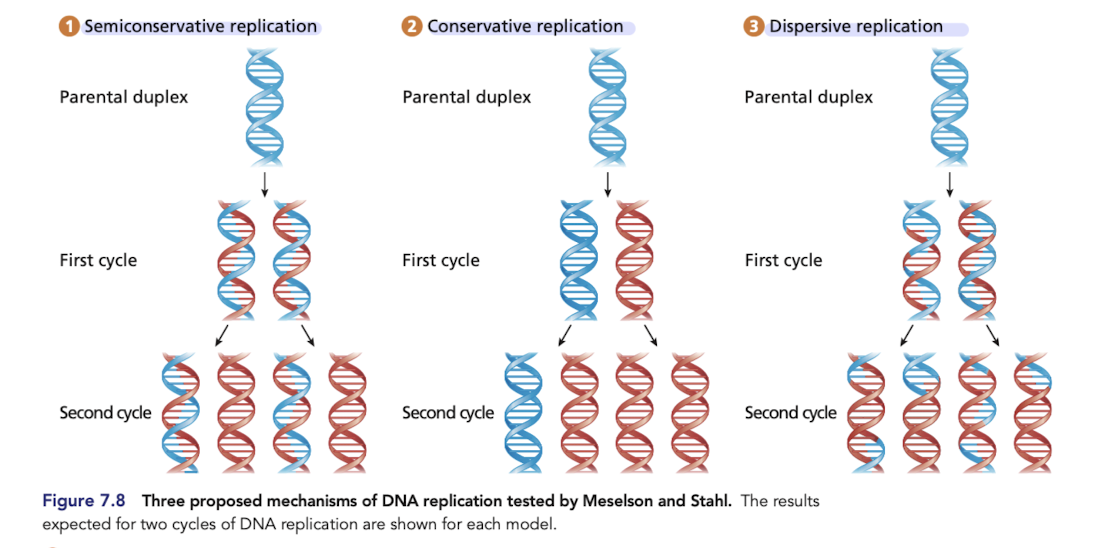
What is conservative replication?
Predicts that one daughter duplex contains the two strands of the parental molecule and the other contains two newly synthesized daughter strands
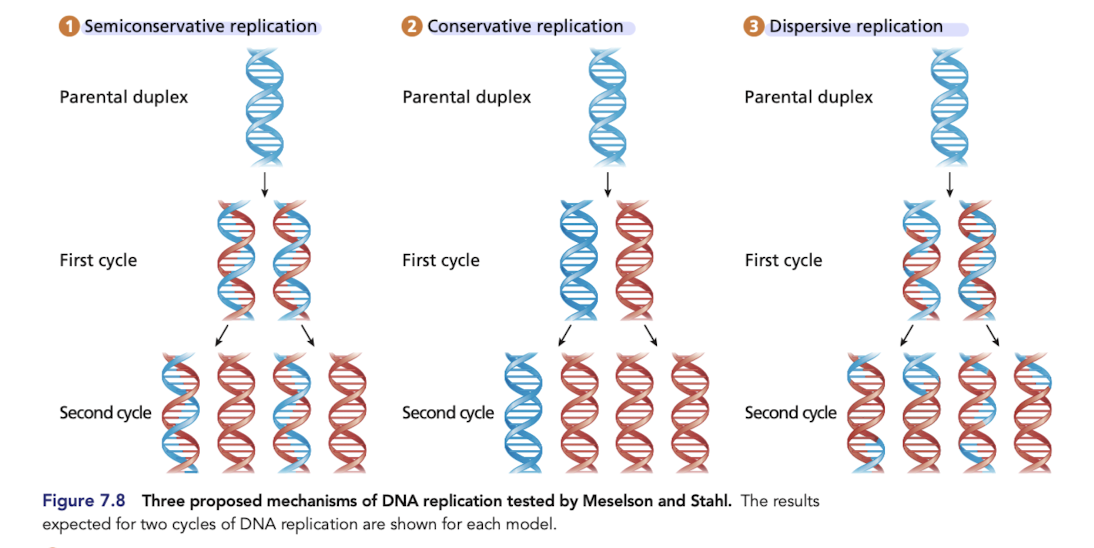
What is semiconservative replication?
Each daughter duplex contains one original parental strand of DNA and one complementary, newly synthesized daughter strand
proved to be correct
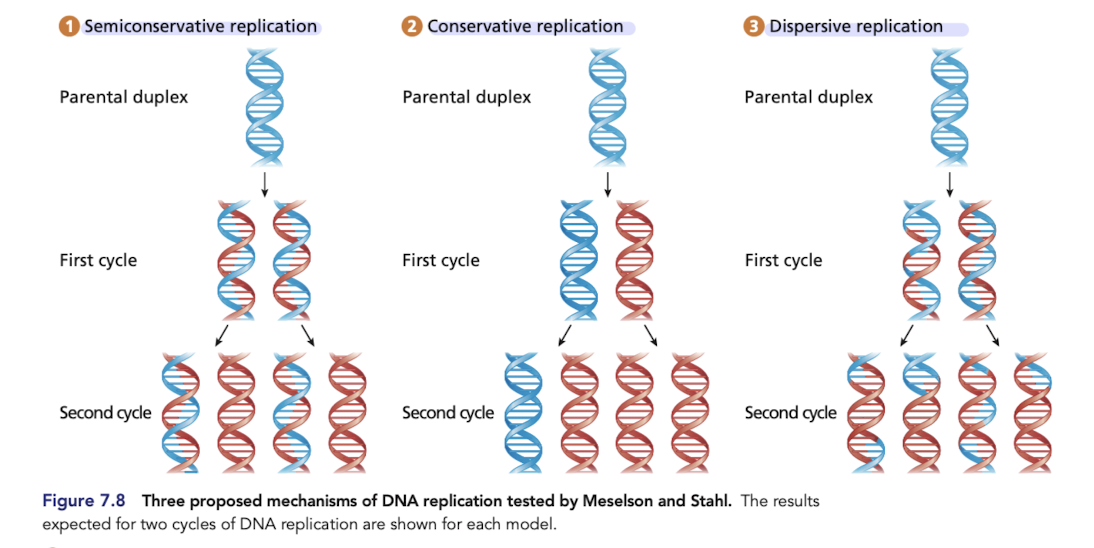
What is dispersive replication
Each daughter duplex is a composite of interspersed parental duplex segments and daughter duplex segments
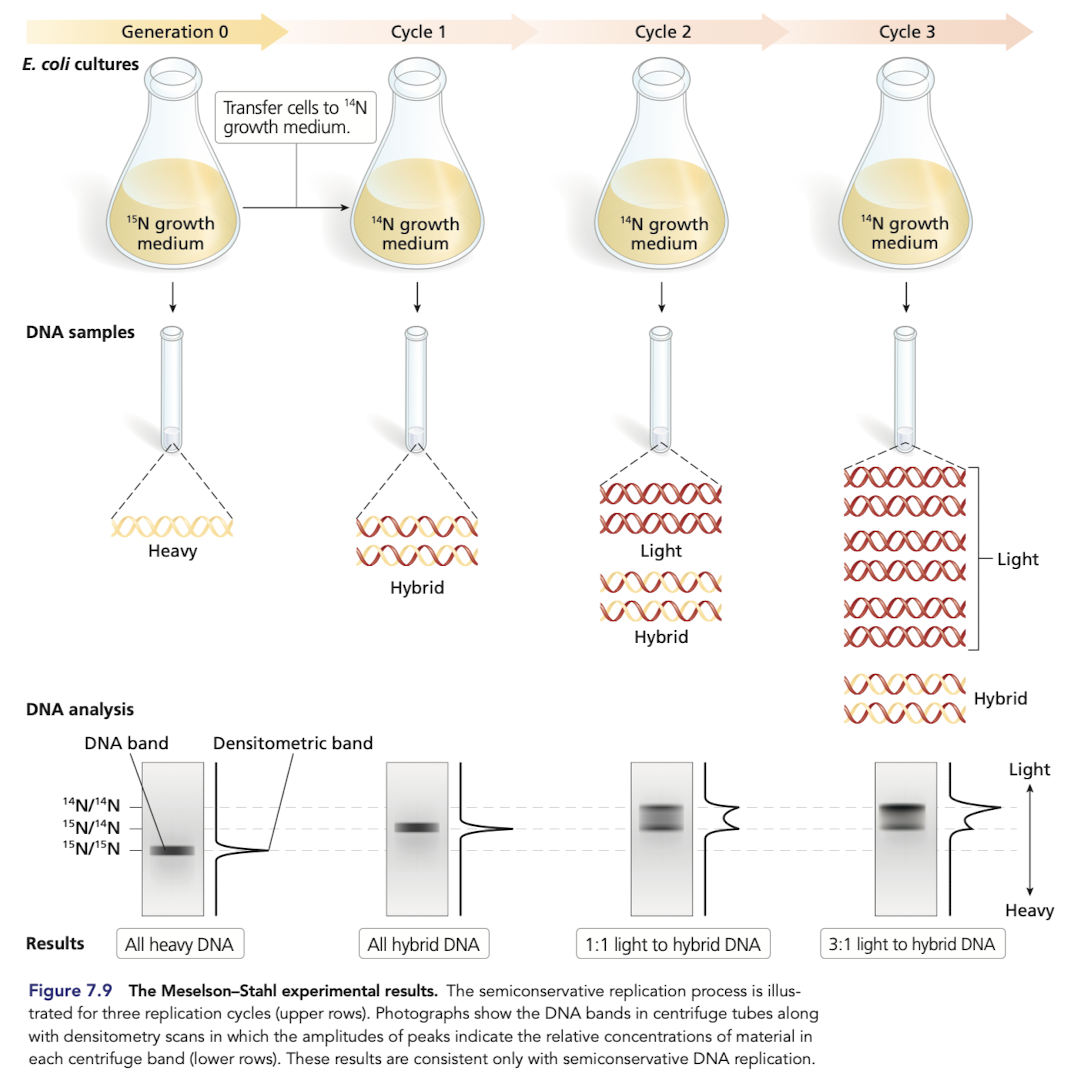
The Meselson-Stahl Experiment
Grew E.coli in a growth medium containing 15N
after transferring to a medium with 14N only, all hybrid DNA was of intermediate weight, rejecting conservative replication
DNA from subsequent generations grown in a medium with 14N only included light and intermediate weights, rejects dipersive replication
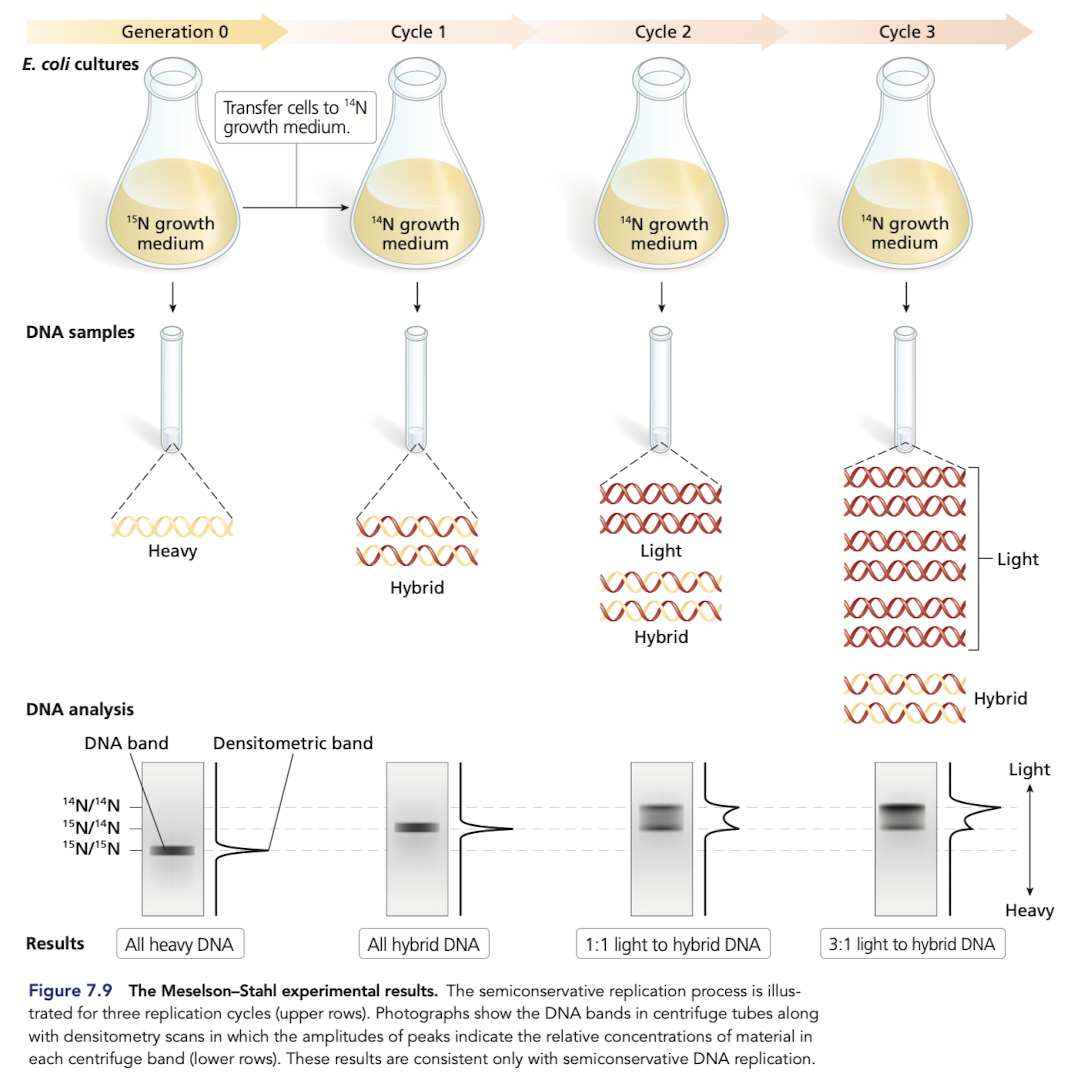
What would you expect if DNA replication was dispersive?
DIspersive model predicts a single density in all generations
in the 2nd generation, expect DNA to be a mixture and show up as one fuzzy band that is not near the oure 14N/14N band
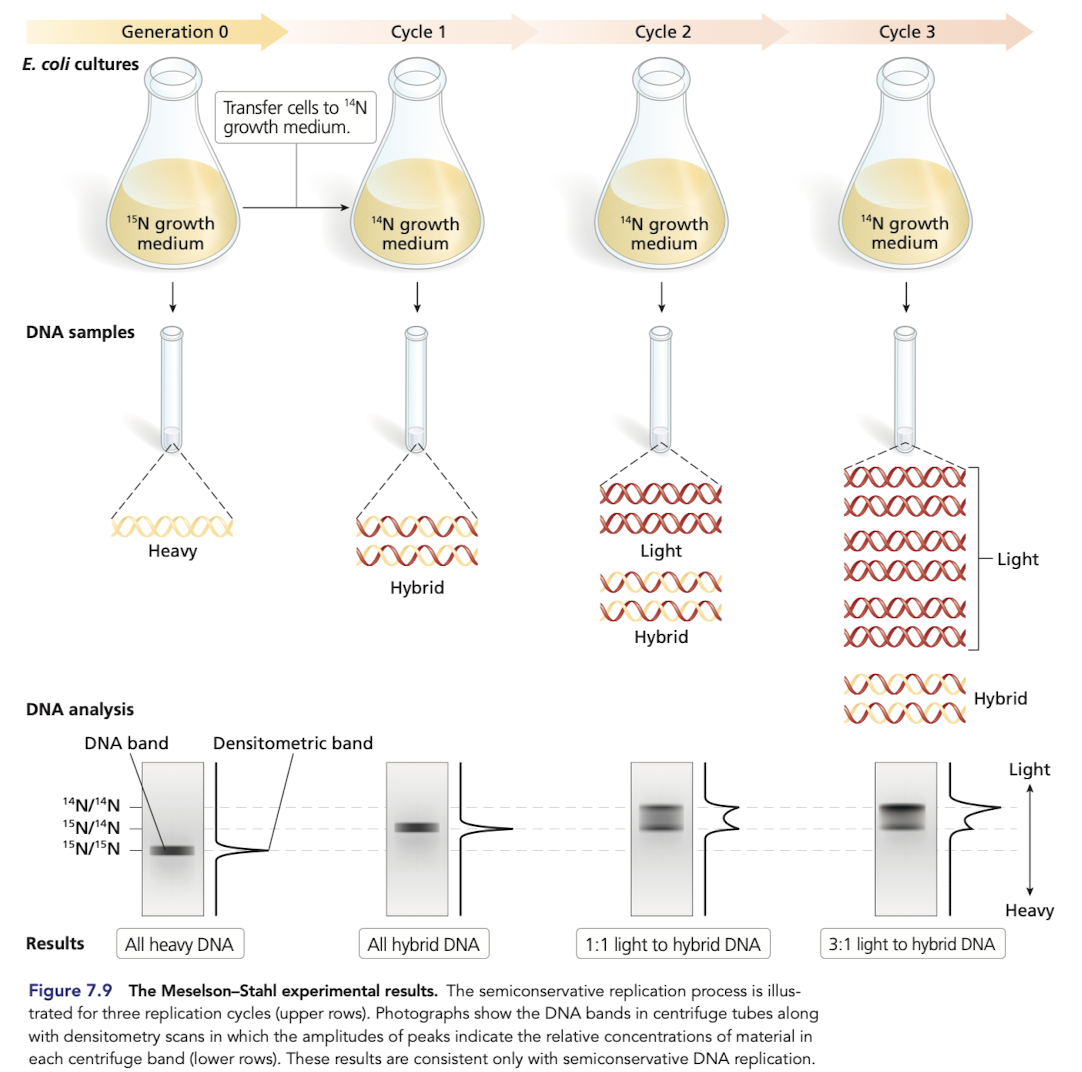
What would you expect if DNA replication was conservative?
Would expect DNA molecules with two distinct densities after generation 1 (15N/15N and 14N/14N)
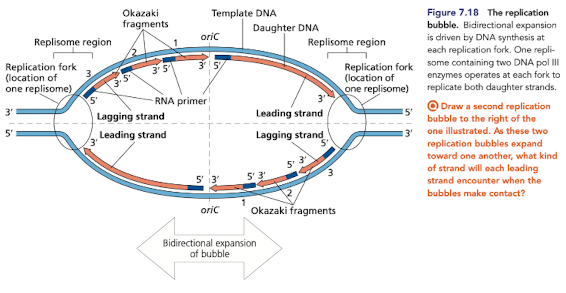
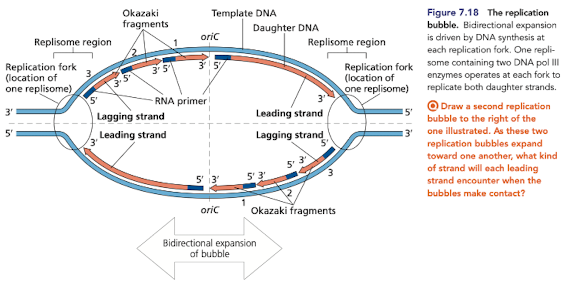
What direcition does DNA replication occur in?
Most often bidirectional, progressing in both directions from a single ori in bacterial chromosomes
from multiple ori in eukaryotic chromosomes
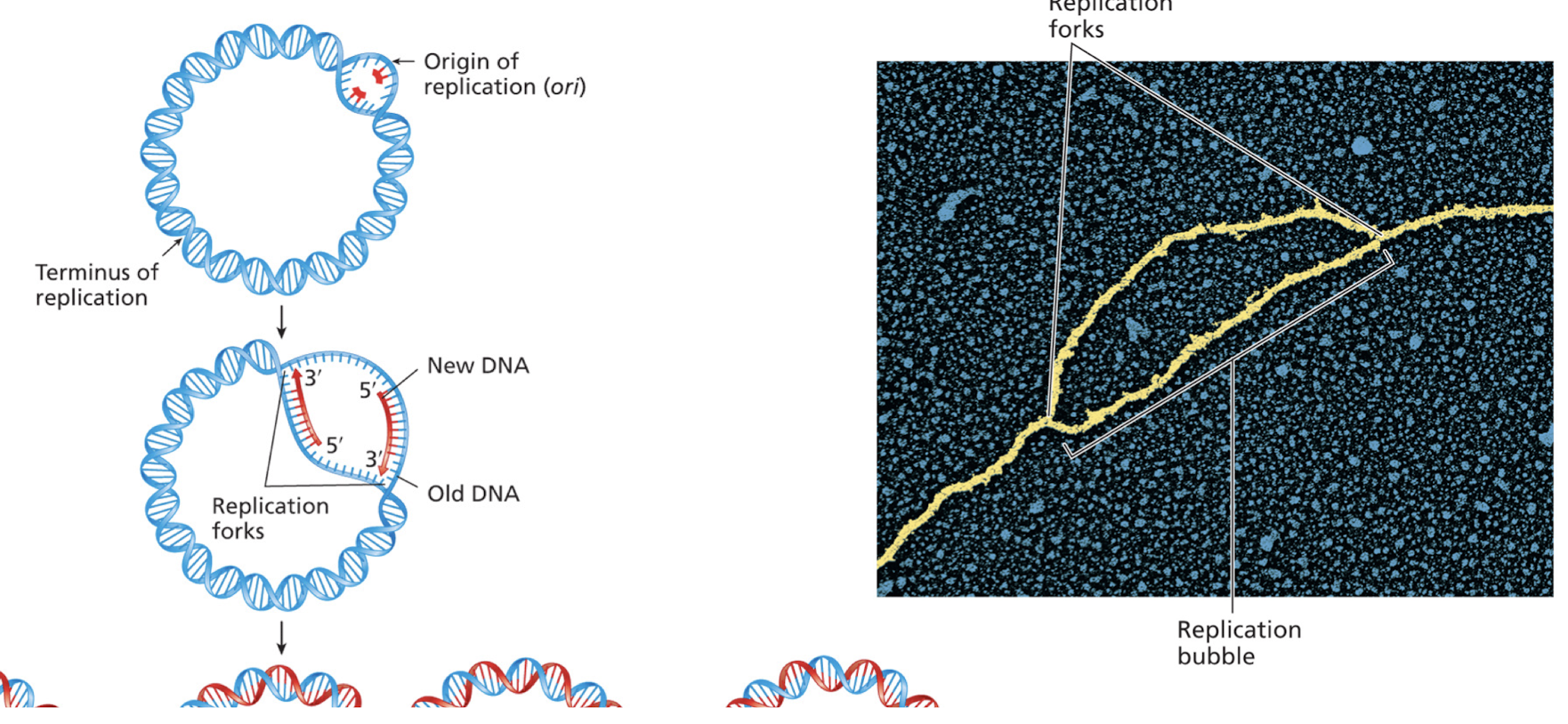
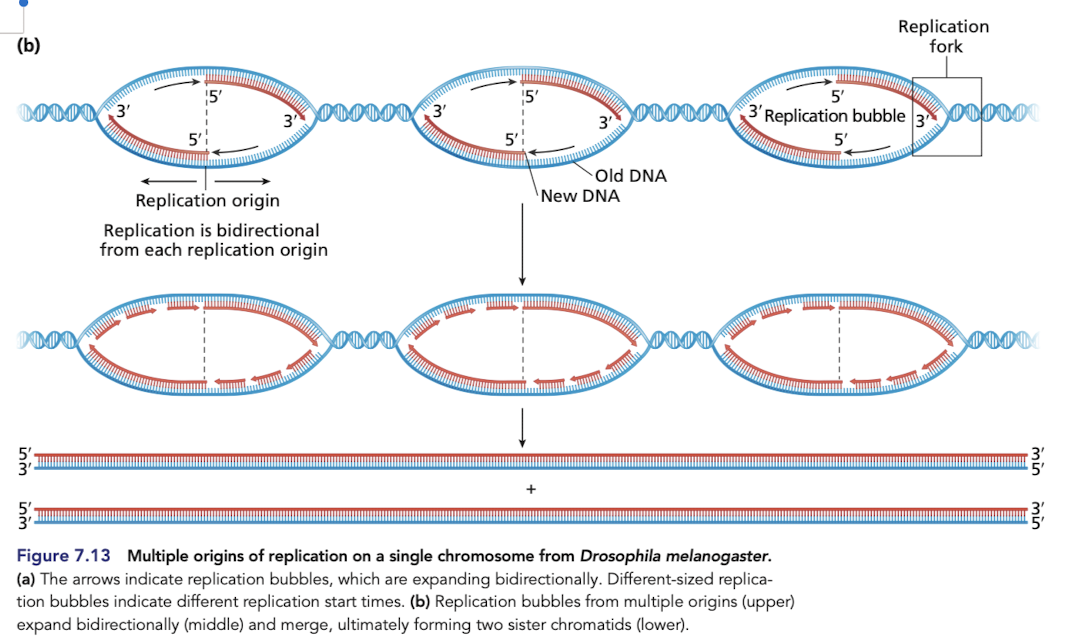
Human origin of replications
DNA replication begins at one or more origin of replications
in eukaryotes, each chromosome typically has multiple ori
human genome contains more than 50,000 ori
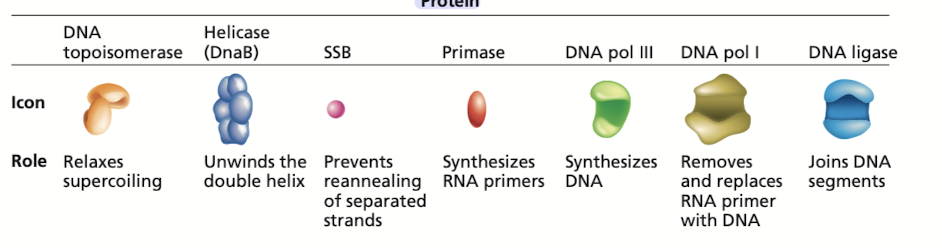
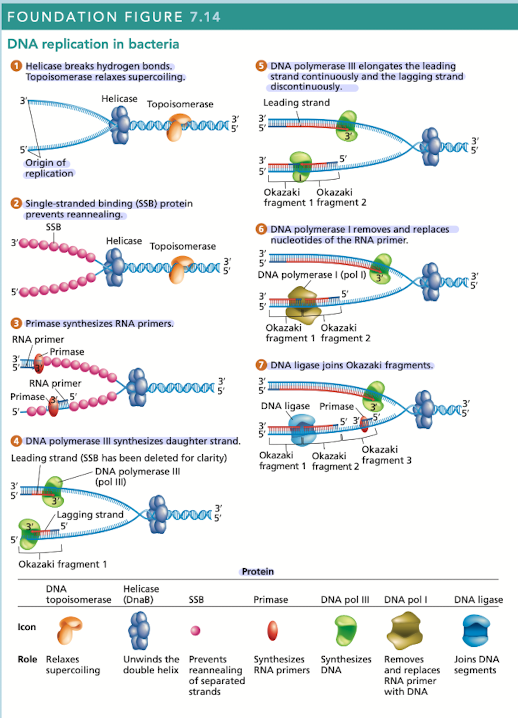
DNA replication is carried out by many proteins
Many of these proteins are part of the “replisome” - complex of proteins at each replication fork
2 DNAP III - one for leading, one for lagging
What does helicase do in DNA replication?
Breaks hydrogen bonds separating the DNA strands, unwinding the double helix ahead of advancing DNA replication
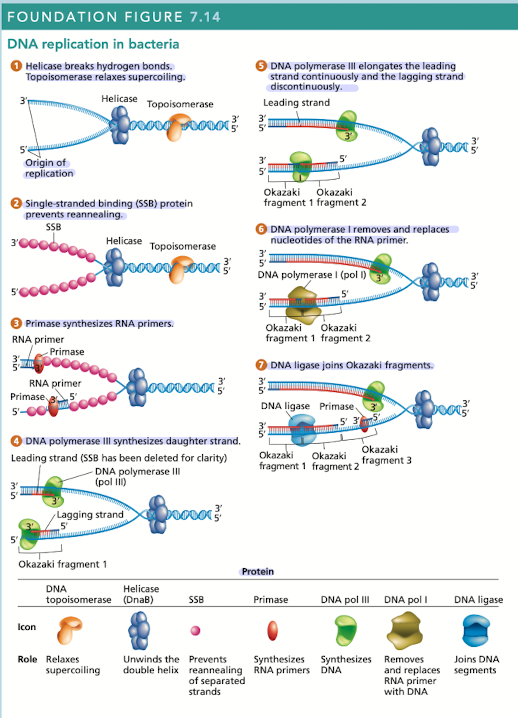
What does topoisomerase do in DNA replication?
Relaxes supercoiling
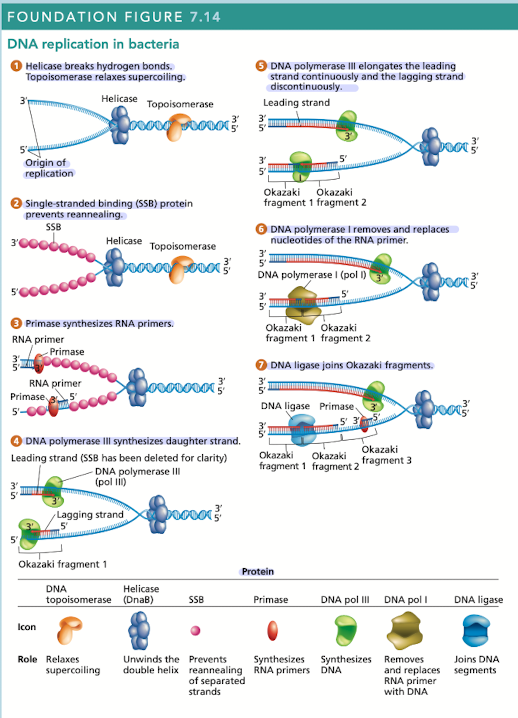
What do single-stranded binding (SSB) proteins do in DNA replication
Prevents unwound strands from reforming a DNA duplex, keeping them available as templates for new DNA synthesis
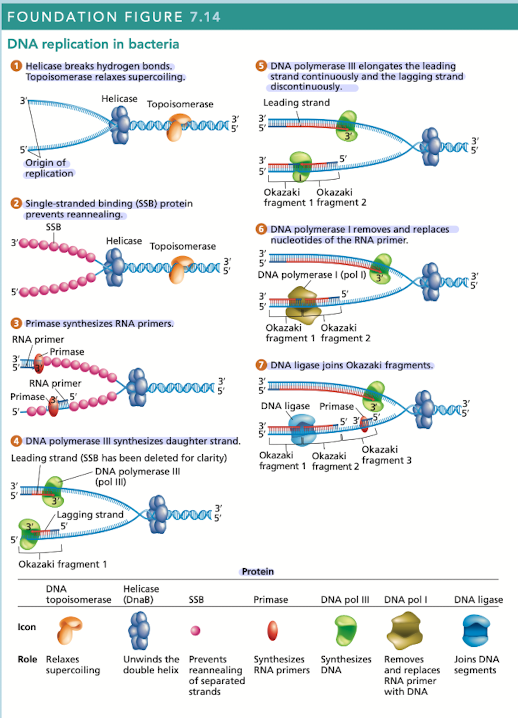
What does primase do in DNA replication?
Synthesizes RNA primers, providing the 3’OH needed for DNA polymerase activity
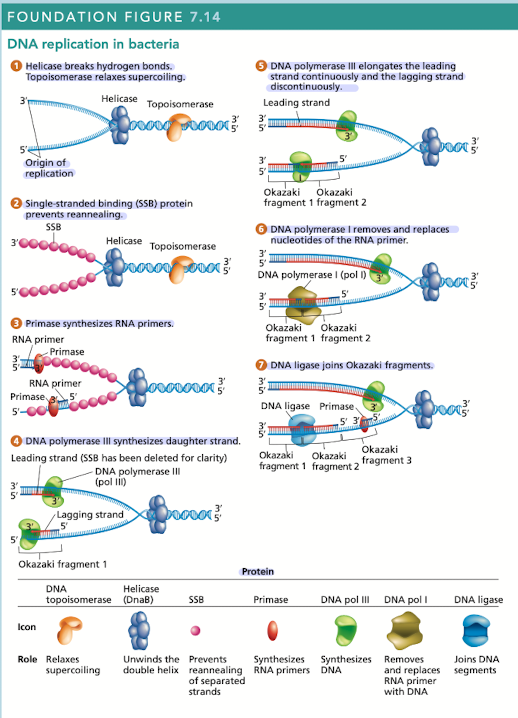
What does DNA polymerase III do in DNA replication?
Synthesizes daughter strand, begins its work at the 3’OH end of an RNA primer
rapidly synthesizes new DNA by adding one nucleotide at a time in a sequence that is complementary/antiparallel to template-strand nucleotides
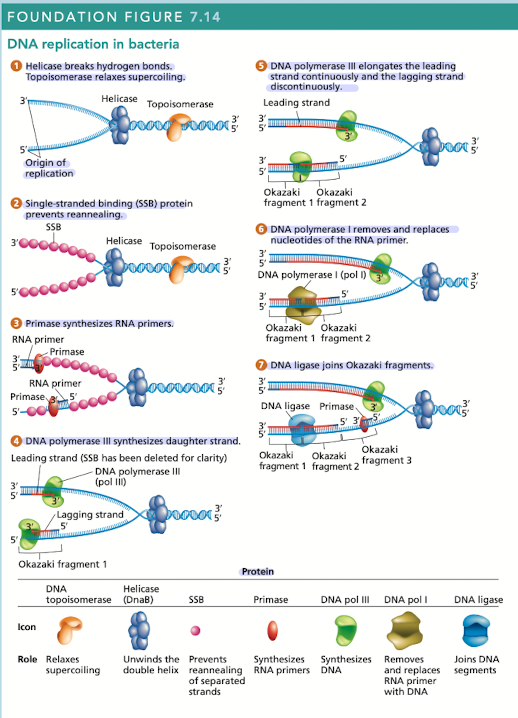
Continuous synthesis
DNA pol III carries out the 5’ to 3’ synthesis of one daughter strand in the same direction, in which the replication fork progresses
Discontinuous synthesis
DNA pol III carries out the 5’ to 3’ synthesis of the other daughter strand in the opposite direction to the movement of the replication fork
strands are elongated, in short segments, each of which is intiated by a RNA primer
What does DNA pol I do in DNA replication?
Has 5’ to 3’ exonuclease activity that removes the RNA primer
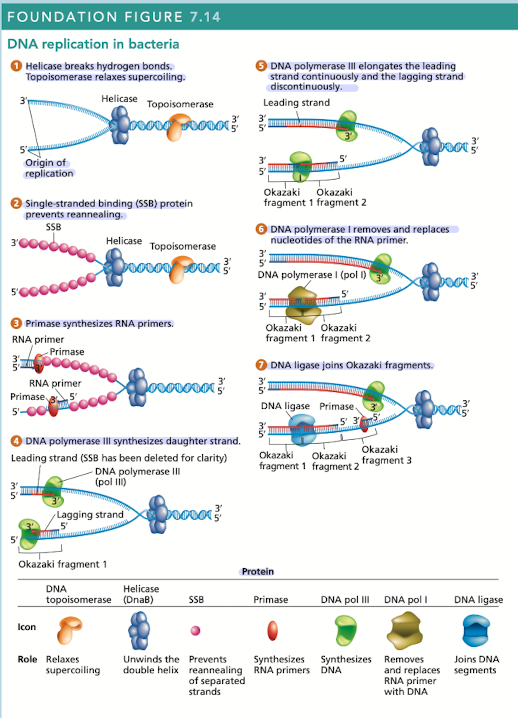
What does DNA ligase do in DNA replication?
Joins okazaki fragments, along with DNA pol I are active on the leading/lagging strand but more so on the lagging strand
Why does DNA pol need a RNA primer to begin synthesizing DNA?
DNA pol is unable to initiate synthesis - only extends the strand
essential role of RNA in DNA replication is consistent with the “RNA world” hypothesis that biology was based on RNA and protein before DNA came along
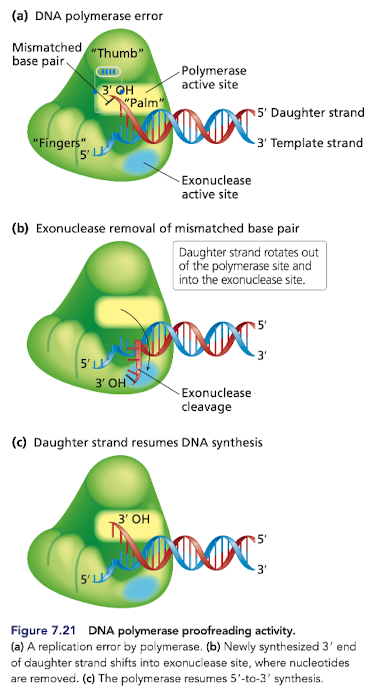
What is DNA proofreading?
Feature of most DNA pols taht momentarily stops and reverses replication to remove an incorrect nucleotide and replace it with the correct nucleotide
many have 3’ to 5’ exonuclease activity
incorrect nucleotide is excised and replication continues
What are mutations?
Result of errors in replication
mutation rate: low - typically 1/10^9bp but this rate varies among species
humans inherit ~60 mutations each generation
most are from our fathers
some parts of the genome mutate more than others (ex. mtDNA)
some types of mutations happen more frequently (ex. microsatellites)
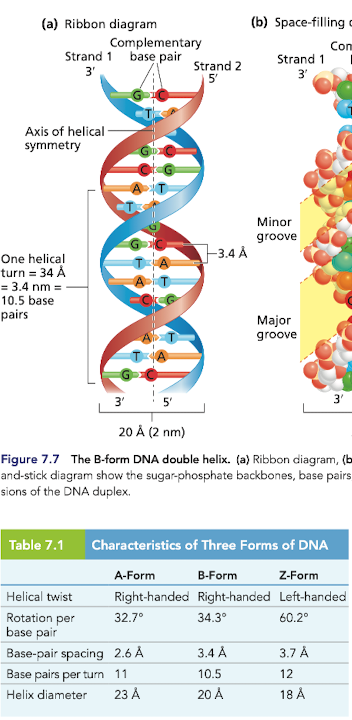
What is the most common 3D form of DNA?
The B-form DNA, has a major and minor groove that is recognized by proteins
helical twist is right-handed
What is the purpose of sex?
Recombination
What are the pros of reproducing sexually?
Male parental care
Sexual selection removes bad alleles
Recombination
What are the cons of reproducing sexually?
Have to make males (2 fold cost of sex)
May disrupt advantageous combinations of alleles
Could be risky (predation, STDs!)
What are hermaphrodites?
Sexually reproducing organisms containing both male/female gametes
Why have separate sexes?
A cost of separate sexes: have to find someone to have sex with
Sexual selection is only possible when you have separate sexes, can be good by purging deleterious alleles carried by males
Species with sexually dimorphic separate sexes may better exploit a complex fitness landscape than one with only one hermaphroditic sex phenotype
Inbreeding may be bad (major issue in hermaphrodites)
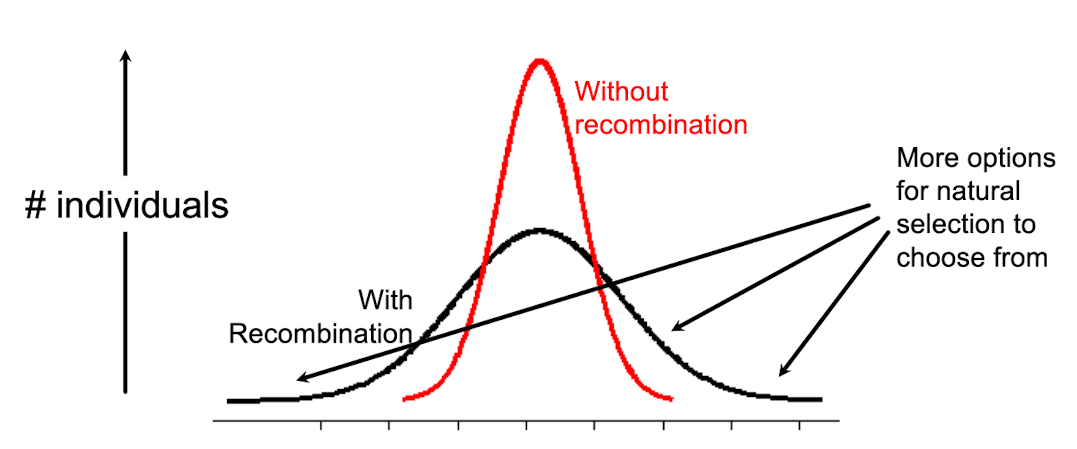
What are the advantages to recombination?
Leads to offspring that have combinations of traits that differ from parents
increases phenotypic value for a polygenic trait by granting more options for natural selction to choose from
What happens when homologous chromsomes are aligned?
Before meiosis, all alleles are linked with other alleles in either one of two phases (here, ABC or abc)
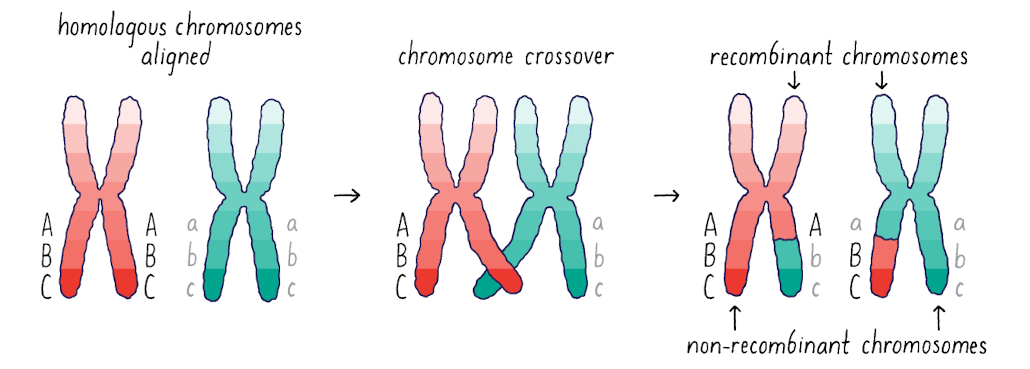
What happens during chromosome crossover?
Recombination associated with synapsis during prophase I of meiosis establishes new allelic phases
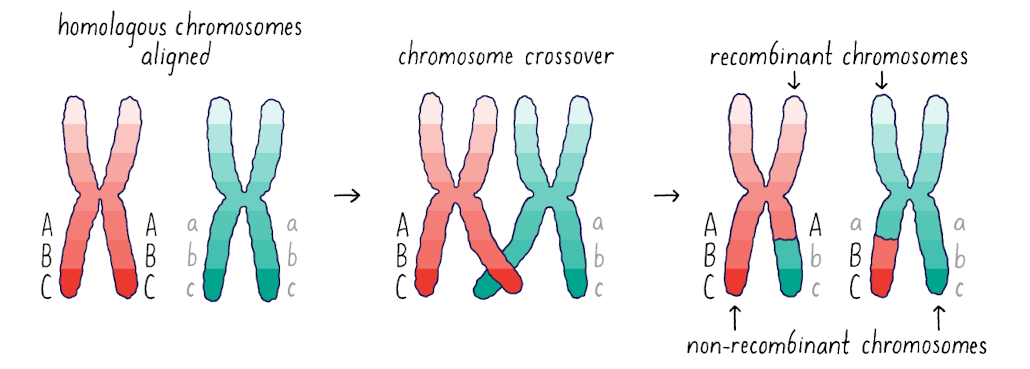
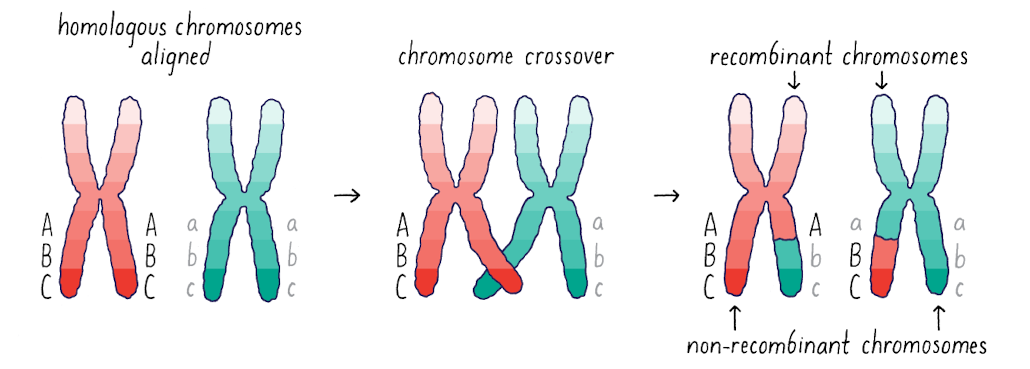
What is observed in recombinant/non-recombinant chromosomes?
Some alleles have novel phases that were not present in the parent (here, Abc and aBC)
What are the 4 possible fates of genomic regions that do not recombine?
Muller’s ratchet
Hill-Robertson effects
Genetic hitchhiking
Background selection
What is Muller’s ratchet?
Decline in fitness due to stochastic (process evolving in random way) loss of the least deleterious allele
without recombination, the best chromosomes can be randomly lost
Example of Muller’s ratchet
In combination with genetic drift, leads to decrease in fitness
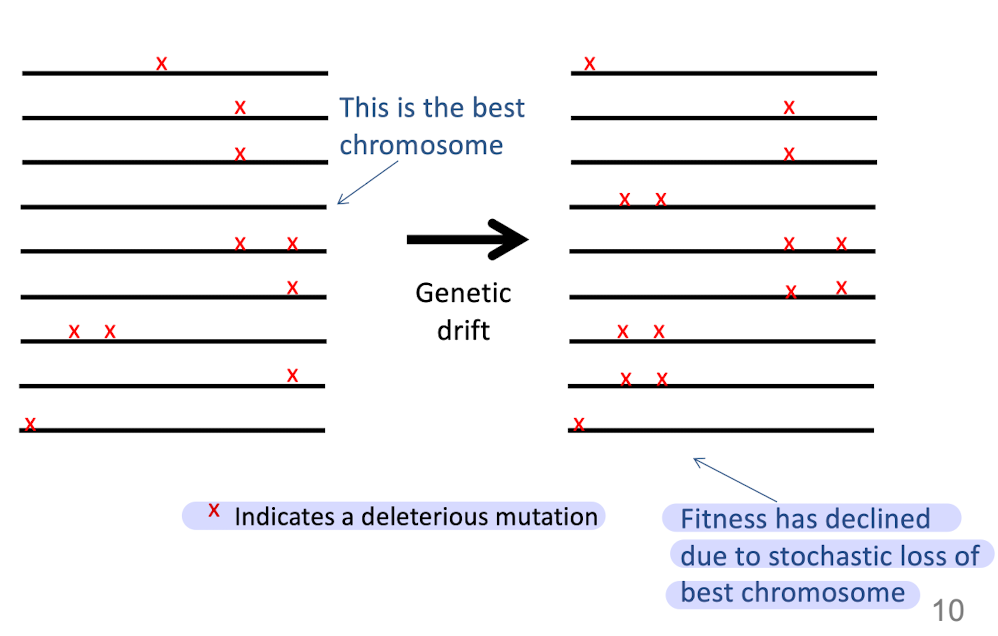
What is the “least deleterious allele”
Gene that decreases the fitness of the organism carrying it
least - would indirectly mean the “best chromosomes”
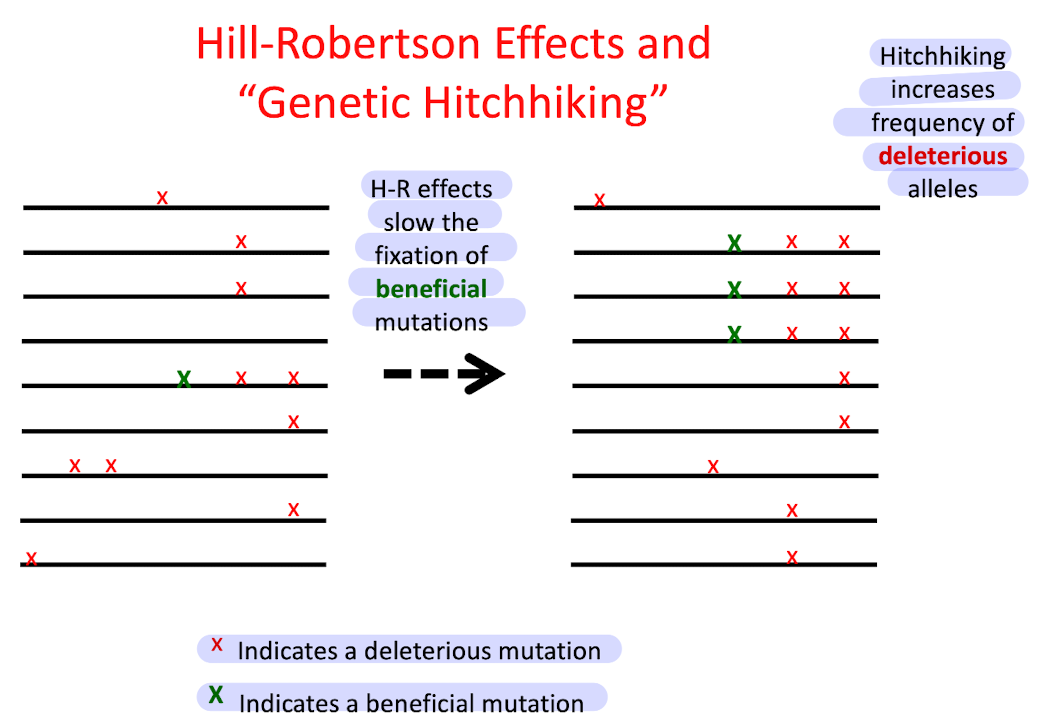
What are Hill-Robertson effects?
Interference between linked beneficial and deleterious mutations which slows fixation of beneficial mutations and slows removal of delterious ones
refers to interference influencing rates of fixiation and extinction
What is genetic hitchhiking?
Some deleterious mutations rise to high frequency or fixation b/c they are linked to advantageous mutations
refers to changes in frequency
Example of HR effects and genetic hitchhiking
Refer to the figure
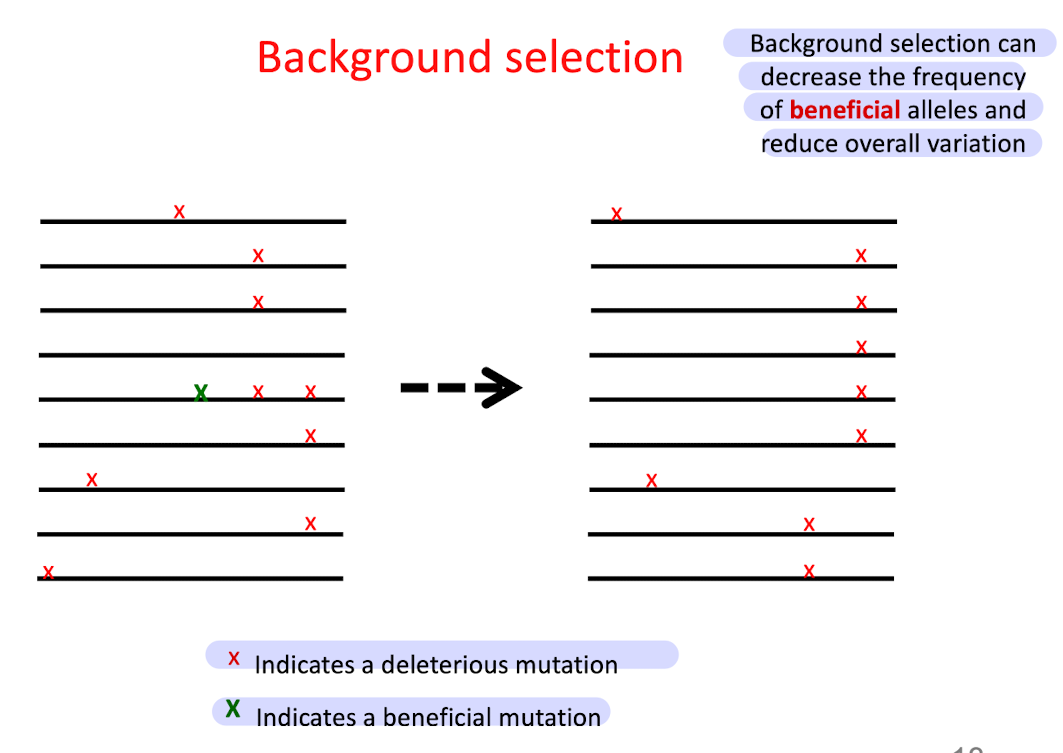
What is background selection?
Some beneficial mutations are removed from a population b/c they are linked to deleterious mutations
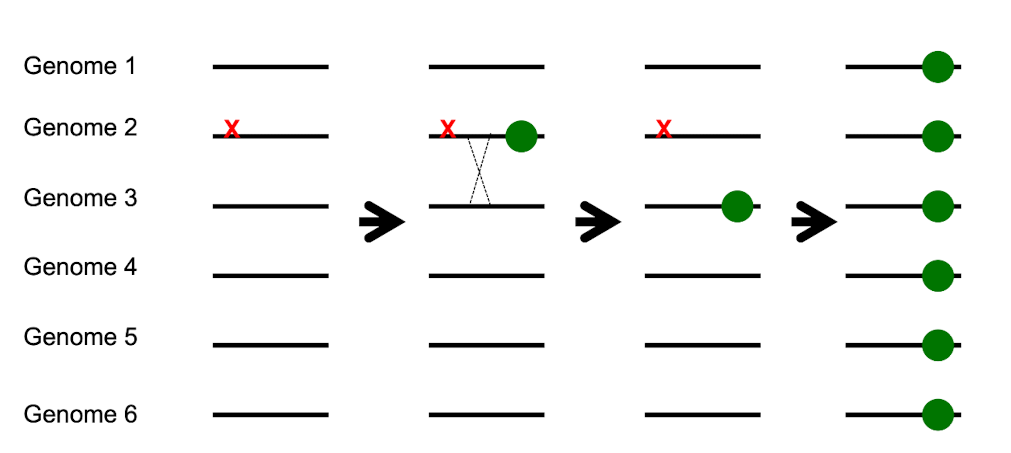
Why is genetic recombination useful? part 1
Can separate good (advantageous) mutations from bad (deleterious) mutations
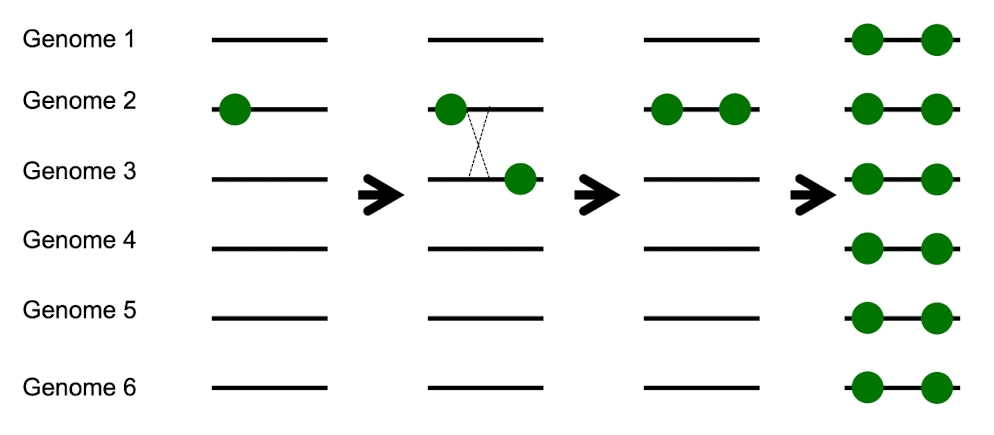
Why is genetic recombination useful? part 2
Can unite multiple advantageous mutations that appeared in different individuals
Replication at the Ends of Linear Chromosomes
Replication process unable to replicate linear chromosomes to the end due to the requirement of a primer
so, chromosomes become shorter with each replication cycle
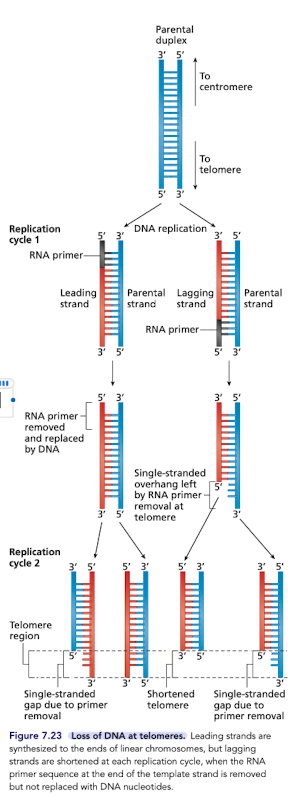
What is the Hayflick limit?
The number of times a normal, differentiated somatic cell can undergo cell division before cell division stops (i.e limit to the length of a cell’s life span)
b/c chromosome shortening (progressive loss of telomere length as cells age)
limit ~50-70 cell divisions
after limit, cell undegrgoes programmed cell death (apoptosis)
What are “exceptional cells”?
Cells with a extended or non-existent limit, such as stem cells, germ cell progenitors, and cancer cells (can divide many more times)
typically have telomerase activity that maintains the chromosome ends
If the bottom of this DNA is the telomere, which strand (red or blue) will not be fully copied at the telomere due to the synthesis of a RNA primer
The red one b/c this is the lagging strand and synthesis must go from 5’ to 3’
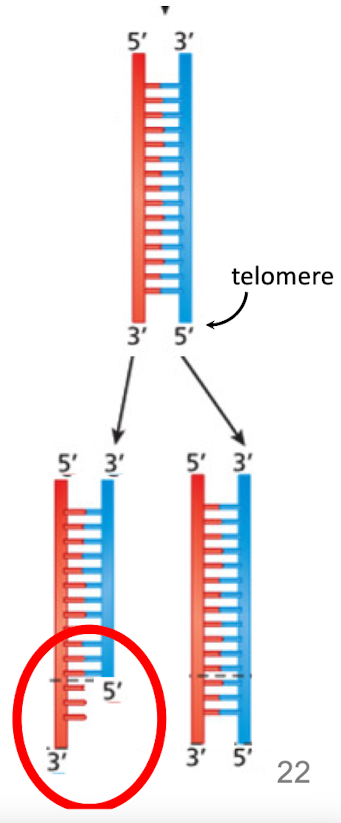
What are telomeres?
Hundred-thousands of copies of telemere repeats at the ends of linear chromosomes to avoid apoptosis
length decreases with age
dysfunctional telomeres associated with genome instability and aneuploidy
low telomerase activity have a variety of disorders
dyskeratosis congenita, aplastic anemia, pulmonary fibrosis, cancer
do not contain coding sequences
What is a ribonucleoprotein?
A enzymactic complex that includes protein and ribonucleic acid
ex. telomerase, ribosomes, and components of the spliceosome
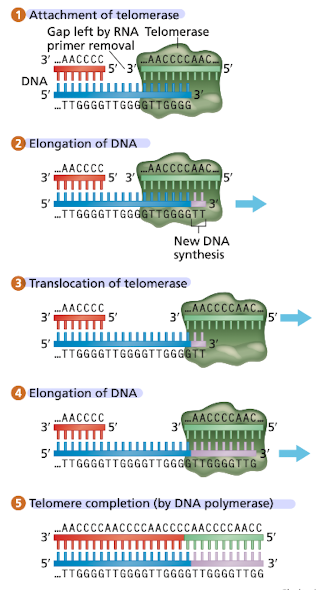
What is telomerase?
Reverse transcriptase, transcribes DNA from a RNA template
depicts the telomeric sequence “synthesis of a telomere” 1
attachment of telomerase
elongation of DNA (using RNA in telomerase as template)
translocation of telomerase
elongation of DNA (create more repeats)
telomere completion (DNA Pol)
Telomerase is usually active only in germ cells and stem cells. Why would evolution not favour all cells to have a high-level of telomerase activity to ensure chromosomal integrity and cell longevity
Most cancers have high telomerase activity so apoptosis of old cells may be favoured to reduce the incidence of cancer
current active area of research
What is PCR (polymerase chain reaction)?
In-vitro DNA replication, copies DNA semiconservatively
Can be done in a very small volume of fluid
Uses temperature instead of helicase to unwind (denature) DNA
Denaturation (~95)
Annealing (~45-68)
Extension (~72)
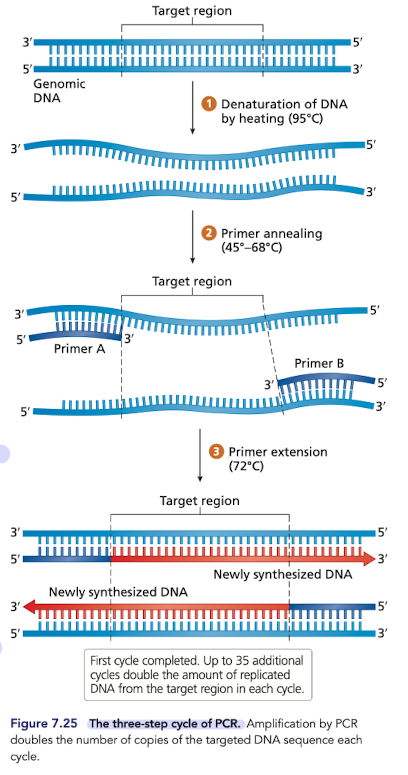
What is needed for PCR?
DNA template
dNTPs (supply of the 4 nucleotides)
heat-stable DNA polymerase (Taq Pol)
forward and reverse DNA primers
a buffer solution
What are the limitations to PCR?
Need to have previous knowledge about the sequence you want to amplify to design the primers
only works for small fragments (<2000 bp, but sometimes up to ~15,000 bp)
What is Thermus aquaticus?
A bacteria that can tolerate high temperatures
lives and undergoes cell division in hot springs
does not denature at high temps (95)
unique property alllows high temp to be used to denature DNA without denaturing the polymerase
Analyses of repetitive regions using PCR amplication and gel electrophoresis part 1
Using one pair of primers that flank a repeat, alleles w differing number of repeats can be amplified
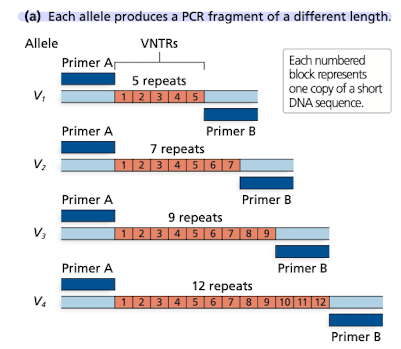
Analyses of repetitive regions using PCR amplication and gel electrophoresis part 2
Amplified alleles can be visualized by separating them by size using gel electrophoresis
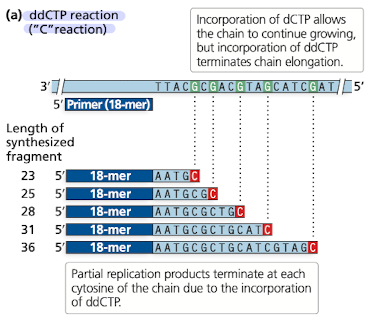
How does Sanger Sequencing work?
4 dNTPs are used in high concentrations, to them is added ddNTPs in a smaller concentration
ddNTPs have absence of 3’OH (carbon)
when ddNTP is incorporated into a growing strand, the synthesis of the strand is terminated at that point
What is transcription
Refers to the biological process where information from DNA is converted to RNA
involves synthesis of RNA from a DNA template by a protein called RNA polymerase
when a gene “expressed” this means it is transcribed (possibly translated too)
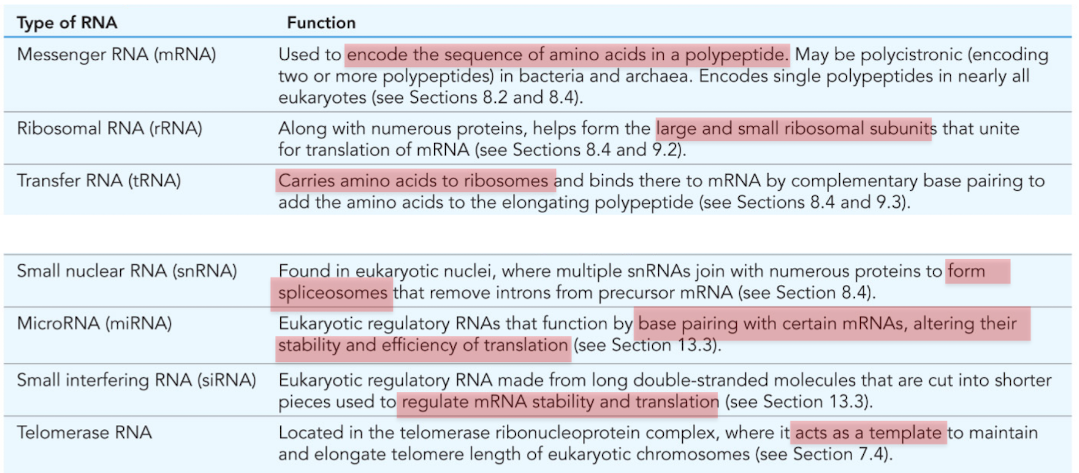
What is mRNA (messenger RNA)
Used to encode the sequence of amino acids in a polypeptide
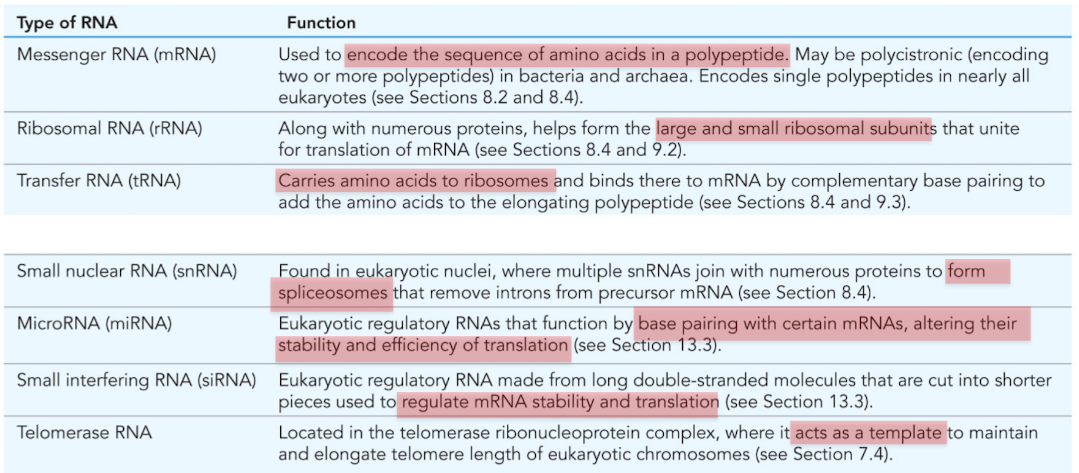
What is ribosomal RNA (rRNA)?
Combines with numerous proteins to form the large/small ribosomal subunits
molecular machine responsible for translation
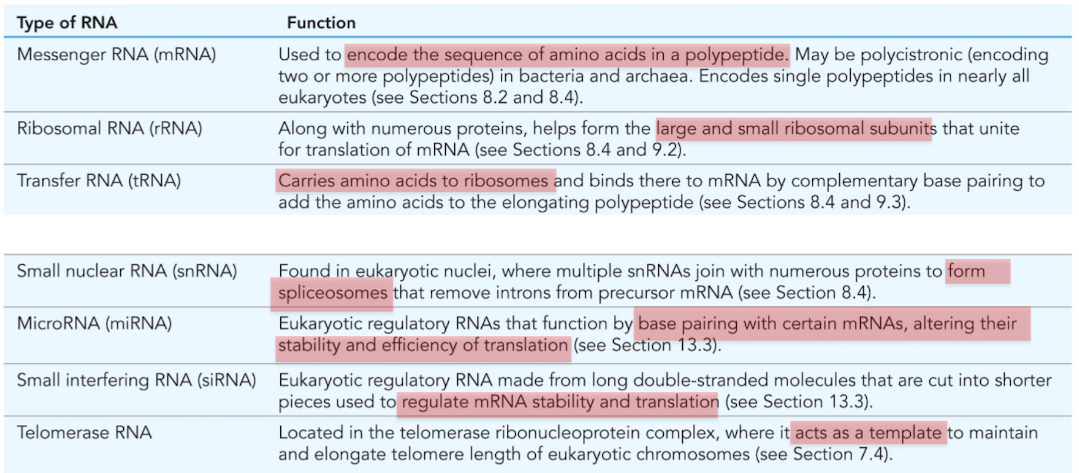
What is tRNA (transfer RNA)?
RNA that carries amino acids to the ribosomes for construction of proteins
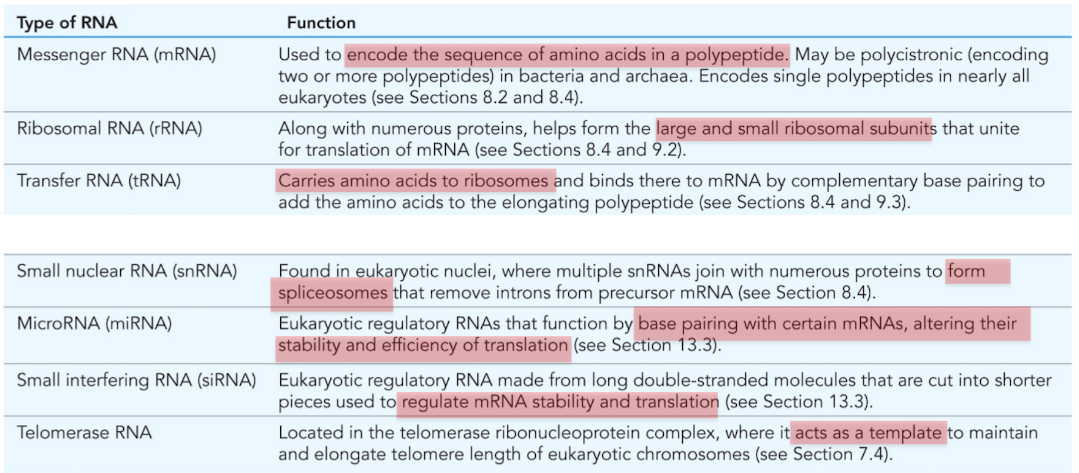
What is snRNA (small nuclear RNA)?
Found in nucleus, forms spliceosomes
mRNA processing/intron removal
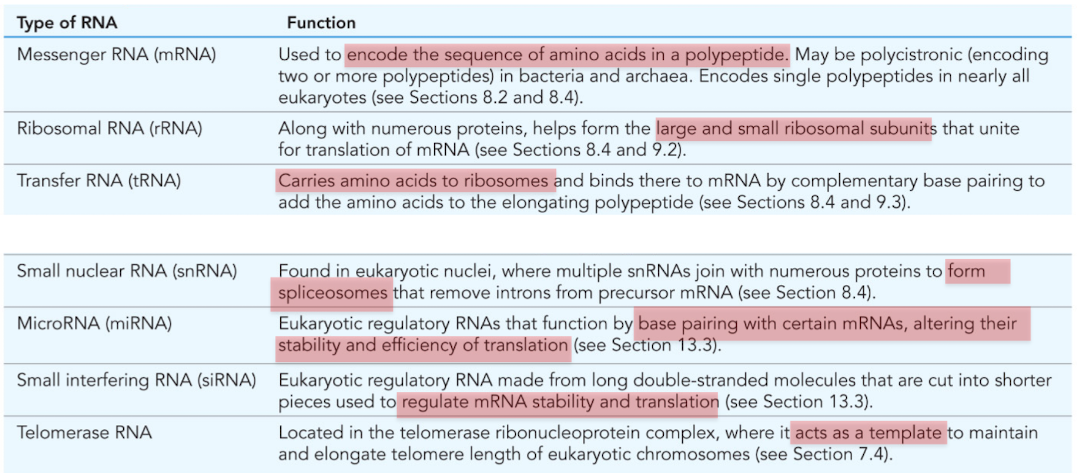
What is miRNA (microRNA)?
Posttranscriptional regulation of gene expression
base pair with certain mRNAs, alter stability and efficiency of translation