Microbio Midterm 2 Study Guide
1/124
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
125 Terms
bacterial cytoplasm
site of metabolism
contains ribosomes
holds genetic material
Bacterial Cytoskeleton (functions)
functions:
cell division (pass on genes)
protein localization (getting things at place at the right time)
cell shape
What are some homologues to eukaryote cytoskeleton components and their functions
MreB is like actin: cell division
Crescentin is like intermediate filaments: shape + hold proteins in place
FtsZ is like tubulin: microtubels
What makes up the eukaryotic cytoskeleton
microfilaments
intermediate
microtubules
microfilaments are?
thin filaments
actin and myosin(motor protein)
what are microfilaments function and bacterial homogloue
movement, endocytosis, cytokinesis, and secretion
bacterial homologue= Mre proteins (Mre B)
Intermediate filaments and bacterial homologue
structural filaments of about 10 nm diameter
structural in function
points of attachment for organalles
highly stable
Bacterial Homologue: crescentin
Microtubles and its bacterial homologue
helical shaped polar cylinders about 25 nm diameter
tubulin subunits
Bacterial homologue FtsZ proteins
dynein and kinesin motor proteins (no known bacterial homologue
Describe the movement of a dynein motor protein on microtubles
turkey legs walking
not always have two legs
subsets of both that can move both ways
Inclusions are in bacterial cells what about them
prokaryotic homologues to membrane bound vesicles
aggregates of organic and inorganic substances: granules, cystals or globules
Storage:
carbon: glycogen and poly beta hydroxybutarate
inorganics: sulfur globules and polyphosphate granules
What is the importance of gas vacuoles
allow microbes to regulate buoyancy
found in cyanobacteria and other aquatic prokaryotes and photosyntetic
protein compartments filled with gas
What are magnetosomes
contain magnetite (Fe3O4)
allow bacteria to orient along magnetic fields
consolidate iron in inclusion bodies
not true endocytic membranes (single lipid membranes)
ruffles that allow invagination of phospholipids
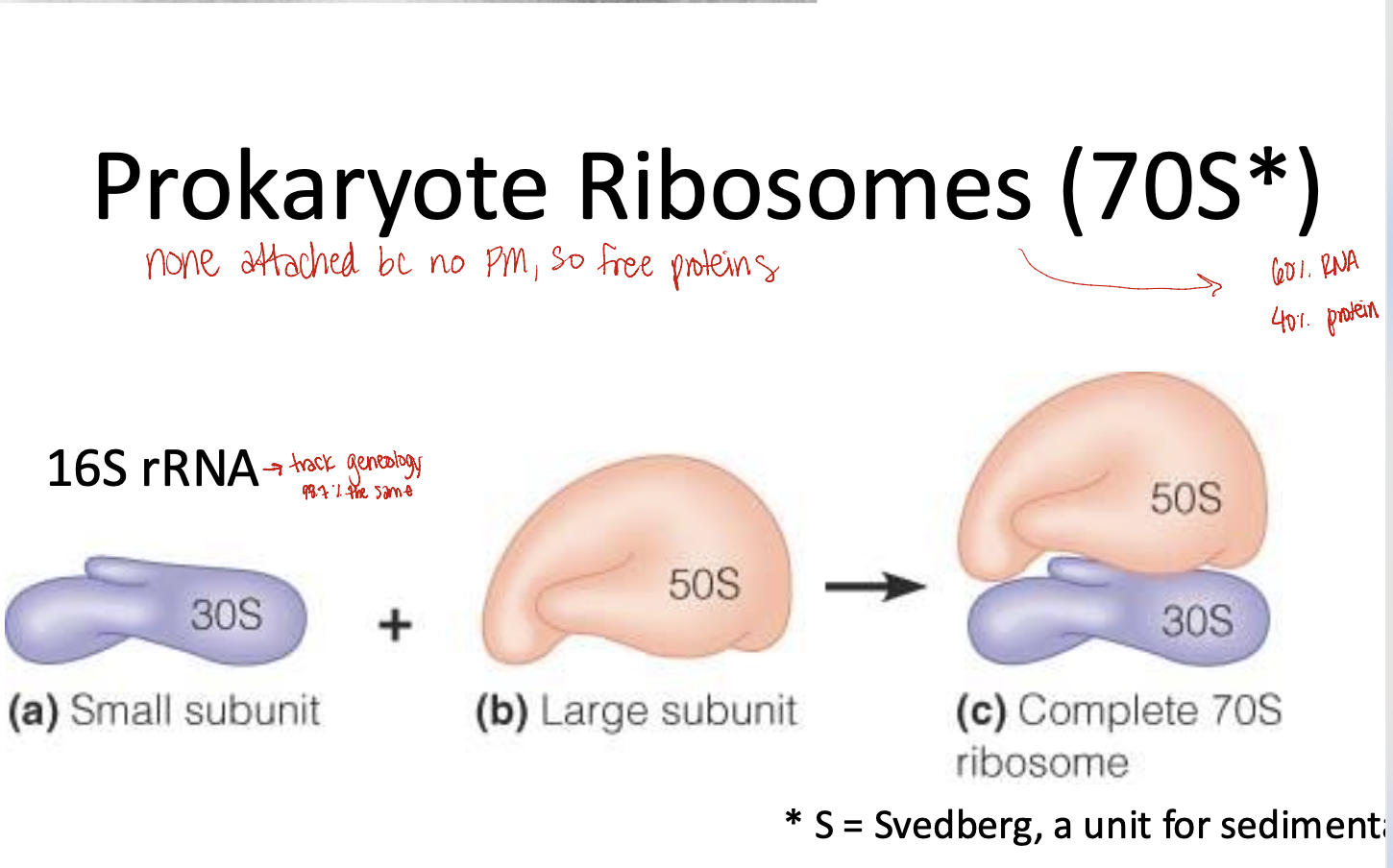
Prokaryote Ribosomes
S*= Svedberg, a unit for sediment
70S= 30S small subunit + 50S large subunit
60% RNA and 40% protein
16S rRNA is found in the 30S small subunit
98.7%= same species
are soluble polysomes: a group of ribosomes reading the same mRNA simultaneously
not attached to PM so free floating protein
Compare and contrast Eukaryotic and Bacterial Ribosomes
Euk.
80S = 60S + 40S
18S rRNA
slightly larger in molecular weight/layer
Bact.
70S = 50S + 30S
16S rRNA
life started int the RNA world
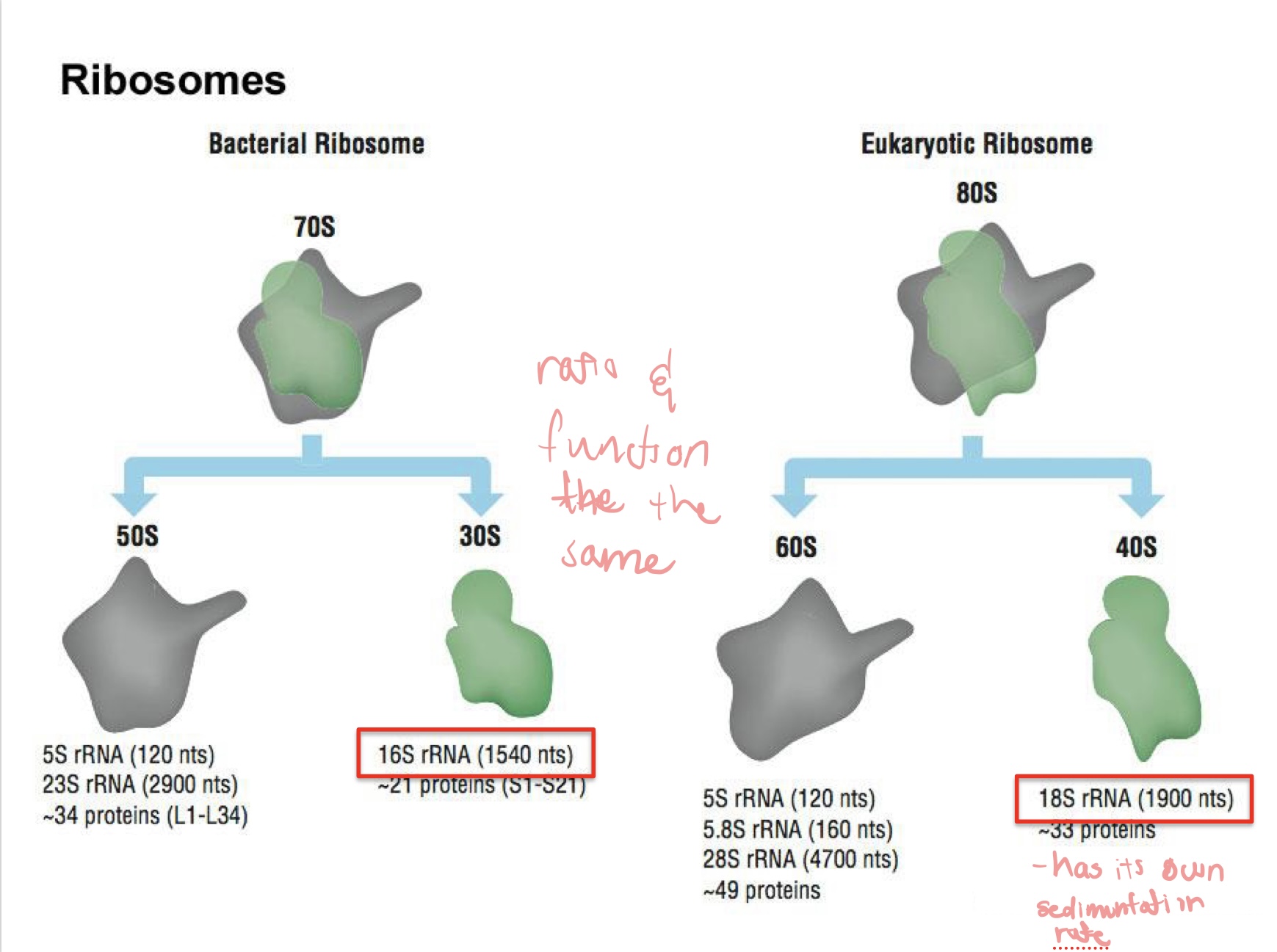
S* = svedberg, a unit for sediment
Both are made of RNA + protein and perform the same core function: protein synthesis — but differ in size, complexity, and evolutionary details.
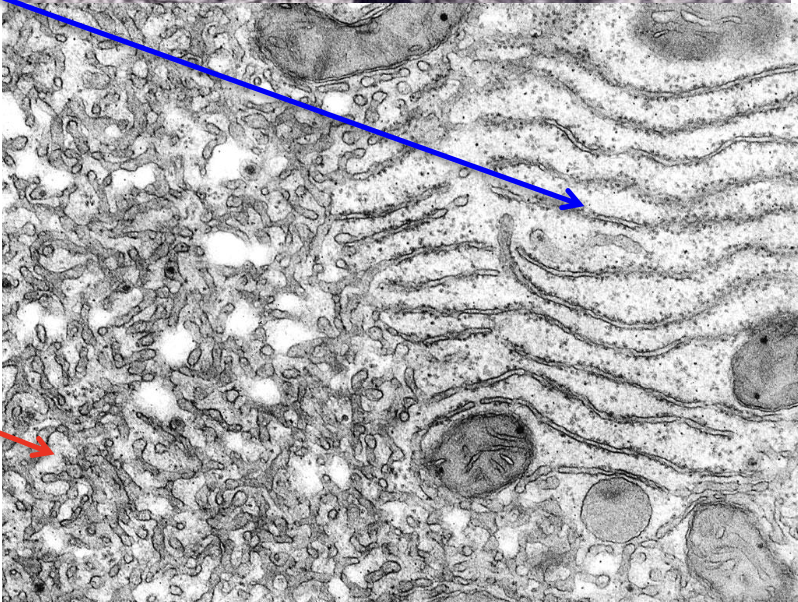
Endoplasmic reticulum (ER)
function: syntesis and transport (shuttling system)
rough (RER): studded w/ ribosomes, sythesis and transport of proteins
smooth (SER): fewer ribosomes, synthesizes lipids and other macromolecules (some proteins but not as many and lots of lipids)
endocytic pathway brings it in
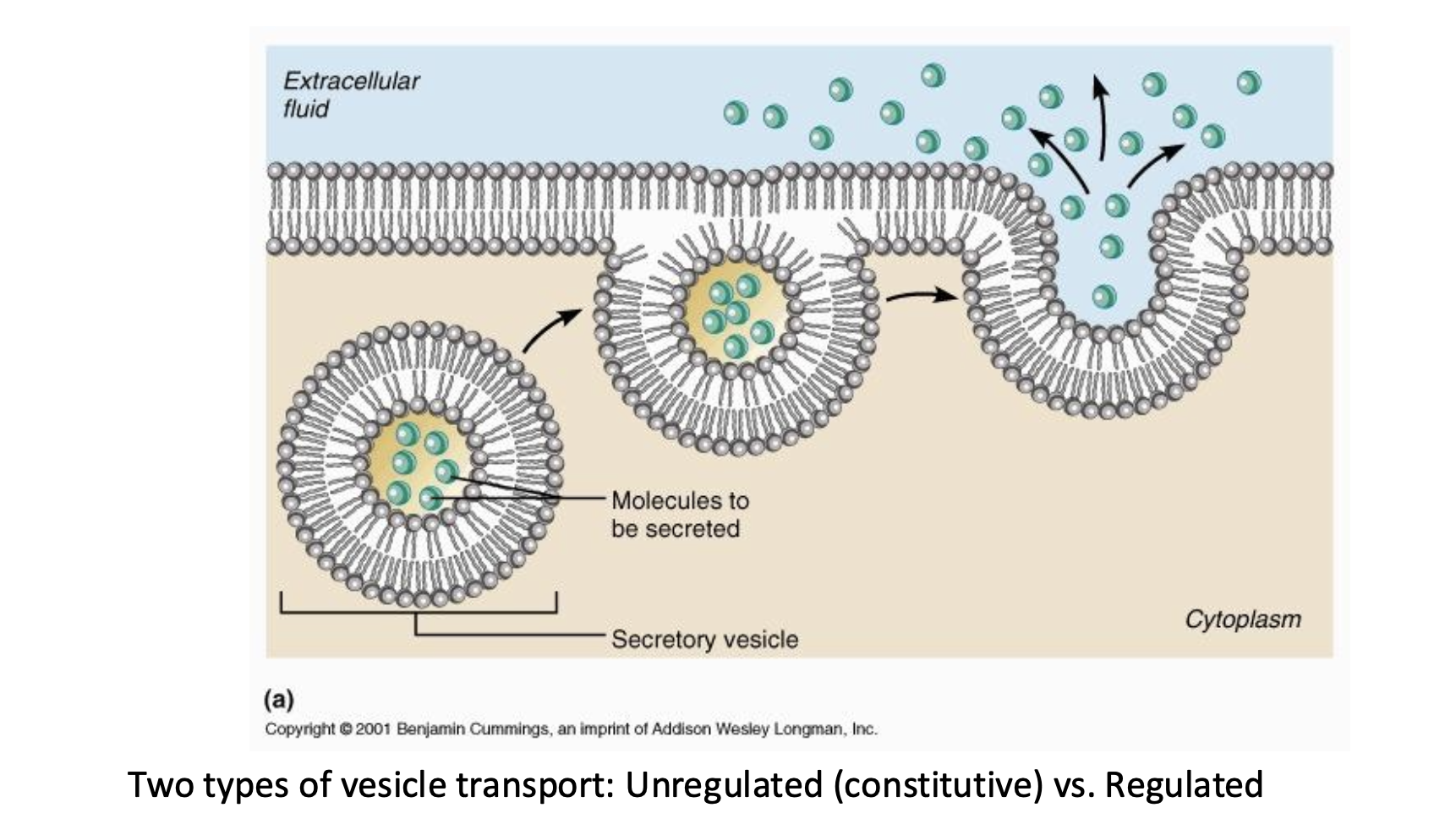
Secretory Pathway
funt: move materials throughout to the PM or to the cell exterior
- a way to get things out eukaryotic cells
- some type of signal or hormone that releases this
- generally coated in the phosphlipid bilyer vesicles
WE DO NOT OBSERVED IN PROKARYOTES
prokaryotes will have some type o channels instead of secretory vesicles
- regulated requires a signal
triggered/controlled
- unregulated does not require signal
continuous
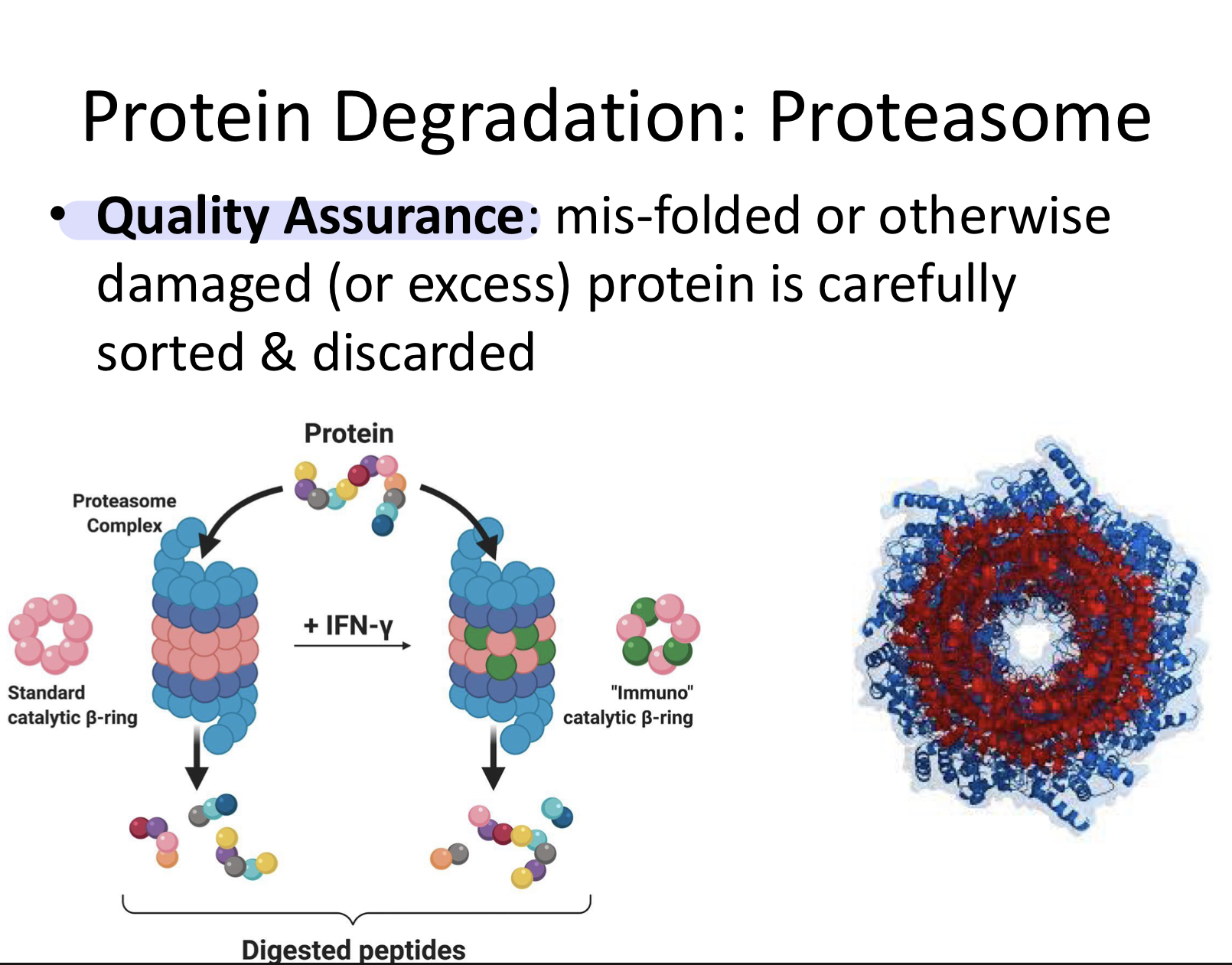
Proteasome
macromolecule
Quality Assurance: misfolded or otherwise damaged (or excess) protein is carefully stored and discarded
basically proteosomes destroy misfolded proteins
Difference btwn prokaryotes and eukaryotes
No phospholipid bilayer membrane compartments in prokaryotic cells
But might have single layer of phospholipid coating
Prokaryotic cells: dont have internal membrane system
Stuck with diffusion or transport across membrane
What do molecule chaperones do
they help proteins fold correctly
What does proteasomal degradation of proteins look like in Pro vs Euk
Pro
Pup proteosome system
PUP
single pup
pupylation
Euk
ubiquitin proteasome system
Poly-Ub
polyubiquitination: protesomeal degredation
ubiquitin links
monoubiquination: transport, signaling, DNA repair
What was the missing link
promodial escort proteins
escort systems important for allowing the plasma membrane to form internal phospholipid bilayer membranes
found in the archea lokiearcheota
Eukaryotic Glycocalyx
outermost boundary of the cell composed of polysaccharides
funct: adherence to surfaces and other cells w/in a biofilm protection
Eukaryotic Plasma Membrane and Cell Wall
PM:
phospholipid bilayer
contains a high proportion of sphingolipids and sterols (cholesterol) for strength and rigidity
selective permeable barrier
CW:
found in fungi and algae
unlike prokaryotes, generally composed of chitin, cellulose or sillica
Sterols(cholesterol) in which biphospholipid membrane
eukaryotic cells
these cholesterol rich regions are a structured system but also regulate exo/endocytosis signal transduction
What are lipid rafts
cholesterol congregates in specalized micodomains
these cholesterol rich microdomains are extremely important for endocyotosis
play critical roles in cell signaling, trafficking, and membrane organization.
What are the proteins called that allow for internalization
escort proteins
allow for channels, secretion
protein with short transmembrane domain cannot enter lipid raft
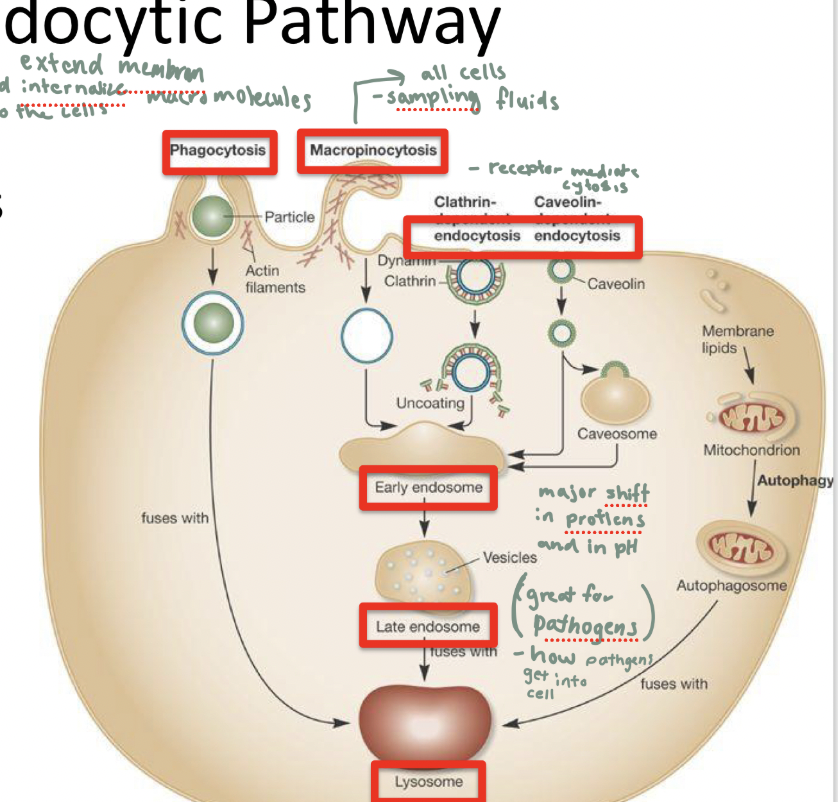
endocytic pathway
observed in all eukaryotic cells
funct: to bring materials into the cell from the outside
endocytosis is nonspecific but ligand specific
what are diff types of endocytic pathways
phagocytosis: extends membrane and internalizes macromolecules into the cells
solid particles
macropinocytosis: sampling fluids (EC fluid)
all cells
clathrin and caveolin endocytocis: receptor mediated cytosis
All pathways endup in a degradative lysosome
pathways 2 and 3 turn into early endosomes which turn into late endosomes and then lysosomes
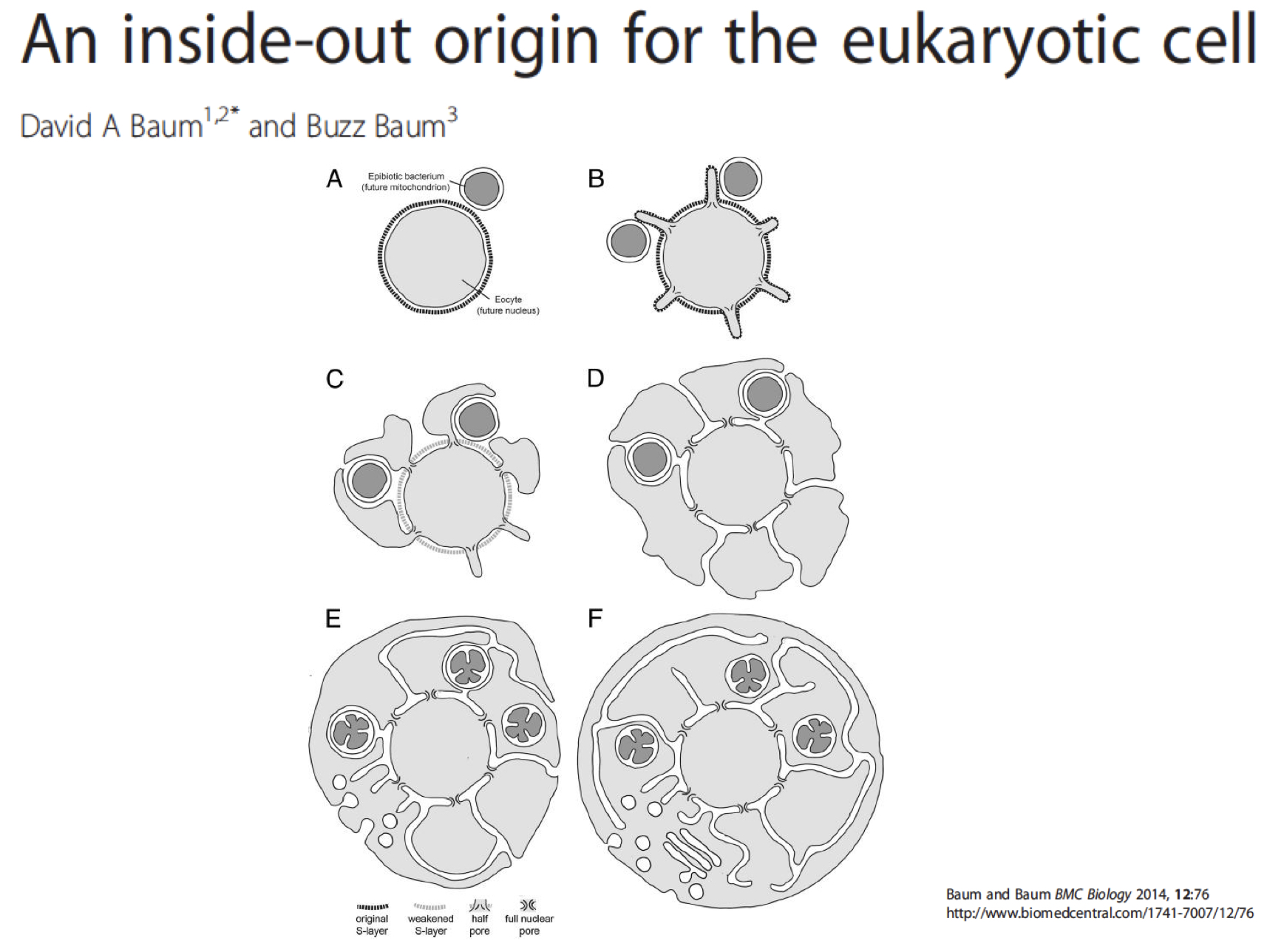
The inside out origin
theory on how the nucleus is acquired: the inside out origin for the eukaryotic ell (typically outside in thought process)
membrane blebs surrond symbiotic prokaryotes such as rickettsia ans through blebs, were able to make phospholipid bilayer needed for the nucleus
nucleus is an extremely protected struture
Prokaryotic Nucleoid
a singular, circular dsDNA
The nucleoid is a non-membrane-bound region in prokaryotic cells (like bacteria) that contains the cell’s DNA.
Unlike eukaryotic nuclei, the nucleoid:
Has no nuclear membrane
Is not surrounded by a nuclear envelope
Is sometimes referred to as “naked DNA”
prokaryotes have histone like homologues called NAP nucleoid-associated proteins
Even though it’s “naked” (no membrane), the DNA is tightly packed and organized: Supercoiling
which twists the DNA into tighter structures.
Eukaryotic Nuclear Envelope
two membranes both consisting of lipid bilayers
nuclear pore complexes allow for transport in and out of the nucleus
requires signals to get in and out
chromosomes released by ER to get packaged and shipped out
have protective endomembrane structure
The outer membrane connects to the ER — allowing for communication and material flow between the nucleus and the rest of the cell.
funct: Protects genetic material, regulates gene expression access
Eukaryotic Nucleus
the repository of genetic info
linear chromosomes: composed of condensed chromatin a complex of DNA, RNA and proteins
DNA is wrapped around histones (protein) in a bead on string format, yielding multiple nucleosomes
1. DNA wraps around histones
2. Nucleosomes coil into chromatin fibers
3. Higher-order folding
4. Supercoiling
plasmids (only prokaryotes, not eukaryotes)
prok. cells carry extra copies of this
not required for replication, recommended not needed
Structure: Small, circular double-stranded DNA molecule
function:
autonomous replication: make multiple copies
confers selective advantages: then shares it w/ others w/o losing it
Impact on host cell:
provides selective advantages
can have a metabolic burden
Plasmid shared via conjugation
"Shared via conjugation" means that a plasmid can be transferred from one bacterium to another through direct contact, allowing rapid spread of useful traits like antibiotic resistance
Typically carries ~30 genes, most of which provide a selective advantage (e.g., antibiotic resistance, virulence, or metabolic functions).
Plasmid vs. Episome vs Vector
episome: integrates w/ genome
goes in and out of cell
all episomes are plasmids but not vv
called vectors in lab
Definition
Plasmid = Independent DNA circle in the cytoplasm
Episome = Special plasmid that can also insert itself into the chromosomal DNA
Vector = A lab-modified plasmid (or virus) used to carry and express foreign DNA in cells.
Mitochondria
semi-independant organelle
funct: energy production and sysnthesis
matrix: DNA (circular) and ribosomes 70S
reproduce by binary fission
Outer Membrane | Smooth and porous; allows small molecules and ions to pass through freely. Contains porins. |
Intermembrane Space | Space between the outer and inner membranes; important for proton gradient in ETC. |
Inner Membrane | Highly folded into cristae to increase surface area; contains ETC complexes and ATP synthase. Selectively permeable. |
Matrix | Innermost compartment; contains enzymes for the Krebs cycle, mitochondrial DNA, and ribosomes |
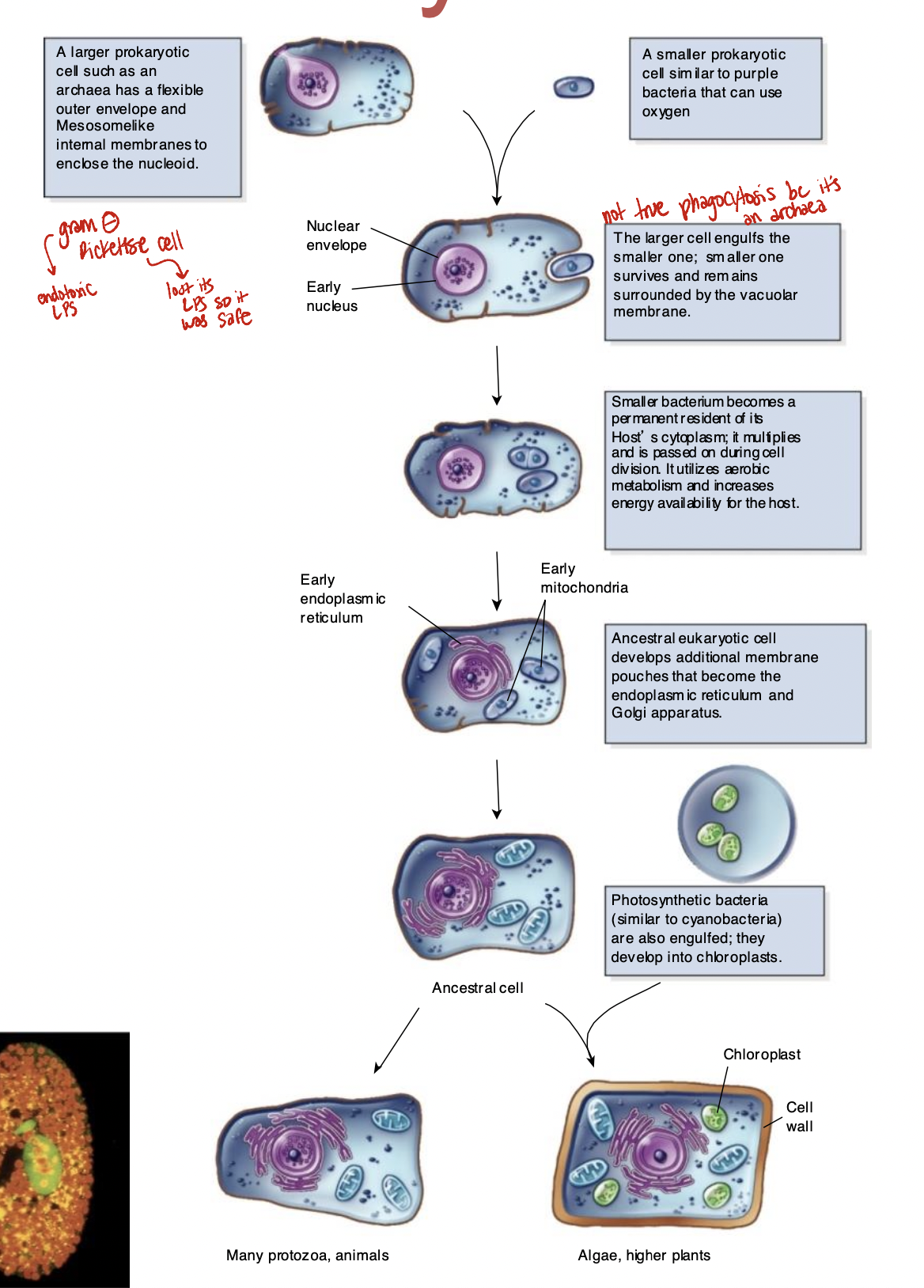
Endosymbiotic Theory
2 billion years ago
evolution from prokaryotic organisms by symbiosis
organelles from prokaryotic cells trapped inside them
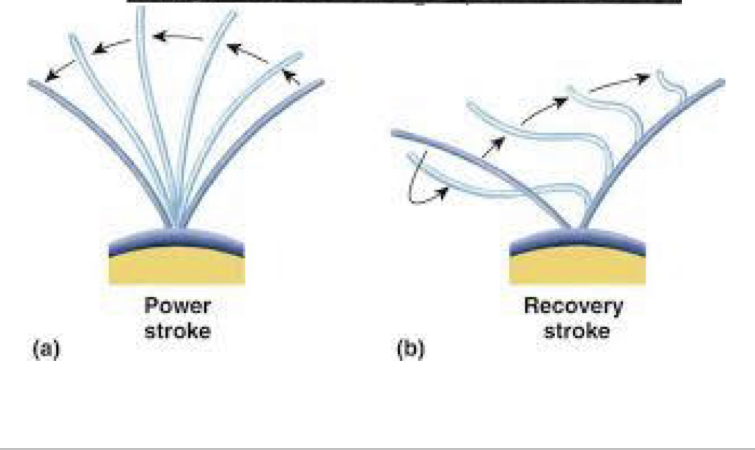
Eukaryotic Cilia
used for locomotion
composed of mircotubles
2 phase stroke: effective (power) stroke and recovery stroke
Eukaryotic Flagella
fewer flagella/organism
longer than cilia
movement is in an undulating motion (base to tip or tip to base)
ultrastructure
membrane bound cylinders
microtubles in a 9+2 pattern
movement and bending is achieved by doublet dynein arms
movement of a dynein motor protein on microtubles
can move both ways up and down
walking motion
Essential Macronutrients
6 essential
CHNOPS
carbon, hydrogen, nitrogen, oxygen, phosphorus, sulfur
CHO
components of important macromolecules
backbone of many biological macromolecules
Elemental makeup of important cell molecules
Proteins —> CHONS
Lipids —> CHOP
Carbohydrates —> CHOP
Nucleic Acids —> CHONP
Why are electrons important
electrons are vehicles for deriving cellular energy, by two important mechanisms:
oxidation-reduction reactions of biomolecules
via the movement of electrons along electron transport chains
classification of microorganisms
Carbon Source: Heterotrophs and Autotrophs
Energy Source: phototrophs and chemotrophs
Electron Source: lithotrophs and organotrophs
Major Nutritional Types of Microorganisms
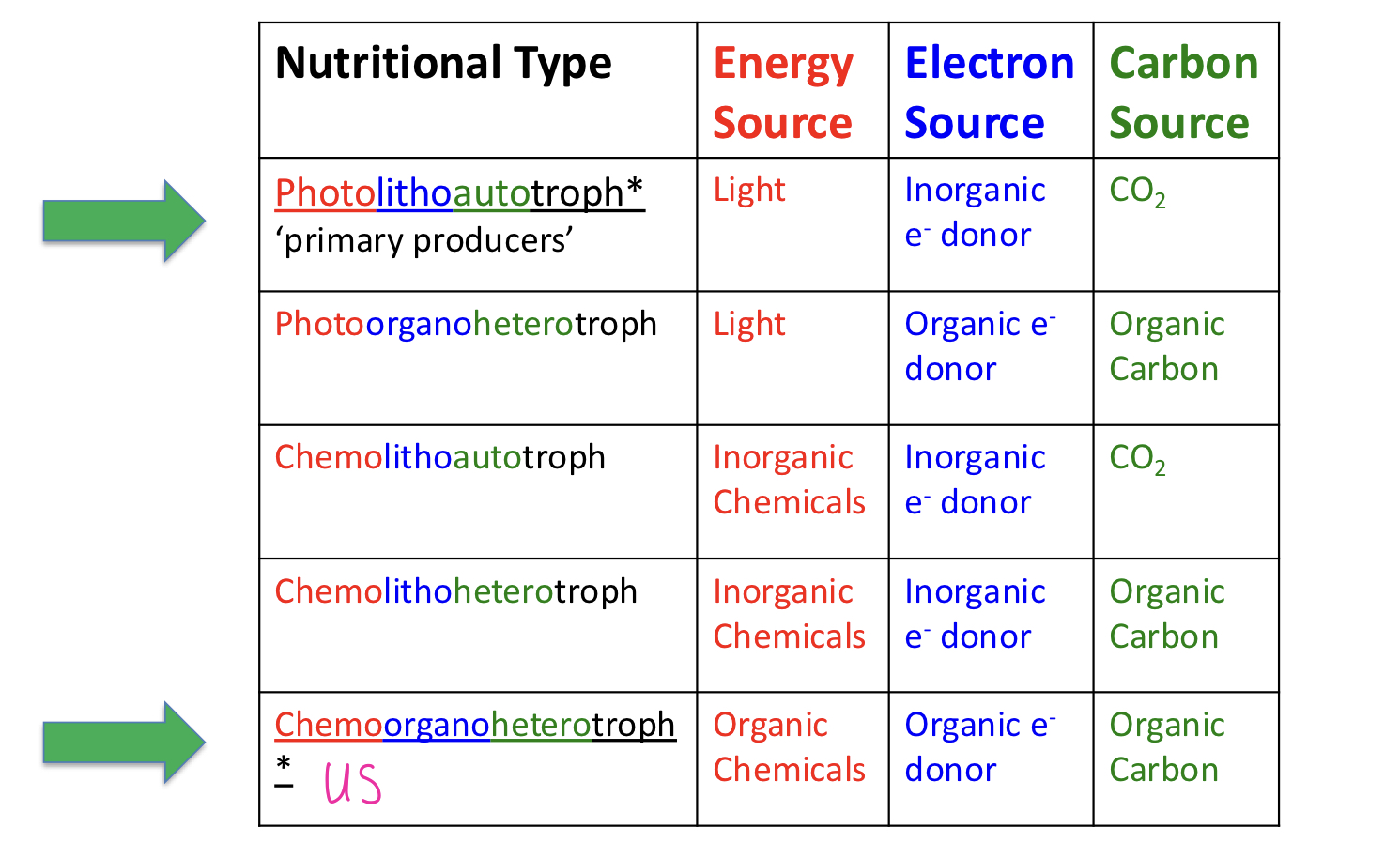
most common *
mutualistic
microorganisms that inhabit a host, yet provide a mutually beneficial element for survival of the host
ex: pineseedling mycorrhizal roots from one tree spread to innoculate other tree roots
commensal microorganisms
microorganisms that derive a benefit from a host, however they may not help nor harm the host
microorganisms that have co-evolved w/ their host (our epithelial body surfaces)
parasitic microorganisms
a microorganism that inhabit a host, derives nutrients and in the process harms the host
pathogens
a parasite that can cause severe harm and death to a host
exceptions are when a commensal/mutulastic bacteria that become opportunistic
saprobic microorganism
organisms that decompose organic matter
generally secrete enzymes to the environment
immobilization
nutrients converted into biomass become tied-up and are unavailable for nutrient cycling
Nitrogen fixation discovered by…
fritz haber
carl bosch
N2 fixation —> NH3
Nitrogen Fixation
requires the synthesis of nitrogenase (enzyme inactivated by oxygen)
organisms that nitrogen fix
rhizobium spp. (uses legume nodule)
bacteria —> infection thread —> early nodule
inside nodules they nitrogen using nitrogenase but they need low oxygen conditions
need a compartment to fix nitrogen
clostridium and azotobacter spp. (free-living soil bacteria)
enviornment is already anerobic
cyanobacteria (heterocysts)
heterocysts: thick walled compartments where nitrogenase converts N2 to NH3 (uses energy)
they also block oxygen
found in water
What is nitrogen fixation
the process of converting atmospheric nitrogen gas (N2) into ammonia (NH3), which plants and microbes can use
done by certain microorganisms not by plants of animals
enzyme that perform this task are called nitrogenous
but its inactivated by oxygen
this is why bacteria need special environments to use it
nitrogenase requires energy
N2 —> NH3
mineralization (decomposition)
the process by which organic matter is decomposed and released as simple, inorganic compounds
aerobic and anaerobic bacteria, fungi
recovering sequestered organic nitrogen into Ammonium (NH4+)
organic N (aa or na) —> NH4+
aka: Ammonification
nitrification
a 2 step process that us carried out by two diff genus of bacteria
step 1: NH4+ —> NO2- carried out by nitrosomonas
step 2: NO2- —> NO3- carried out by nitrobacter
long membrane tublues: generate a H+ gradient from the oxidation of Ammonium
assimilatory reduction
taking (NH4+) into a readily usable oxidized nitrate into usable oxidized (NO3-)
assimilatory reduction
incorporation of an oxidized (useable) form of molecule into microbial biomass via reduction
NO₃⁻→NO₂⁻→NH₄⁺→Amino acids
consumes energy
Build proteins, nucleic acids, etc.
reduction of nitrate (NO3-)
denitrification
uses dissimilatory reduction
occurs in many bacteria
dissimilatory reduction: NO3- serves as a terminal electron acceptor during anaerobic respiration; when gases N2 and N2O are produced and released into atmosphere, process called denitrification
N2O —> N2
DR:
NO3- —> N2
NO3- —> N2O
NO3- —> NO2-
wastewater
any water that has been adversely affected by humans
sewage
subset of water contaminated by feces or urine
whitewater
potable water
greywater
household water that has no fecal matter
blackwater
fecal contaminated water
goals of wastewater treatment
return to then environment free of pathogens
decrease the load of nutrients (carbon and nitrogen)
the goal is NOT to clean wastewater to drinking water levels
oxygen depletion
Oxygen depletion in water refers to a reduction in the amount of dissolved oxygen (DO) available in a body of water
caused by things like thermal pollution, runoff, sewage pollution, algae growth
Waste Water Treatment Stages
Headworks: removes bulk inorganic solid
Primary: removal of bulk organic solid material
Secondary: removal of dissolved organic matter by conversion into microbial biomass and CO2 (activated sludge)
Tertiary: Final purification and removal of : nitrogen and phosphorous compounds, heavy metals, biodegradable organics and remaining microbes, including viruses
Primary Treatment
Screening: use of fine screens to remove smaller debris
Sedimentation/clarification: large, still tanks of water allow for removal of about 90% of settleable solids, about 55% suspended solid removed (sludge) oil and grease skimmed from top
Secondary treatment
*nutrient removal
aeration basin: O2 and activated sludge is mixed w/ clarified water (mixed liquor)
allows aerobic bacteria to grow and remove organic compounds
2ary treatment reduces dissolved organics available to microorganisms by about 90-95%
activated sludge
first described in the 1914 by E. Ardern and W.T Locket (great britain)
activated sludge flocs are complex system composed of microbial cells and minerals in a polymeric matrix
saprophytic bacteria
archea
protoza: amoebae, spirotrichs, peritrichs
rotifers
a fraction of the activated sludge is returned to the aeration basin. Excess is removed for further treatment
healthy flocs are dense, large particulate that settle during sedimentation
healthy sludge must be maintained and can be disrupted by influent contaminants:
toxins, medications, antifreeze, floods
Sludge bulking
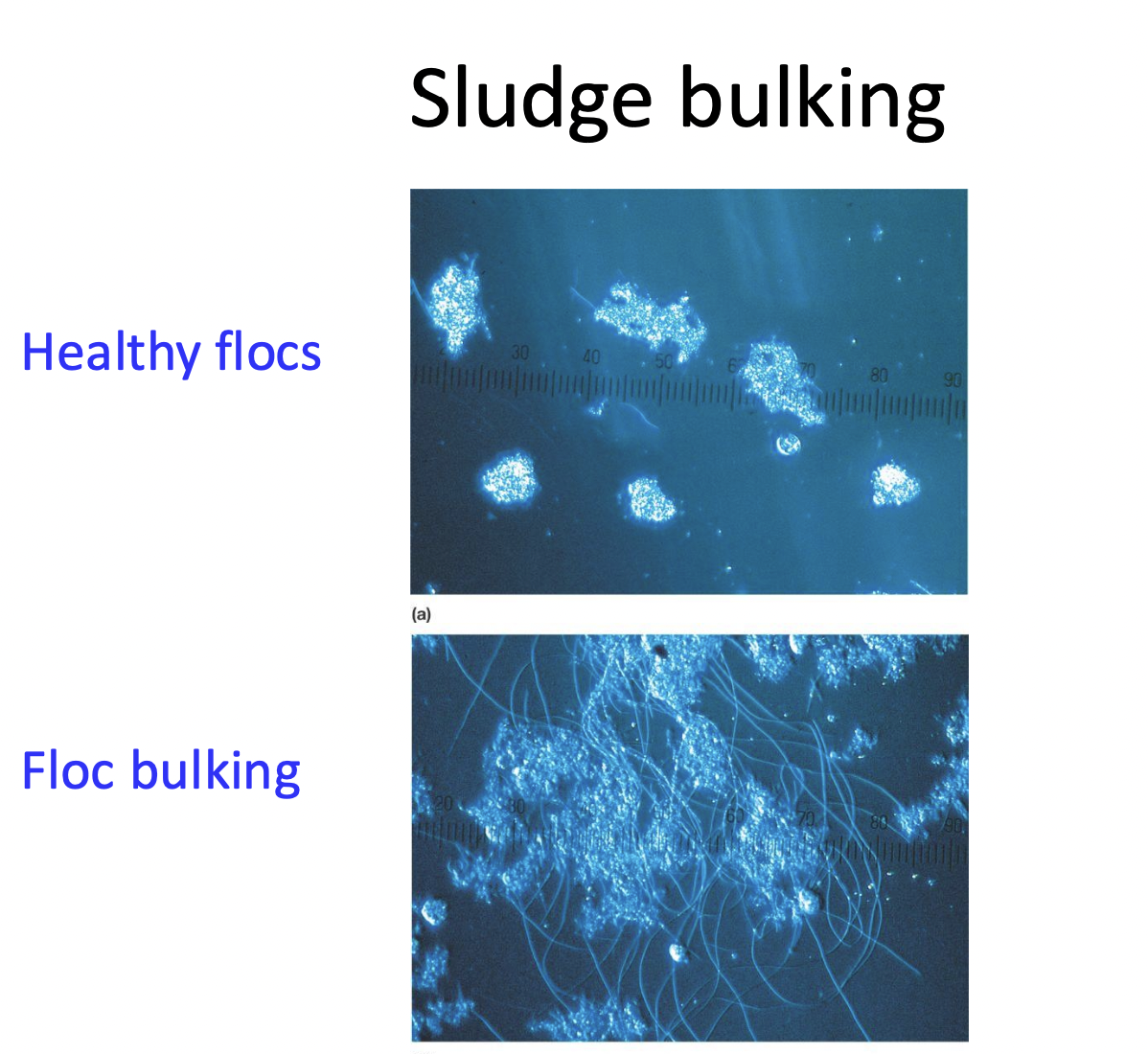
the failure of sludge to seperate during sedimentation
generally a result of poor aeration, inappropriate microbial communities in the activated sludge. Overgrowth of filamentous bacteria: Sphaerotilus, Thiothrix
filamentous bacteria have a greater volume and surface areas, resulting in open/porus flocs that dont settle
Secondary Treatment
*microbial removal
secondary sedimentation/clarification: same as primary, large particulate settle are removed
other secondary treatment: trickling filter
Tertiary treatment
wetlands as tertiary treatment

Sludge Treatment
reduce pathogens, odors and lower concentrations of metals
Step 1: Dewatering (centrifuge)
Step 2: Anaerobic digestion
Step 3: final dehydration and removal
Anaerobic Digestion
fermentation: big molecules to small molecules (generally anerobic)
acetogenic: makes acetate, H2, CO2 (takes another week)
Methanogenic reaction: archea breaks into single carbon molecules
what is a dehydrater
pressing the remainder of waste
Baby blue syndrome
when there are inc levels of nitrates in water met hemoglobin goes up
methemoglobin (ferric)
does not bind to oxygen as well
3 bound oxygens instead of 4 bound oxygens
Methemoglobin cannot carry oxygen, leading to tissue hypoxia (lack of oxygen)
adults have the enzyme cytochrome b reductase that converts it back but babies dont have it
energy
the ability to do work
displayed in many forms and is transferable, thus converted in form
thermodynamics is the study of energy
major types of energy
thermal energy (heat)
rediant energy (waves)
electrical energy (flow of e-)
mechanical energy (physical change in position)
atomic energy (stored energy in an atomic nucleus)
chemical energy (stored energy in the bonds of molecules
what we focus on
catabolism
the process and pathways by which larger molecules are broken down and in turn release energy
anabolism
the process by which larger molecules are built from smaller units, using energy in the process
exergonic reactions
Products have less free energy than reactants
energy released is used by the cell
spontaneous
endergonic
Products have more free energy than reactants
energy is supplied (enters) for reaction to occur
nonspontaneous
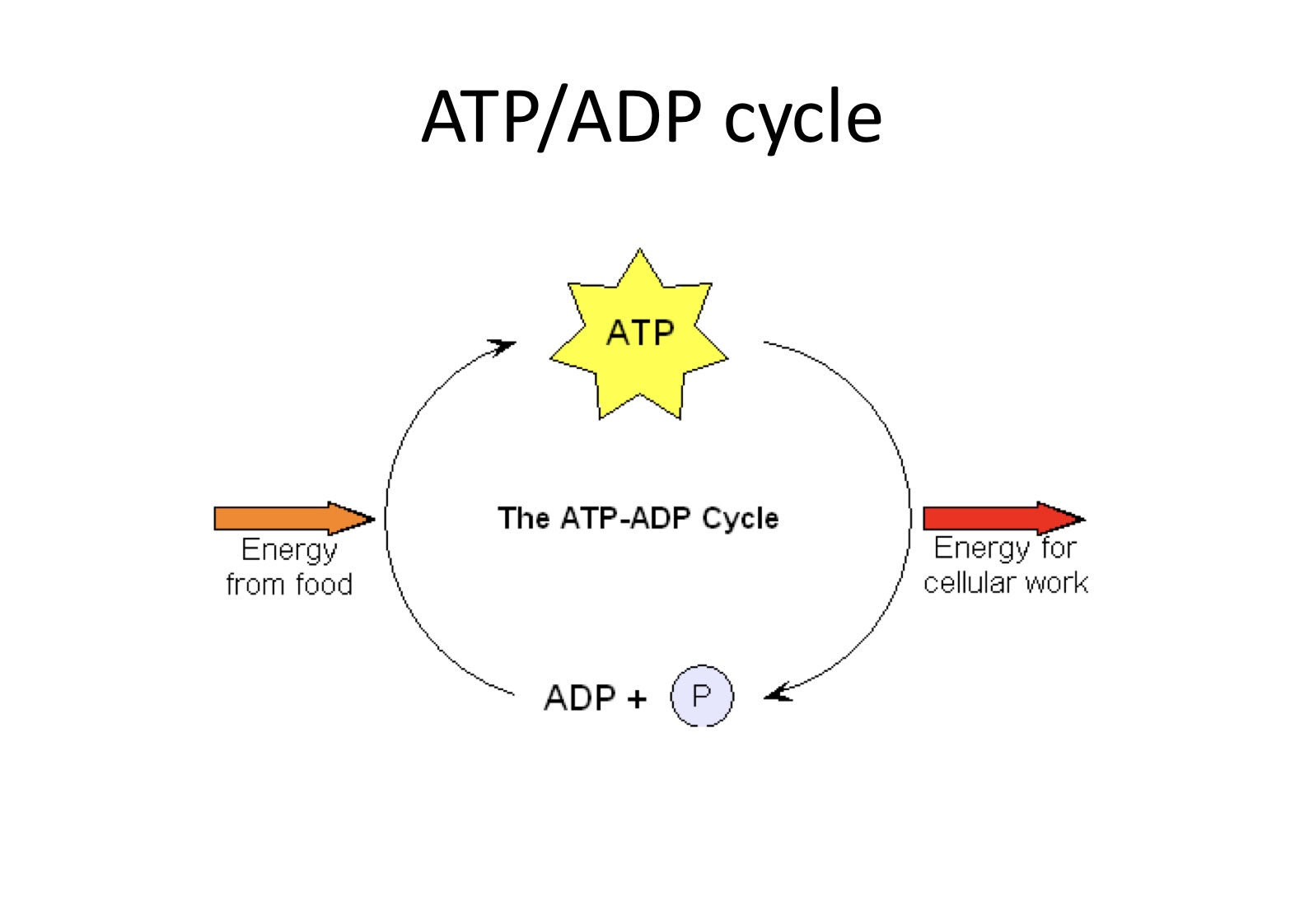
Why is ATP important
ATP: cellular energy storage to carry out metabolic reactions
ATP: is a high-energy molecule
ATP: almost completely hydrlyzes to ADP
ATP/ADP cycle
Other important cellular energy storage
GTP
NADH
NADPH
FADH2
they all have a similar backbone
adenine or gaunine
NAD+
gets reduced to NADH
they are limiting reagent (especially NAD+)
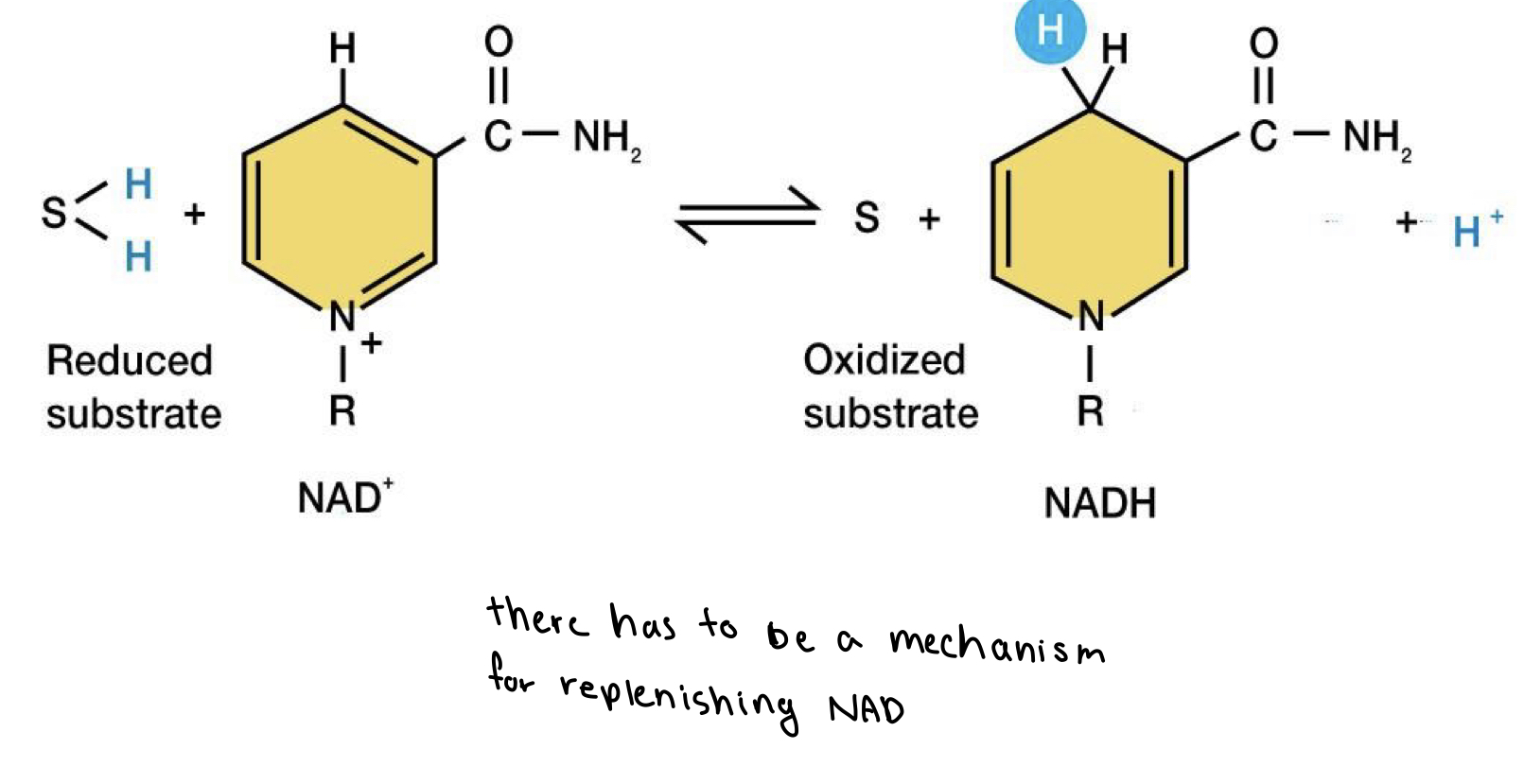
recycling NAD+ is much faster and efficient compared to de-novo biosynthesis (generating from scratch)
common monosaccharides
glucose
fructose
galactose
glucose is the most common
Glycolysis
breaking sugar
taking a high energy potential molecule and breaking it to get that energy
glucose is a good e donor
breaking glucose= few ATP and a lot of potential energy
NAD+ is a limiting agent
yields 2 pyruvate
what is required: ATP, NAD+, glucose, enzymes
Fates of Pyruvate
anaerobic respiration: nonoxygen electron acceptors (ex: SO4²-, NO3-, CO3²-, )
aerobic respiration: O2 is final electron acceptor
fermentation: an organic molecules is final electron acceptor (pyruvate, acetaldehyde, etc.)
yields the least amount of ATP
Exactly the same till the last step
aerobic and anaerobic are exactly the same till the last step (different electron donor)
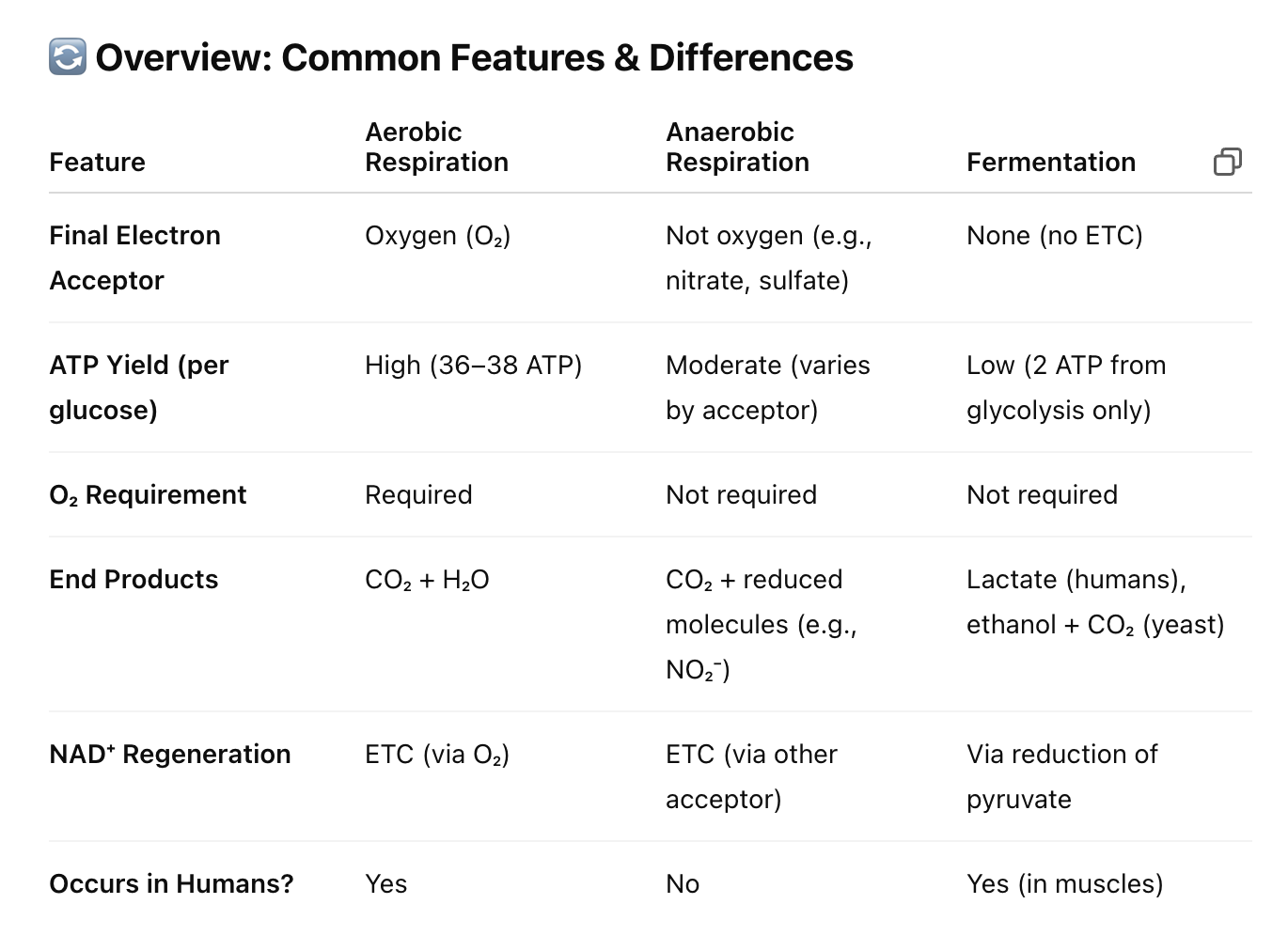
Common features and differences (aerobic respiration, anaerobic respiration, and fermentation
In aerobic respiration there are 4 complexes and O2 is the final e- acceptor making water
yields about > 30 ATPs
In anaerobic respiration (obligate anaerobes)
complex 4’s are different
instead of oxygen: Nitrate, Sulfate, Fumarate are the final electron acceptors
Fermentation (most different)
for glycolysis to continue, a limiting reagent must be regenerated (NAD)
uses pyruvate as an electron acceptor in the absence of the ETC
this is what maintains glycolysis
is diverse (soy sauce, vinegar, cheese)
all cells; but not all cells carryout for a long time
locations of pathways in Eukaryotes
External to mitochondrion
glycolysis
fermentation
Inside mitochondrion inner membrane
respiratory chain
Matrix
citric acid cycle
Locations of pathways in Prokaryotes
Cytoplasm
glycolysis
fermentation
citric acid cycle
Inner Face of Plasma Membrane
respiratory chain
Where is the ETC located
Prokaryotes: along the plasma membrane
Eukaryotes: inner membrane of the mitochondria
H+ concentration gradient role of ATP synthase
complexes accept electrons gaining energy to pump H+ which creates a high concentration gradient in the inter membrane space and a low concentration gradient in the mitochondrial matrix
this concentration gradient powers ATP synthase allowing it to generated many ATP molecules
Describe H+ concentration gradient (proton motive force/stored potential energy) and role of ATP synthase
the proton motive force causes protons to diffuse back into the mitochondrial matrix through the membrane channel protein ATP synthase
Aerobic vs. anaerobic fermentation (Pasteur vs Crabtree effect)
Aerobic Fermentation
Crabtree Effect
abnormal but an increase in sugar concentration pushes the pathway through fermentation to make ethanol
seen in organisms such as yeast
Anaerobic Fermentation
Pasteur Effect
if O2 is present, most organisms use aerobic respiration
Four Important Observations about Fermentation
NADH is oxidized to NAD+
The electron acceptor is pyruvate or pyruvate derivative (Fermentation, ETC are respiration)
An ETC cannot operate = decreased ATP yield (in general not always)
O2 generally not present (where pasteur vs crabtree comes into play)
What must pyruvate be modified to, to enter the TCA cycle?
pyruvate —> acetyl CoA
Binary Fission
one cell divides into two cells
specific to prokaryotes and archaea
asexual reproduction and cell division
rapid process about 20 min @ RT (not for all microbes)
some eukaryotes (some yeast) and eukaryotic organelles (mitochondria) divide by binary fission