Control of gene expression by epigenetics and RNA interference
1/25
Earn XP
Description and Tags
These flashcards cover key concepts on gene expression control mechanisms via epigenetics and RNA interference, as discussed in SCMOL 3001 Lecture 4.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
26 Terms
Ch6 - How Cells Read the Genome: From DNA to Protein
From DNA to RNA
RNA molecules are single stranded
Transcription produces RNA complementary to one strand of DNA
RNA polymerase enzymes carry out DNA transcription
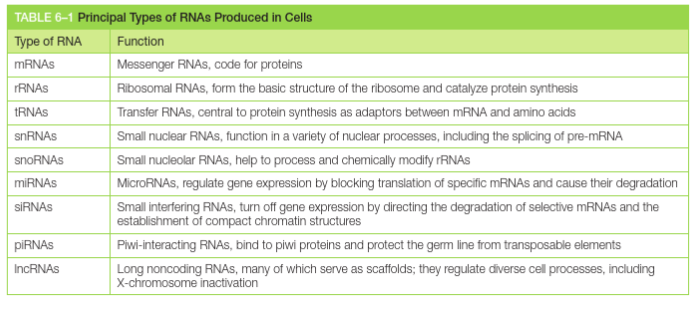
Cells produce different categories of RNA molecules
Signals encoded in DNA tell RNA polymerase where to start and stop
Transcription start and stop signals have nucleotide sequences that are quite varied
Transcription initiation in Eukaryotes requires many proteins - Bacteria have a single type of polymerase, eukaryotic nuclei have 3.
RNA polymerase II requires a set of general transcription factors - bacterial RNA polymerase has single sigma factor, complex eukaryotes require multiple. RNA polymerase II has a ‘general’ cohort required at nearly all promotors.
RNA polymerase II requires activator mediator and chromatin modifying proteins
What is the flow of biological information in a cell?
DNA to RNA to Protein.
DNA does not synthesise protein directly, first the portion of DNA required is transcribed into RNA.
RNA is then used as a template to direct the synthesis of proteins, this is called translation.
What are nucleosomes made of?
Nucleosomes consist of DNA and histone proteins. Includes about 200 nucleotide pairs of DNA. (slide: 147 nucleotide pais of dsDNA)
Chromatin is DNA bound to histone and non-histone proteins.
Nucleosome core particle is made up of DNA wrapped around a histone core.
Octomeric histone core = 2 of each histone H2A H2B H3 and H4. First forms H3-4 and H2A-B dimers, H3-4 dimers form tetramer, and then associate with the 2 H2A-B dimers, forming the octameric core.
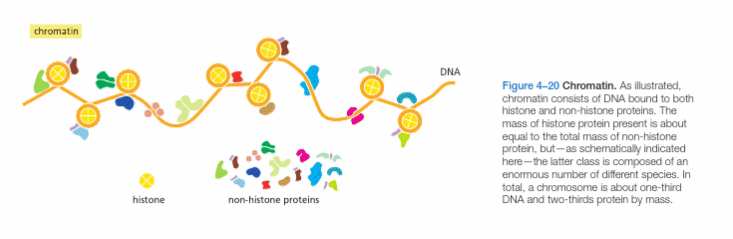
What is chromatin, and its different types?
DNA is compressed into chromatin.
Euchromatin is relaxed, more transcriptionally active and makes up most of the human genome (90%)
Heterochromatin is tightly packed, transcriptionally less active (10%), and contains histone H1 “linker histone” which binds to each nuclosome, changing the path of the protein exiting the nucleosome.
What are histone modifications?
Histones amino acid side chains can undergo several types of covalent modifications: acetylation of lysines, mono- di- and tri- methylation of lysines, serine phorphorylation.
These modifications can alter chromatin structure, allowing for binding of transcription factors, and gene expression. They can also recruit other proteins that can act to determine how and when genes are expressed, and other chromosome functions.
What is the function of histone acetyltransferases (HATs)?
Participate in chromatin remodeling: HATs transfer acetyl groups to histones, which allow the binding of specific transcription factors, promoting gene transcription.
What is the function of histone deacetylases (HDACs)?
Participate in chromatin remodeling: Histone deacetylases (HDACs) remove acetyl groups from histones, leading to chromatin compaction and repression of gene transcription.
How are histone modifications spread along the chromosome?
They are propagated through nucleosome remodeling and recruitment of specific proteins that recognize modified histones, leading to further modifications in neighboring histones.
What is the role of maintenance methyltransferases in DNA methylation?
They recognize hemi-methylated DNA and methylate the corresponding unmethylated strand.
What is genomic imprinting?
Genomic imprinting is a phenomenon where the expression of a gene depends on the parent of origin.
What is mRNA?
mRNA is coding RNA, for creating proteins. It makes up about 1% of the RNA pool at any one time. A large concentration of the RNA pool are non-coding RNAs
Non-coding RNAs include:
rRNA, tRNA
short non-coding RNAs (RNAi, miRNA, siRNA)
long non-coding RNAs
What is RNA interference (RNAi)?
RNA interference (RNAi) is a biological process in which short 20-30 nucleotide, single-stranded RNAs (small non-coding RNAs) act as guide RNAs that reorganize and bind other RNAs in the cell. They can inhibit gene expression by degrading target messenger RNA (mRNA) or preventing its translation.
They all work by RNA-RNA base pairing, and generally cause reduced gene expression.
What are the main classes of small non-coding RNAs that regulate gene expression via RNAi?
Small interfering RNAs (siRNAs), microRNAs (miRNAs), and Piwi-interacting RNAs (piRNAs).
How are miRNAs formed?
double-stranded primary microRNA (pri-miRNAs) are transcribed from miRNA gene by RNA pol II and III, and are capped and poly-adenylated.
They are cleaved by the enzyme Drosha (ds-RNA specific endo-ribonuclease enzyme) in the nucleus to become pre-miRNAs.
After exportation, pre-miRNAs are further processed by Dicer endo-ribonuclease in the cytoplasm to produce duplex (ds) miRNAs.
This mature miRNA is bound by argonaute and other proteins, this forms the RNA-induced silencing complex (RISC). Argonaute is a helicase protein, this unwinds the ds-miRNA and one strand of the miRNA is discarded, the other strand acts as a guide that leads the RISC to bind to the 3 prime untranslated region (UTR) of the target mRNA.
How do micro RNAs (miRNAs) work?
miRNAs can act as guide sequences that bring destructive nucleases into contact with specific mRNAs
If the base pairing of the miRNA and mRNA is extensive, the mRNA is cleaved by the Argonaute protein, and remove the poly-A tail, and exonucleases degrade the mRNA. Otherwise in less extensive pairing, the mRNA is repressed transcriptionally, and sequestered to eventually be degraded.
How do small interfering RNAs (siRNAs) work?
Similar to the formation of miRNA from mRNA.
Many viruses produce double stranded (ds)RNA, RNAi occurs via the attraction of a dicer containing protein, which then cleaves the dsRNA into ds-siRNAs (23np).
Like miRNAs, the ds-siRNAs are bound by the argonaute and RISC proteins, unwound, and a single strand retained as a guide to target viral RNA. RISC carrying the ss-siRNA binds complementary RNA in viral RNA molecules and targets them for destruction, or represses the transcription until the RNA can be degraded.
siRNAs can also assemble into RNA-induced transcriptional silencing (RITS) complex. ss-siRNA still acts as a guide, but the RITS complex binds as the complementary RNA emerges from the RNA polymerase II
RITS attracts histone modification proteins, which direct the formation of heterochromatin, and supressing further transcription initication.
How do siRNAs function in antiviral defense?
siRNAs can bind to viral RNA, leading to its degradation.
What is gene silencing?
Gene silencing is a mechanism ensuring only required genes are transcribed.
Reasons may include:
defence - protection against viruses and transposons.
regulate gene expression
downregulate expression of target genes.
Methods:
Methylation - permanent inactivation of transcription
Chromatin structure repression - no mRNA produced
Post-transcription - mRNA degraded, or not translated.
How are piwi-interacting RNAs (pi-RNAs) formed?
Made in the germ line, piRNAs block the moevment of transposable elements.
Genes coding for piRNAs are made up of sequence fragments of transposable elements (transposons, DNA sequences that move from one location on the genome to another). The clusters of fragments are transcribed and then broken into short piRNAs, which are actually longer than miRNAs and siRNAs. Also unlike miRNA and siRNA, the process involves Piwi, a class of proteins related to Argonaute, and there is no Dicer enzyme involved.
Once complexed with Piwi, the piRNAs target via base pairing, silence intact transposon genes, and destroy any RNA (incl mRNA) produced by them.
What distinguishes piRNAs from miRNAs and siRNAs?
piRNAs do not involve the Dicer enzyme in their processing. They are also slightly longer than miRNA and siRNA, and complex with Piwi rather than Argonaute.
What are long non-coding RNAs (lncRNAs)?
LncRNAs are RNAs longer than 200 nucleotides that do not code for proteins. Many non-coding RNA's with unknown function fall under this heading.
What is the main molecular mechanism behind epigenetic changes?
DNA methylation and histone modifications.
What is the consequence of 5-methylcytosine in DNA?
It can repress transcription by interfering with the binding of transcription regulators.
What are imprinting disorders associated with DNA methylation?
Prader-Willi syndrome and Angelman syndrome.
Both involve deletion of the chr#15p, but one inherited from the father, the other from the mother.
How can identical twins exhibit different phenotypes despite having the same genetic sequence?
Due to differences in DNA methylation patterns and histone modifications related to environmental factors.