WEEK 8-Lac Operon, Mutations, Mutagens
1/26
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
27 Terms
The Lac Operon
First operon discovered in E. coli
Encodes enzymes for lactose metabolism
Polycistronic mRNA with 3 genes:
lacZ: Encodes B-galactosidase (LacZ), breaks lactose into glucose and galactose
lacY: Encodes lactose permease (LacY), transports lactose into the cell
lacA: Encodes B-galactosidase transacetylase (LacA)
LacY (Permease) and LacZ (B-galactosidase)
LacY allows lactose entry into the cell
LacZ converts lactose into glucose and galactose
Induced in gram-negative cells when lactose is present
Control of Gene Expression in Bacteria
Sigma factors control multiple genes
Gene expression controlled by repressors and inducers
Small molecules can activate gene expression
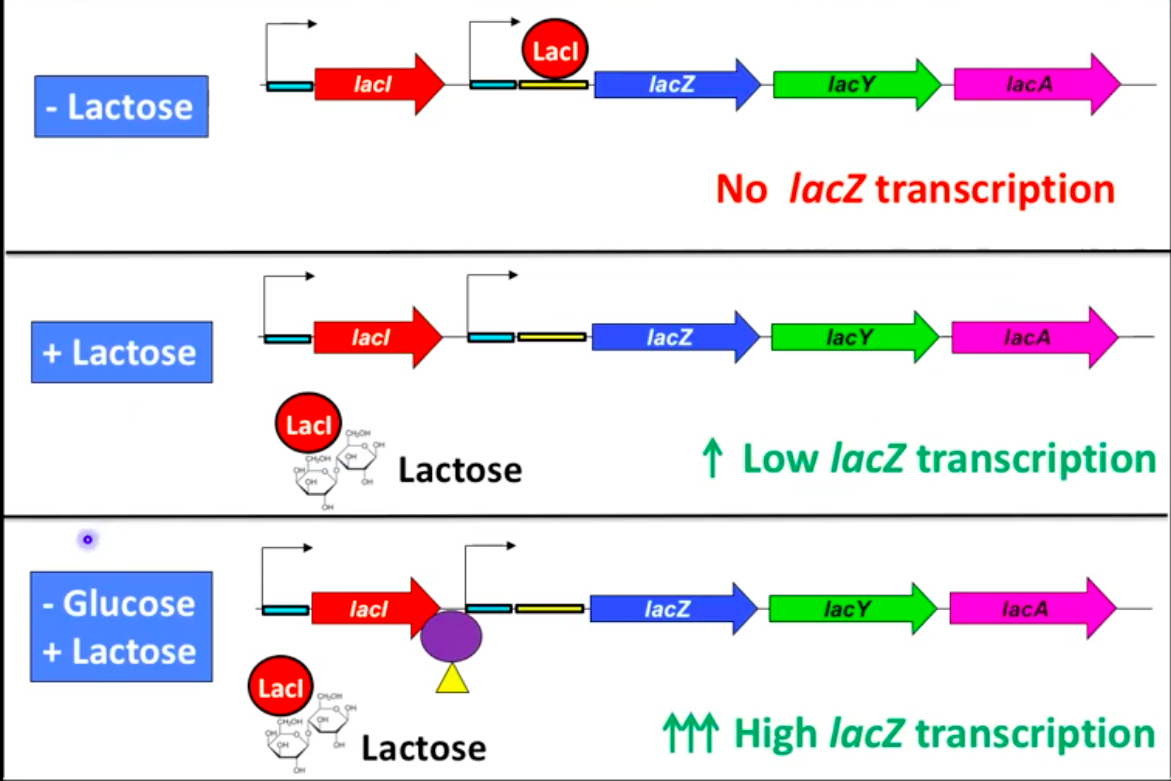
Regulation of the Lac Operon
Highly regulated; only transcribed when lactose is present
Turned on in presence of lactose, turned off without lactose
LacI (repressor) binds operator and blocks transcription in the absence of lactose
LacI binding prevents RNA polymerase from initiating transcription
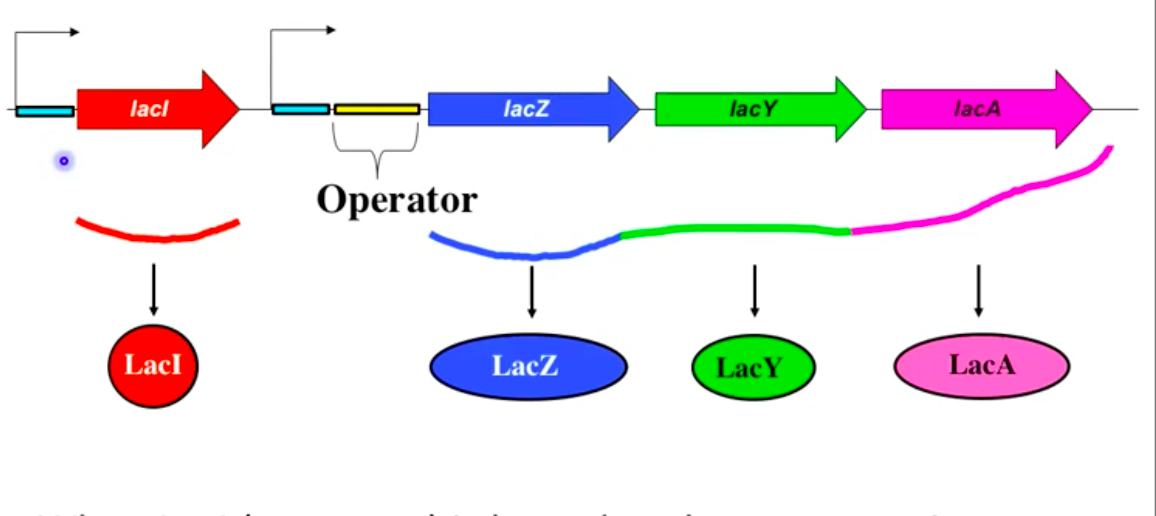
Repressor (LacI)
Protein that prevents gene expression by binding the operator
Blocks RNA polymerase from initiating transcription
Bends DNA to prevent RNA polymerase access to lac promoter
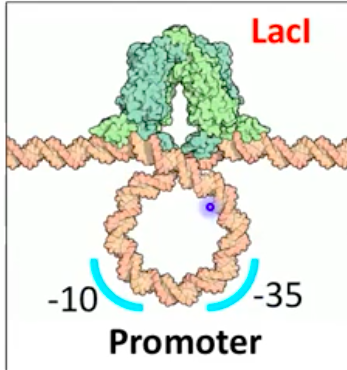
Lac Operon in the Presence of Lactose
Low or absent glucose with lactose induces operon
Modified lactose binds repressor protein, changing its shape
Repressor cannot bind the operator, allowing low-level transcription
Inducer: substrate that induces expression of genes for its metabolism
Apoinducer and CAP
Apoinducer: Protein that enhances gene expression
Catabolite Activator Protein (CAP) binds promoter with cAMP
cAMP (cyclic AMP) levels vary with nutrient availability:
Low glucose → high cAMP (forms CAP-cAMP complex)
High glucose → low cAMP (no CAP-cAMP complex)
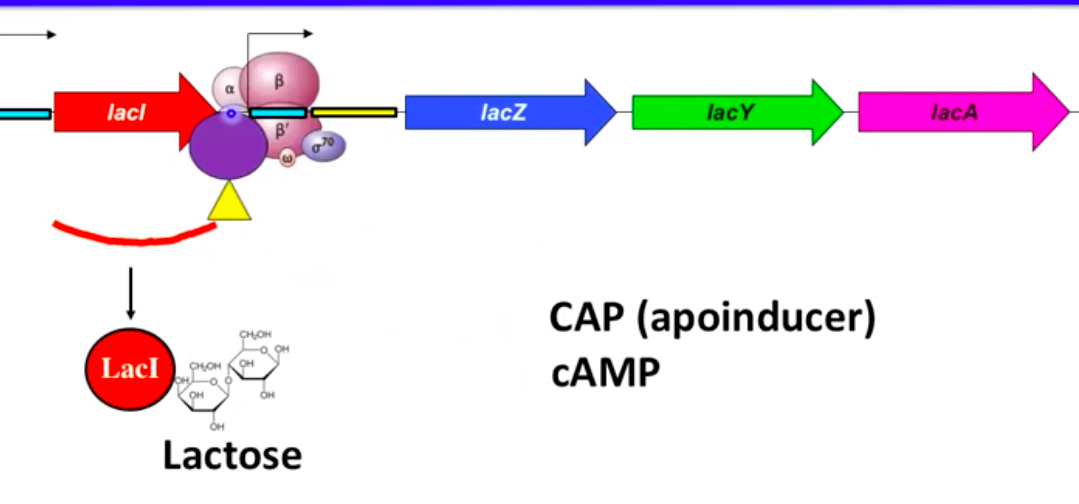
Lac Operon Control
Gene expression can be controlled by:
Repressors and inducers
Small molecule activation
Two systems work together in Lac operon activation:
Inducer control: Lactose binds to repressor, disabling it to allow transcription
Small molecule activation: cAMP binds to CAP, enhancing transcription
Role of cAMP in Lac Operon
cAMP levels inversely related to glucose levels
Low glucose = high cAMP (CAP-cAMP enhances transcription)
High glucose = low cAMP (no enhancement of transcription)
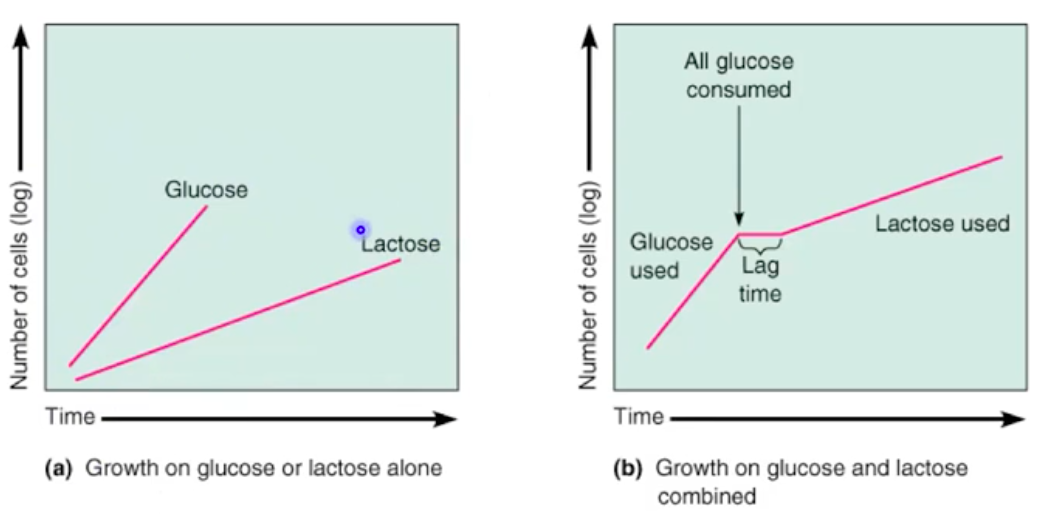
Catabolite Repression and Diauxic Growth
Catabolite repression causes bacteria to preferentially use glucose
Diauxic growth: Glucose metabolism first, followed by lactose metabolism
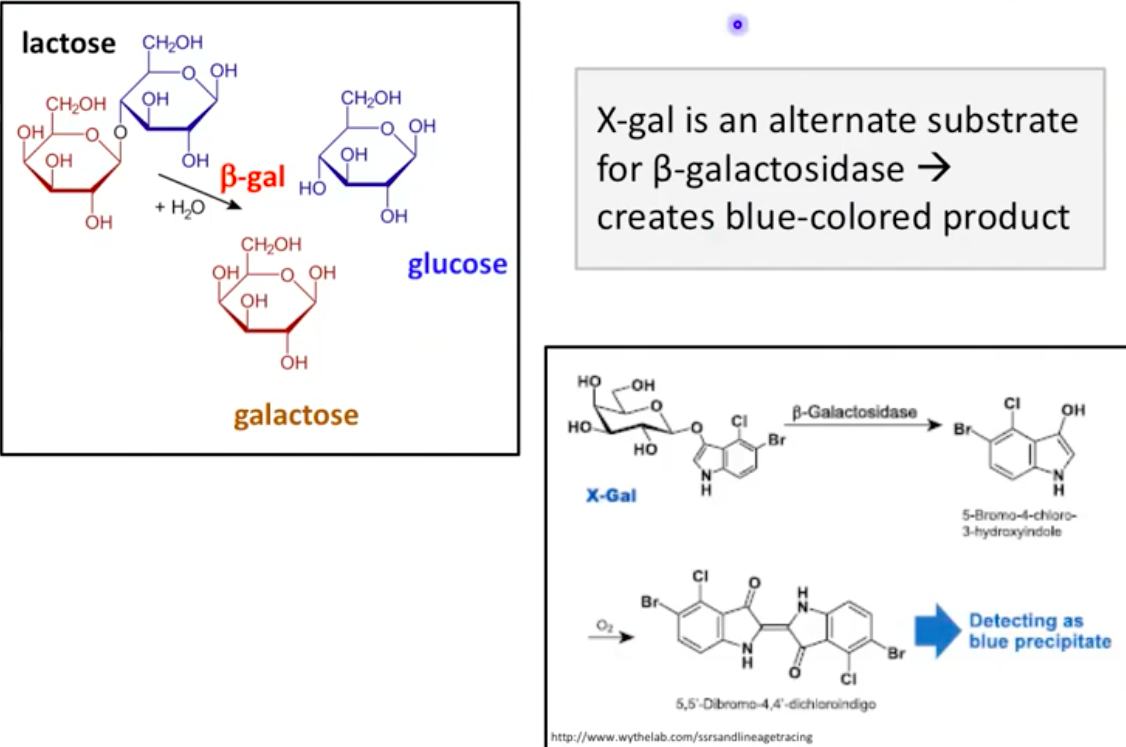
X-gal as a Substrate for B-gal
X-gal: alternative substrate for B-galactosidase
Creates a blue-colored product indicating B-gal activity
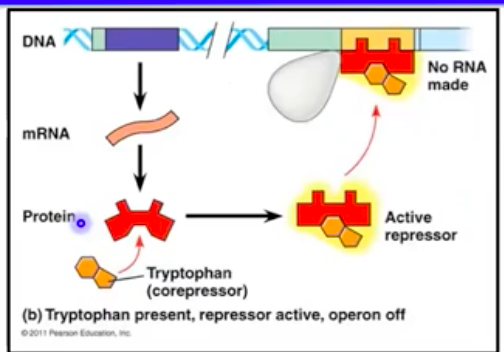
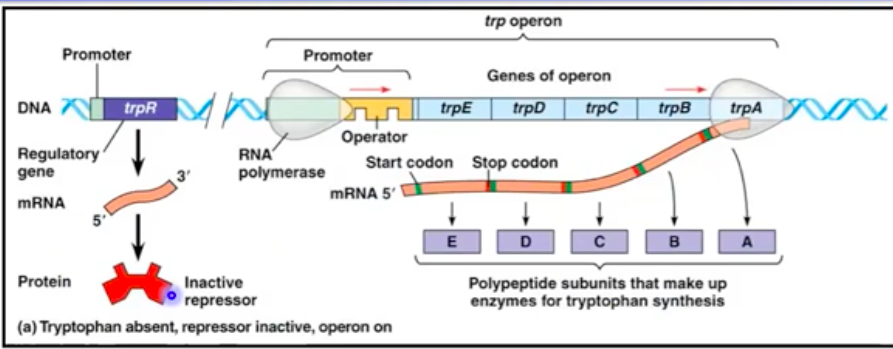
The trp Operon (Repressible Operon)
Encodes enzymes for tryptophan biosynthesis
Repressor protein binds tryptophan to inactivate the operon
High tryptophan levels inhibit the operon, preventing excess
Mutations
Heritable changes in the DNA sequence
Effects on the organism:
Almost always deleterious
Sometimes neutral (no effect)
Rarely beneficial (new property)
When you still see bacteria in zone of inhibition it is because those colonies have undergone a mutation that is resistant.
Heritability of Mutations
Changes in DNA sequence become heritable through replication
Mutant: cell line inheriting a specific mutation
Points Mutations
Single nucleotide base pair mutation
Types:
Substitution: e.g., "THE CAT ATE ELK" -> "THE RAT ATE ELK"
Insertion: e.g., "THE CAT ATE ELK" -> "TRH ECA TAT EEL K"
Deletion: e.g., "THE CAT ATE ELK" -> "TEC ATA TEE LK"
Genotype vs Phenotype
Genotype: DNA sequence
Phenotype: Observable trait (e.g., protein function)
Wild-type (Genotype)-> mRNA-> Protein (Phenotype)
Silent Mutations
Genotype changes, phenotype unchanged
DNA is mutated -> genotype changes
Protein is NOT changed-> phenotype doesn't change
Example: CAA -> CAG (same amino acid)
Missense Mutation
Amino acid substitution
May affect phenotype, depending on location
DNA is mutated-> genotype changes
Protein is changed -> phenotype changes may occur depending on location
Example: CAA -> CAT
Nonsense Mutations
Amino acid changed to stop codon
Typically causes loss of function
DNA mutated → genotype changes
Protein changed=loss of function in addition to phenotypic change
Example: CAG -> TAG
Physical Mutagens
UV light: Causes C=C or T=T dimers
X-rays: Break DNA backbone bonds
Chemical Mutagens
Nucleotide Analogs: Resemble nucleotides, cause replication errors and mispairing
Nucleotide-Altering Chemicals: Alter base structure to a different base or base analog
Framsift Mutagens
Frameshift Mutagens
Insert between DNA bases, causing insertions or deletions during replication
DNA Repair Mechanisms
Direct Repair, Single Strand Repair, Error-prone repair
Direct Repair
Small damage on one strand
Examples:
Base-Excision Repair: Removes mutated base, replaces it
Light Repair: Photolyase enzyme fixes C=C or T=T dimers
Single Strand Repair
Nucleotide Excision Repair: Cuts out damaged section, uses template for repair
Mismatch Repair: Removes mismatched bases
Error-Prone Repair
SOS Response: Fills gaps with random sequences as a last resort
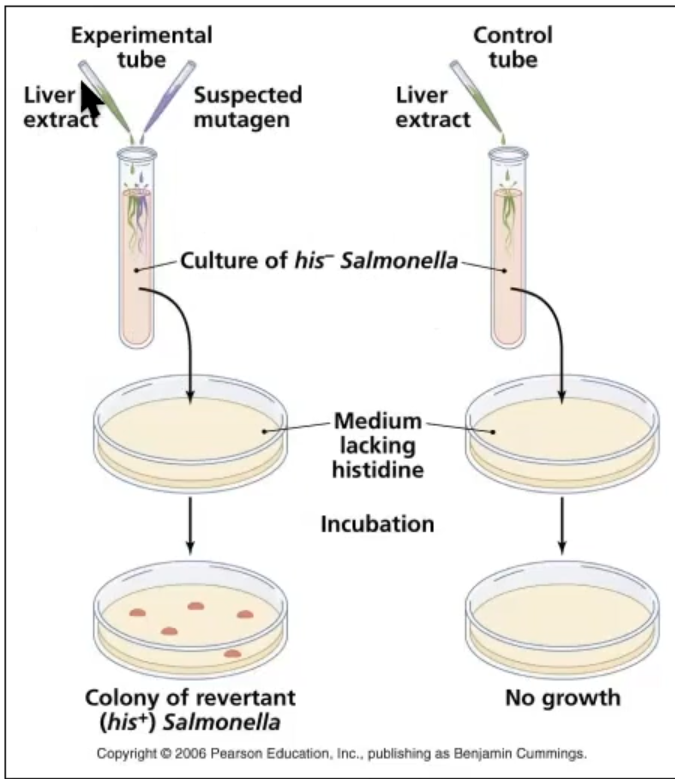
Ames Tests
Used to identify mutagens
Uses Salmonella bacteria to test if a substance is a mutagen
Presence of colonies indicates mutagenic potential