Microbiology Exam 5
1/97
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
98 Terms
genotype
the nucleotide sequence of the genome
phenotype
the physical properties of the organism
mutation
a heritable change in the base sequence of the genome
wild-type strain
the strain isolated from nature
mutant
a strain carrying a change in its nucleotide sequence
mutant phenotype
an altered phenotype relative to the parental strain
selectable mutations
mutant has an advantage under certain environmental conditions and will outcompete and replace the parent (wild type)
non-selectable mutations
mutation does not provide an advantage so it will remain rare in the population
selectable marker gene
intentionally add certain gene to acquire certain trait for easy screening of mutants
point mutations
chemical changes in just one base pair of a gene (silent, nonsense, or missense)
silent mutation
does not affect amino acid sequence
nonsense mutation
a codon becomes a stop codon; short and incomplete polypeptide
missense mutation
an amino acid changed; altered polypeptide
frameshift mutations
mutation that shifts the "reading" frame of the genetic message by inserting or deleting a nucleotide
-DISASTROUS effect
back mutation (reversion)
when a mutated gene reverses to its original base composition (original sequence is restored)
-often occur in point mutations
mutagenesis
the production of mutations; usually very low rate in spontaneous mutations so process can be sped up
ames test
simple test to screen chemicals for potential mutagenicity
-screen for an increase in the rate of back mutations in auxotrophic bacteria in the presence of the chemical disc
transversions
a purine and a pyrimidine are interchanged
transitions
a purine or pyrimidine is swapped for the other (A to G, G to A, C to T, or T to C)
transferring genetic material
two ways to do it:
-vertical gene transfer (VGT)
-horizontal (lateral) gene transfer (HGT or LGT)
genetic exchange in prokaryotes
3 mechanisms of genetic exchange:
1. transformation
2. transduction
3. conjugation
transformation
free DNA is incorporated into a recipient cell and brings about genetic change
transduction
a bacterial virus (bacteriophage) transfers DNA from one cell to another
conjugation
genetic transfer from a donor cell to a recipient cell requiring cell-to-cell contact (mating)
rolling circle replication
transfer plasmid DNA from donor to recipient cell (happens during conjugation); some viruses also use the same mechanism
F (fertility) plasmid
circular DNA that contains genes that regulate DNA replication and contains several transposable elements
-contains tra genes that encode transfer functions
-is an episome
F-
cell without F plasmid
F+
cell with non-integrated F plasmid
HFr (high frequency og recombination)
cell with integrated F plasmid
-high rates of genetic recombination occurred between genes on the donor chromosome and those of the recipient
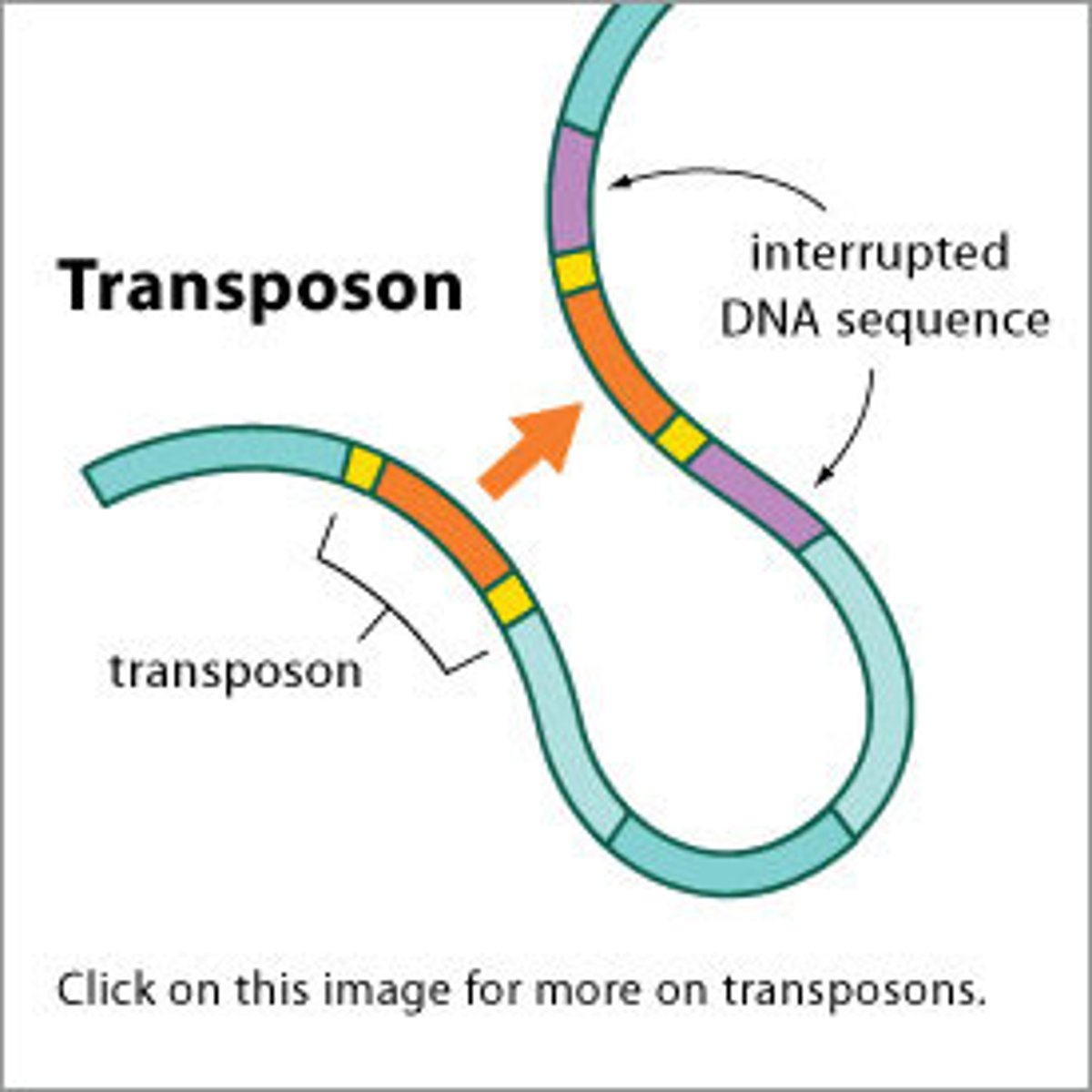
transposable elements
DNA fragment that move (or copy) as a unit from one location to another
-found in all 3 domains of life
-move by a process called transposition
-two main types in Bacteria: insertion sequence and transposons
transposition
movement of DNA segments within and between DNA molecules
insertion sequence
2 inverted repeats (IR) + transposase gene
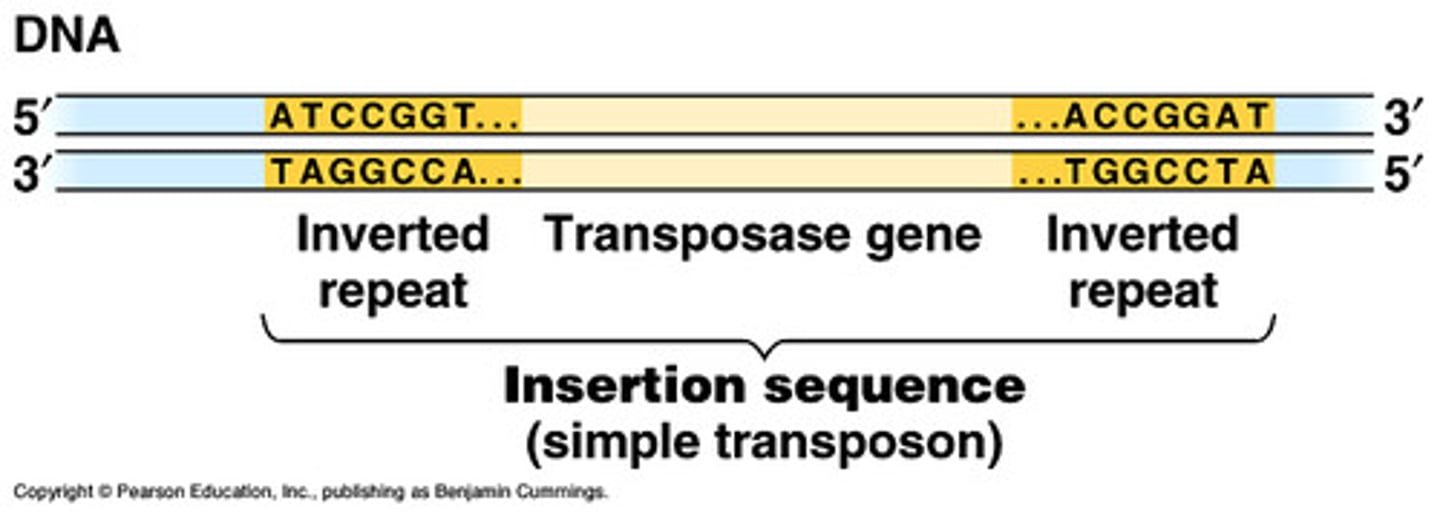
transposons
2 IR + transposase gene + other genes
(e.g. antibiotic resistant gene)
transposase gene
codes for enzyme that catalyzes transposition
restriction enzymes
recognize specific base sequences within DNA (recognition sequences) and cut the DNA at specific sites; common in prokaryotes
-type of defense mechanism against hostile foreign DNA
palindromes
inverted repeat sequences; used in restriction enzymes
type II restriction enzymes
cleavage site located within the recognition sequence; form three different ends
sticky (cohesive) ends
5'-overhang and 3'-overhang
blunt ends
cuts are straight through both DNA strands at the line of symmetry
CRISPR (clustered regularly interspaced short palindromic repeats)
segments of prokaryotic DNA containing short repetitions of base sequences; each repetition is followed by short segments of "spacer DNA" from previous exposures to a bacterial virus or plasmid
CRISPR-Cas9
binds to a target viral dsDNA and cuts both strands; essentially like magic scissors
gel electrophoresis
separate DNA fragments by their size and shape; the phosphodiester bond in DNA is - charged and it migrates towards + pole; fragments are stained for observation
-larger fragments move slowly
-compact shape move fast
nucleic acid hybridization
single stranded DNA or RNA can be used to screen for homologous sequences by utilizing complementary base pairing (hybridization)
procedure of hybridization
label known nucleic acid (DNA or RNA) with radioactivity or a fluorescent dye, hybridize probes with unknown samples, if probe sticks then this indicates a match in the sequence
southern blotting
hybridization technique where DNA in the gel is labeled with DNA or RNA probe
northern blotting
hybridization technique where RNA in the gel is labeled with DNA or RNA probe
western blotting
hybridization technique where protein in the gel is labeled with antibody
splicing
introns are removed and exons are joined by spliceosome
-spliceosome contains small nuclear ribonucleoproteins (snRNPs)
introns
junk DNA sequences
exons
expressed sequence of DNA; codes for a protein
reverse transcriptase
makes a DNA copy of RNA (cDNA); cDNA can be made from mRNA, tRNA, or rRna; provides a means of synthesizing eukaryotic genes from mRNA transcripts (synthesized gene is free of introns)
PCR (polymerase chain reaction)
method to amplify DNA; rapidly increases the amount of DNA in a sample
process of PCR
primers are known sequence are added, to indicate where amplification will begin, along with special heat tolerant DNA polymerase and nucleotides; repetitively cycled through denaturation, priming, and extension; each subsequence cycle DOUBLES the # of copies for analysis
denaturation, priming, and extension
otherwise known as melting, annealing, and synthesis; 3 basic steps of PCR
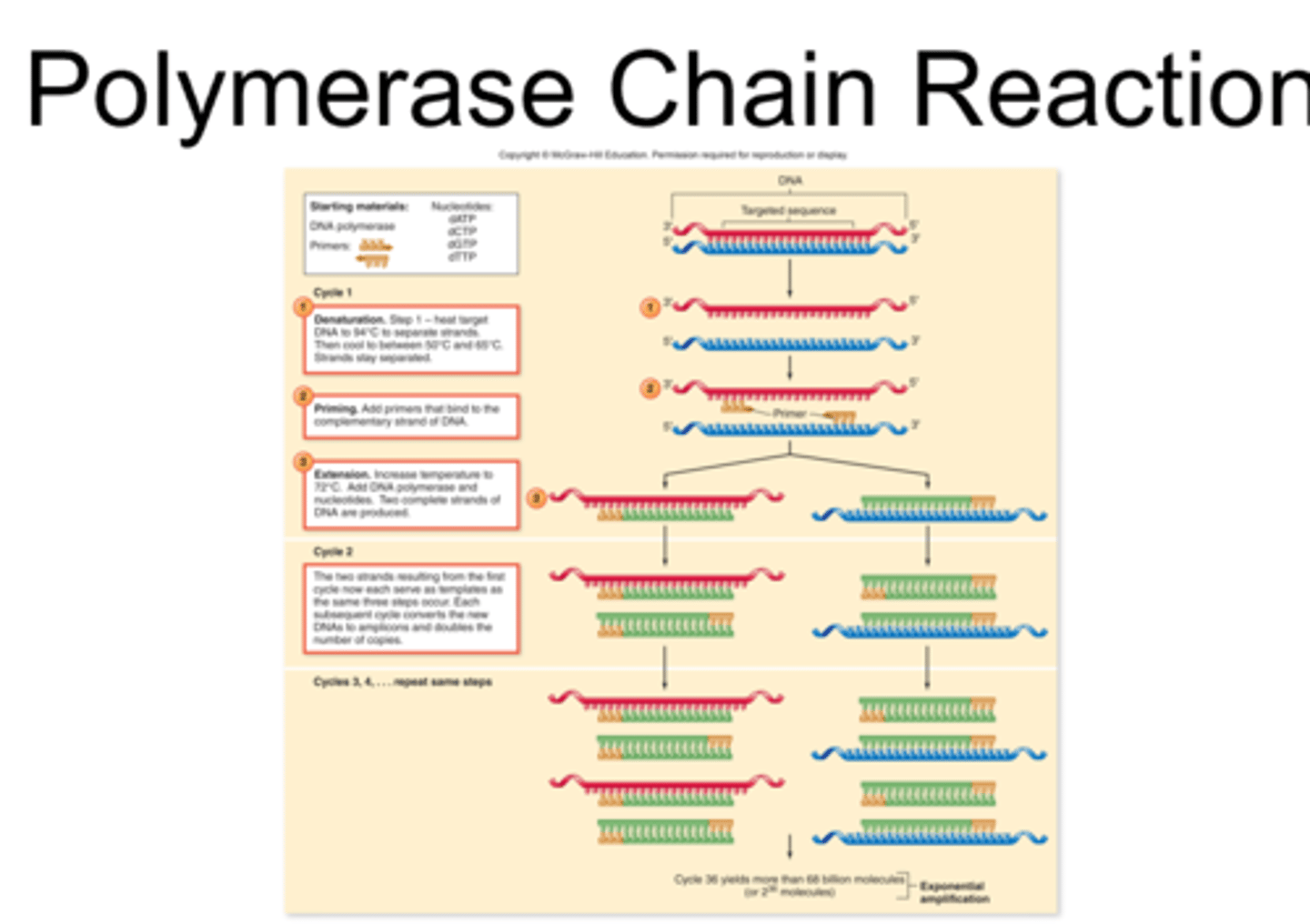
what is PCR used for?
gene mapping, studying genetic defects and cancer, forensics, taxonomy, and evolutionary studies
**amplifies certain strands of genes
shotgun sequencing
sequences are assembled by pairing their overlaps to generate the long, continuous sequence
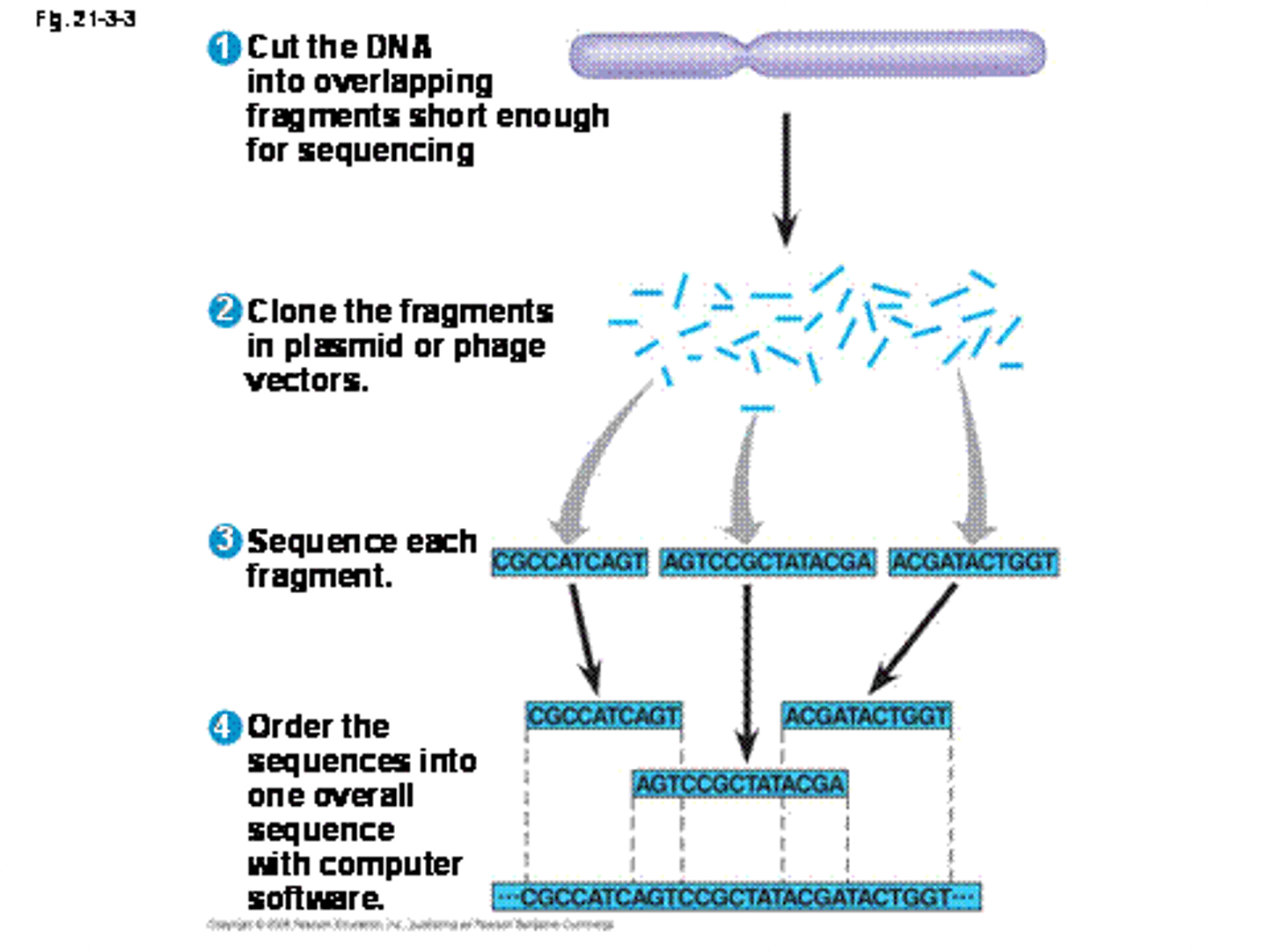
pyrosequencing
DNA sequencing technology that is used for whole-genome shotgun sequencing; fast and cost-effective analysis
procedure of pyrosequencing
1. DNA fragmentation
2. Separate and sequence small DNA fragments
3. Assemble pieces of DNA sequences together
4. Annotation; finding ORF, putative regulator sequences, etc.
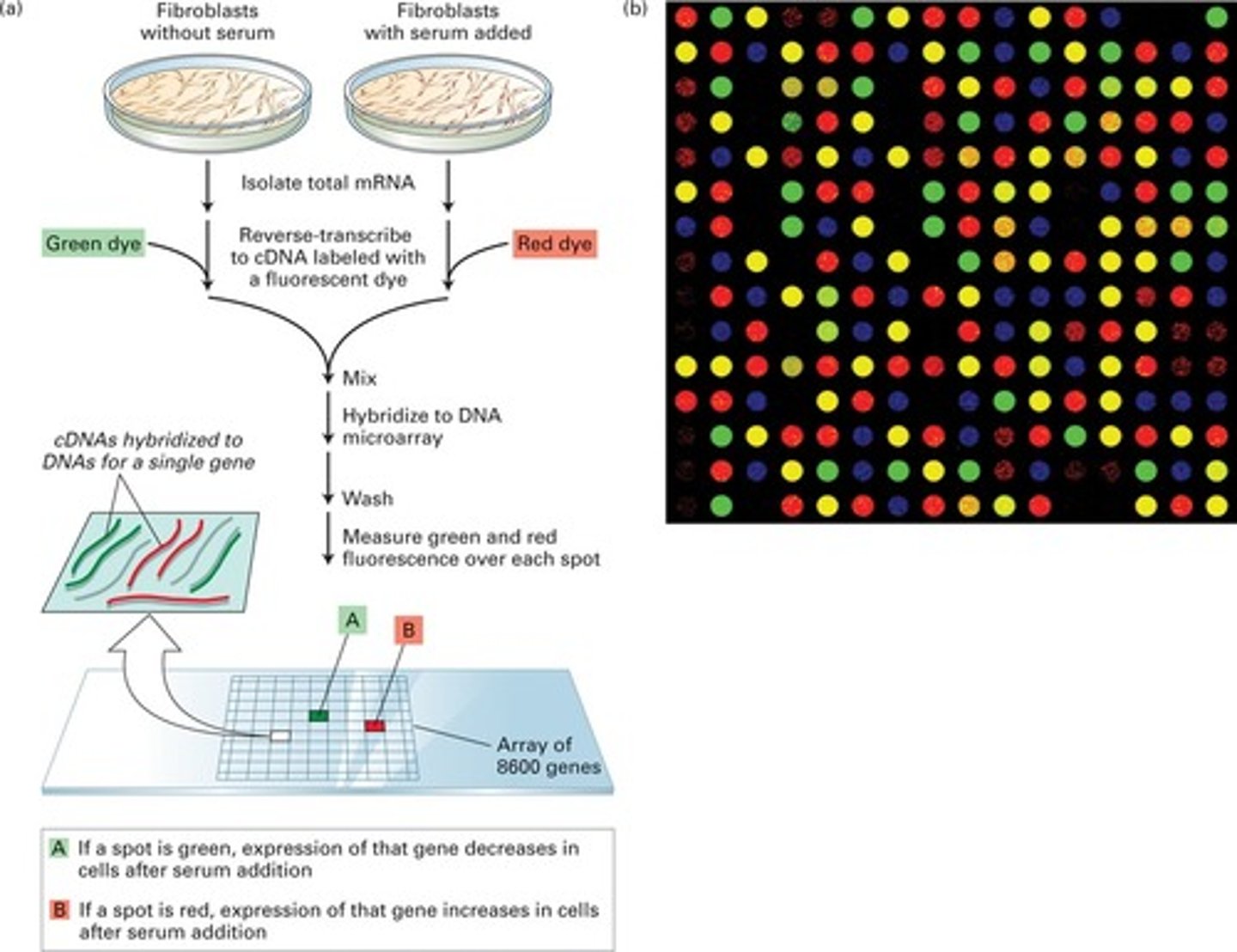
microarray
track the expression of thousands of genes; used to identify and devise treatments for diseases based on the genetic profile of the disease
plasmids as cloning vectors
requirements to use:
-small size; easy to isolate DNA
-independent origin of replication
-presence of selectable markers
-polylinker (multiple cloning site)
-vector transfer carried out normally by chemical transformation or electroporation
desirable features in cloning host:
1. fast growth rate
2. can be grown in large quantities using ordinary culture methods
3. nonpathogenic
4. genome that is well delineated
5. capable of accepting plasmid or bacteriophage vectors
6. maintains foreign genes through multiple generations
7. will secrete a high yield of proteins from expressed foreign genes
competence
capable of accepting plasmid or bacteriophage vectors
selectable markers
vector genes that make it possible to pick out cells harboring a recombinant DNA molecule constructed with that vector
shuttle vectors
vectors that are stably maintained in two or more unrelated host organisms; able to easily move genes between multiple hosts
-usually requires multiple selection markers for the different hosts
-usually requires multiple origin of replications for the different hosts
expression vectors
allow experimenter to control the expression of cloned genes; multiple cloning site is located immediately downstream of a promoter region
-allow high levels of protein expression
-promoter is repressed unless activated by an inducer
-strong transcription terminator sites prevent read-through
Ti plasmid vector
used to introduce foreign DNA into plants; contains genes that mobilize DNA for transfer to the plant; segment of the Ti plasmid that is transferred to the plant is called the T-DNA
Agrobacterium tumefacines
plant pathogen with the plasmid used in Ti plasmid vectors
crown gall
caused by Ti plasmid vectors

3 steps in gene cloning
1. Isolation and fragmentation of source DNA
2. Insertion of DNA fragment into cloning vector
3. Introduction of cloned DNA into host organism
isolation & fragmentation of source DNA
-isolation of source DNA (e.g. genomic DNA, cDNA from RNA, or PCR products)
-fragmentation of source DNA by restriction enzymes
***most vectors are derived from plasmids or viruses
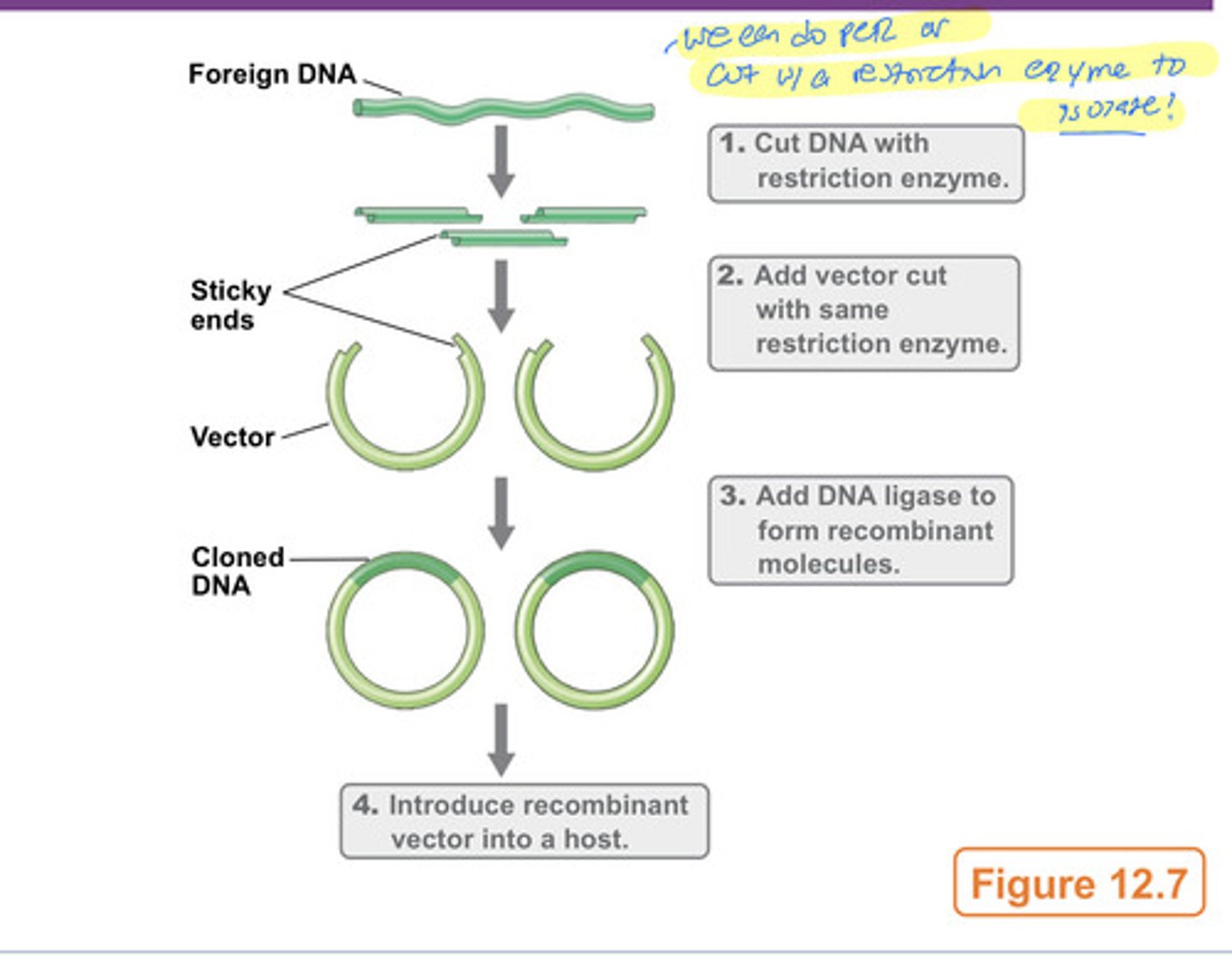
inserting DNA fragment into cloning vector
-DNA is generally inserted in vitro
-DNA ligase works with sticky or blunt ends
DNA ligase
enzyme that joins two DNA molecules (either with sticky or blunt ends)
introduction of cloned DNA into host organism
-transformation is often used to get recombinant DNA into host
-some cells will contain desired cloned gene, while other cells will have other cloned genes
electroporation
a technique to introduce recombinant DNA into cells by applying a brief electrical pulse to a solution containing the cells
lac Z gene in cloning
polylinker (multiple cloning site) inside gene that allows for screening
-no foreign DNA inserted = blue colonies
-foreign DNA inserted = white colonies (lacZ gene is inactivated)
inactivated lacZ gene
cannot process X-gal (galactose homolog) so no color develops during the screening process
barriers of expression in mammalian genes
-differences in codon usage
-presence of introns in eukaryotes
-degradation by host intracellular proteases
-toxicity to prokaryotic host
-formation of inclusion bodies
-improper protein folding
-lack of post-translational modification
advantages of using bacterial system
-quick replication (mass production)
-cheap production cost
codon usage
differences in use between prokaryotes and eukaryotes for codons
inclusion bodies
granules of storage material such as sulfur that accumulate within some bacterial cells
post-translational modification
changes made to polypeptides following translation (disulfide bridge or glycosylation)
industrial microbes
usually are fungi (yeast) or streptomyces (bacteria)
properties of useful industrial microbes
-high yield of desired products
-grow rapidly
-easy to inoculate
-large scale liquid culture
-inexpensive medium
-non-pathogenic
-amenable to genetic manipulation (genetic engineering)
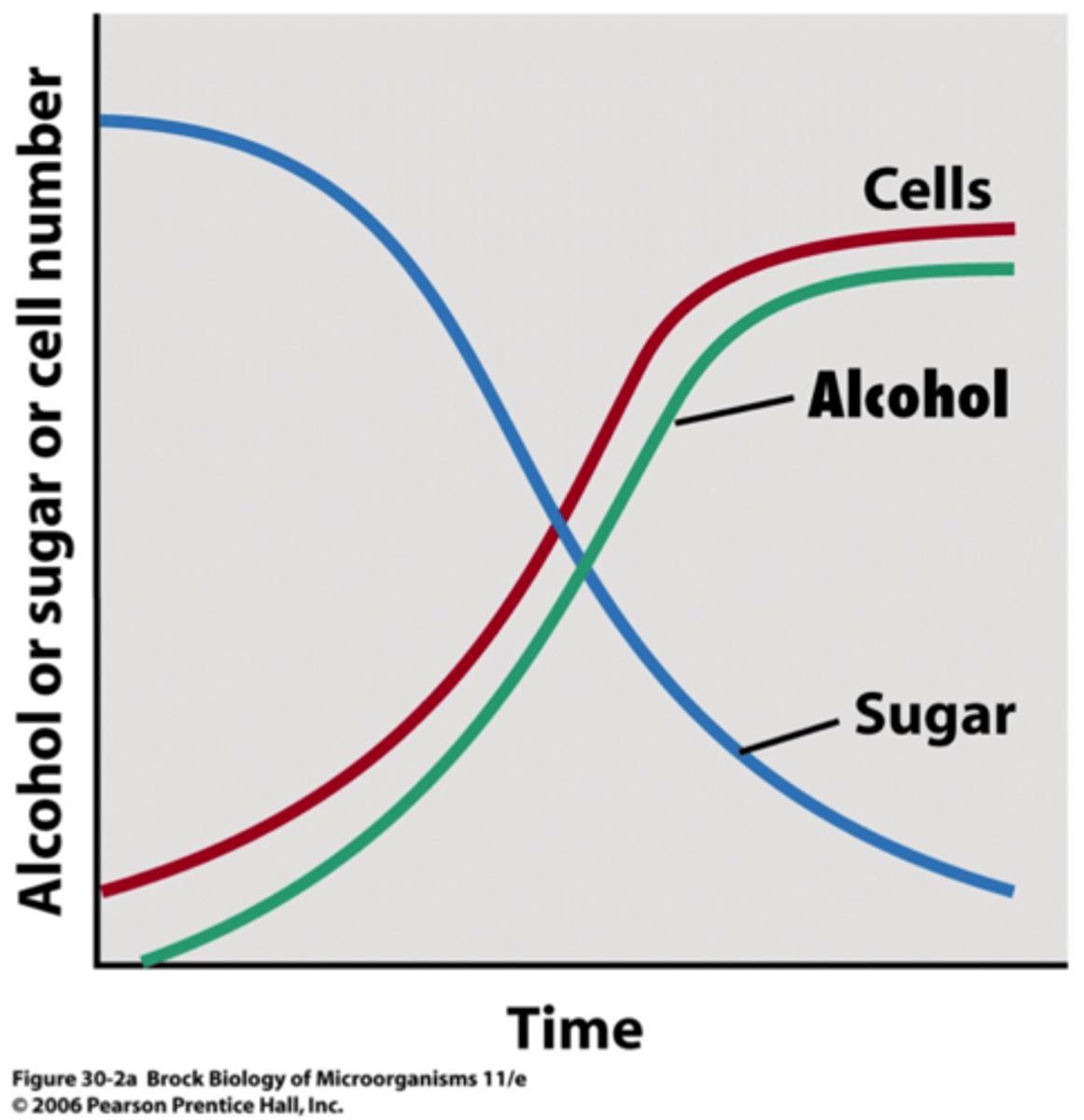
primary metabolites
produced during the exponential growth phase
-alcohol is an example; formed as part of energy metabolism
-alcohol production is parallel with growth curve; CANNOT be significantly overproduced (usually the waste product)
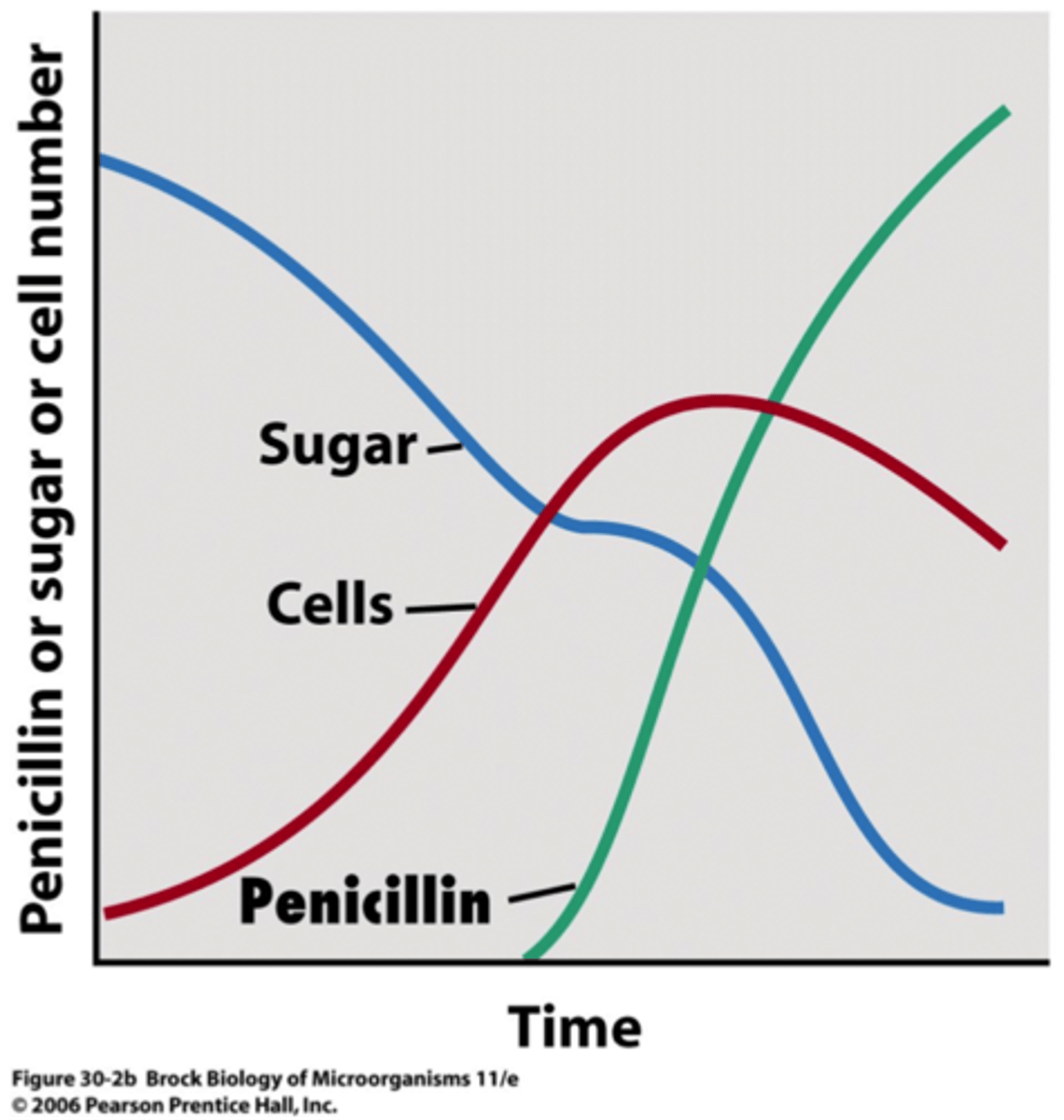
secondary metabolites
produced near the end of growth phase or in the stationary phase; not essential for growth and reproduction
-formation highly depends on growth conditions
-often produced as a group of related compounds
-often significantly overproduced
-often produced by spore-forming microbes during sporulation
fermenter
vessel where the microbiology process takes place; most are aerobic processes
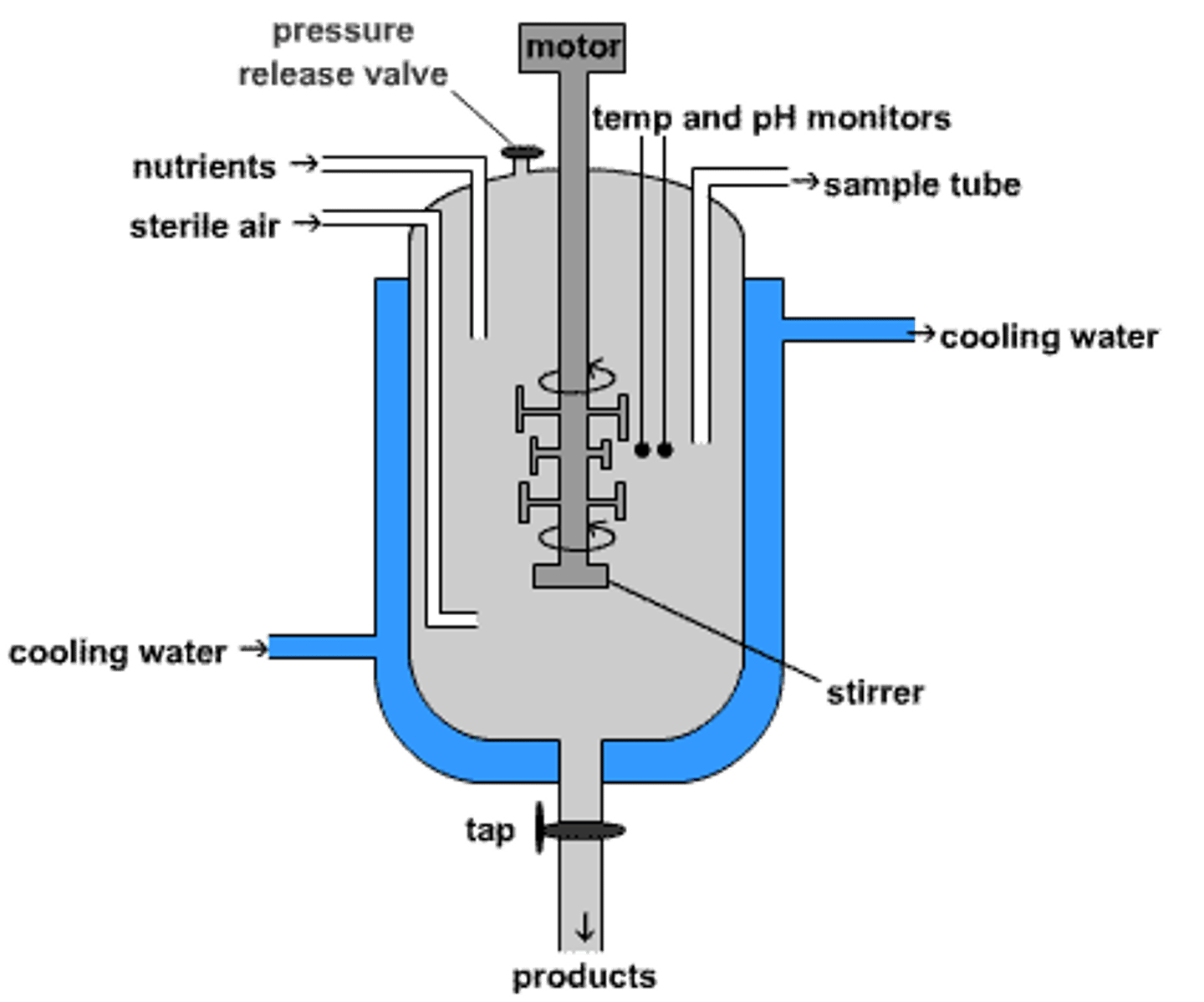
impeller
helps supply fermenter with O2 by mixing the culture
sparger
helps supply fermenter with O2 by pumping in high pressure air for aeration
antibiotics
compounds that kill or inhibit the growth of other microbes
-typically secondary metabolites
antibiotic producers
filamentous fungi or bacteria of the Actinomycetes group
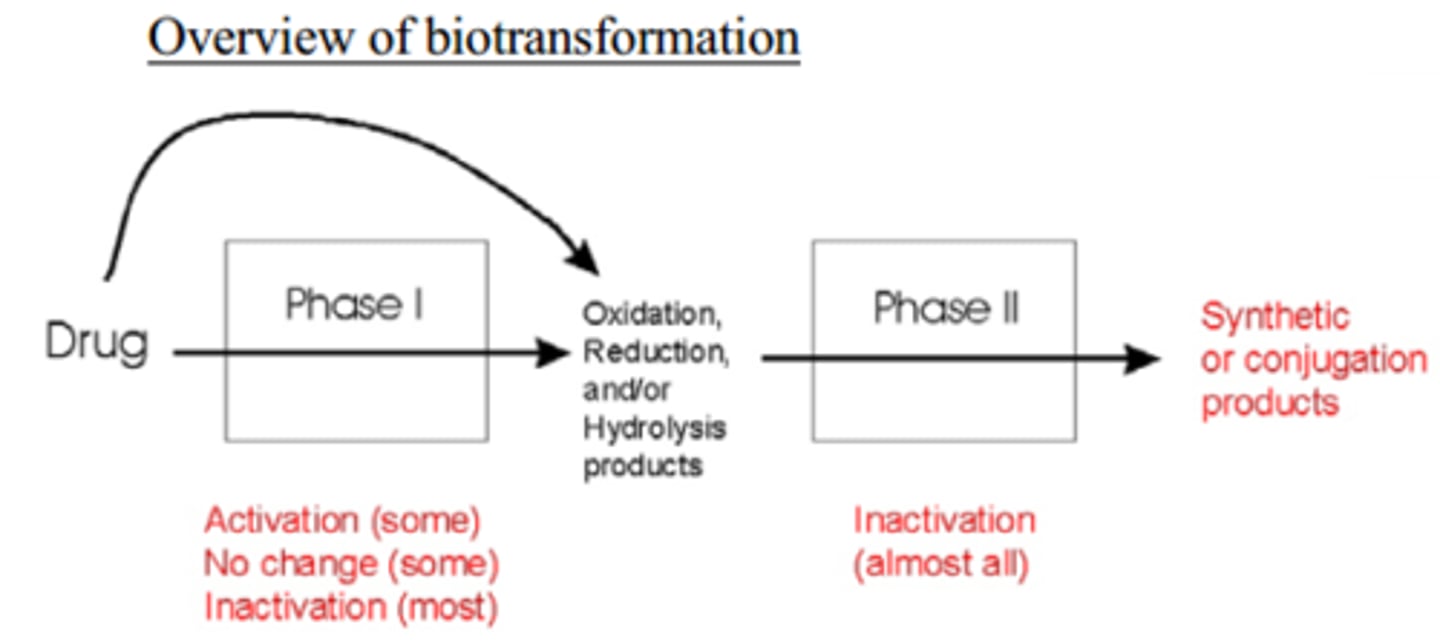
biotransformation
the process in which enzymes convert a drug into a metabolite that is itself active, possibly in ways that are substantially different from the actions of the original substance
extremozymes
enzymes that function at some extreme environment (i.e. pH or temperature)
-produced by extremophiles
carrier enzyme
bonding of enzyme to a carrier, such as bead, fiber, film, or membrane
cross-linking enzyme
cross-linking of enzyme molecules
commodity ethanol
glucose obtained from cornstarch and fermented mostly by yeast (Saccharomyces cerevisiae)
-used as an industrial solvent, a gasoline supplement, etc.
malolactic fermentation
secondary fermentation by lactic acid bacteria (primarily fermentation by yeast)
-high quality dry red wines and a few white wines use this process
vinegar
results from conversion of ethanol to acetic acid
-performed by the acetic acid bacteria (Acetobacter and Gluconobacter)
citric acid
a widely used food industry additive and in the leavening of bread; produced by the mold Aspergillus niger
-citrate acts as a iron chelator to scavenge iron for uptake into the cell
-purified by addition of lime (CaO)