L9 DNA Recombination and Genome Integrity 2/2
1/19
Earn XP
Description and Tags
Lecture 9
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
20 Terms
What are sources of breaks in DNA
DNA replication (endogenous source)
ionizing radiation and chemicals (exogenous source)
What type of breaks is most common in DNA?
single strand breaks
how are single strand breaks repaired
they are repaired by the ligase
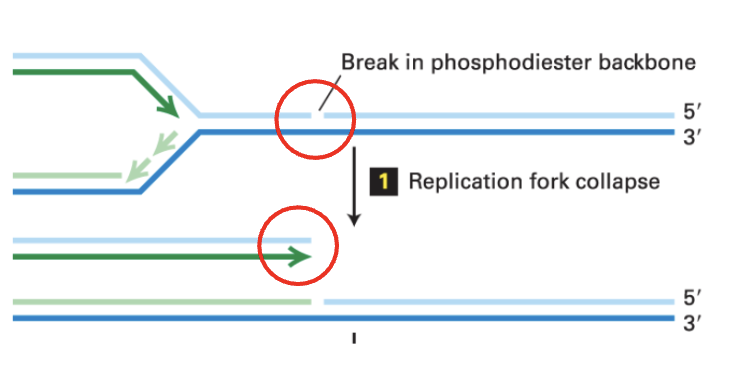
How can a single strand break lead to double strand break?
Collision with a pre-existing single strand break during DNA unwinding because of replication can cause the replication fork to collapse, generating a double strand break
What is the “Effect” and “Repair” for the Mutagens: ionizing radiation or anticancer drugs like bleomycin
Effect:
single-strand break
(or)
double-strand break
=> both leading to genomic rearrangements or translocations
Repair:
repair by non-homologous end-joining (NHEJ)
directly ligating the broken ends without a template
error prone
(or)
repair by homologous recombination (HR)
uses a homolgous template to fix double-strand breaks with high accuracy
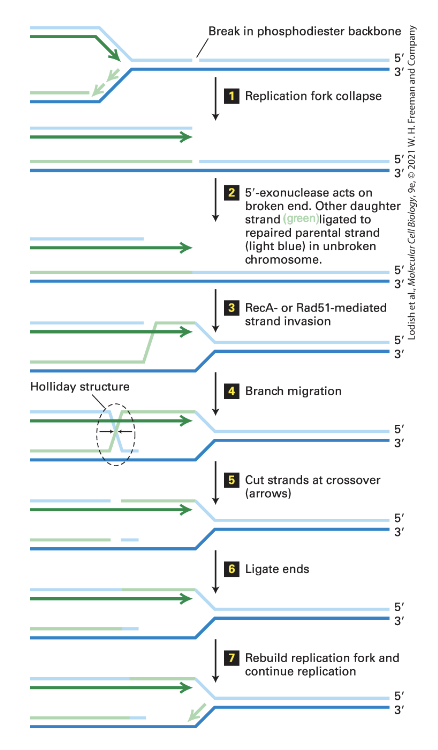
how is a double strand break fixed when there is a replication fork by homologous recombination? (7 steps)
(1) Replication fork collapse
DNA synthesis could not occur at this point because of the break
(2)
5’ exonuclease acts on the broken end (5’ end) without removing the other strand with the 3’ end
The other daughter strand is ligated to the repaired parental strand in the unbroken chromosome
(3) RecA- or Rad51-mediated strand invasion
recombinases helps by binding to single to single strand DNA (in green)
somehow are going to find a complementary strand for this broken strand and are going to pair with it
(4) Branch Migration
Holiday structure formed from the recombinations (4-way DNA structure)
reannealing of the parental strands
annealing of the two nascent strands
(5) Cut strands at crossover
There are endonucleases specific for this
(6) Ligate the ends
(7) DNA replication can continue
Rebuilding of the replication fork and proceeding with DNA replication
double strand break repair by Homologous Recombination (HR) advantage, what does it need, how does it work (quick step explanation)?
is a higher fidelity mechanism to repair double strand breaks than NHEJ
requires an undamaged copy of the chromosome to be repaired
the process required:
processing breaks to expose ssDNA seq.
conduct a search for identical sequences
use them as a template of DNA synthesis
and repair the break
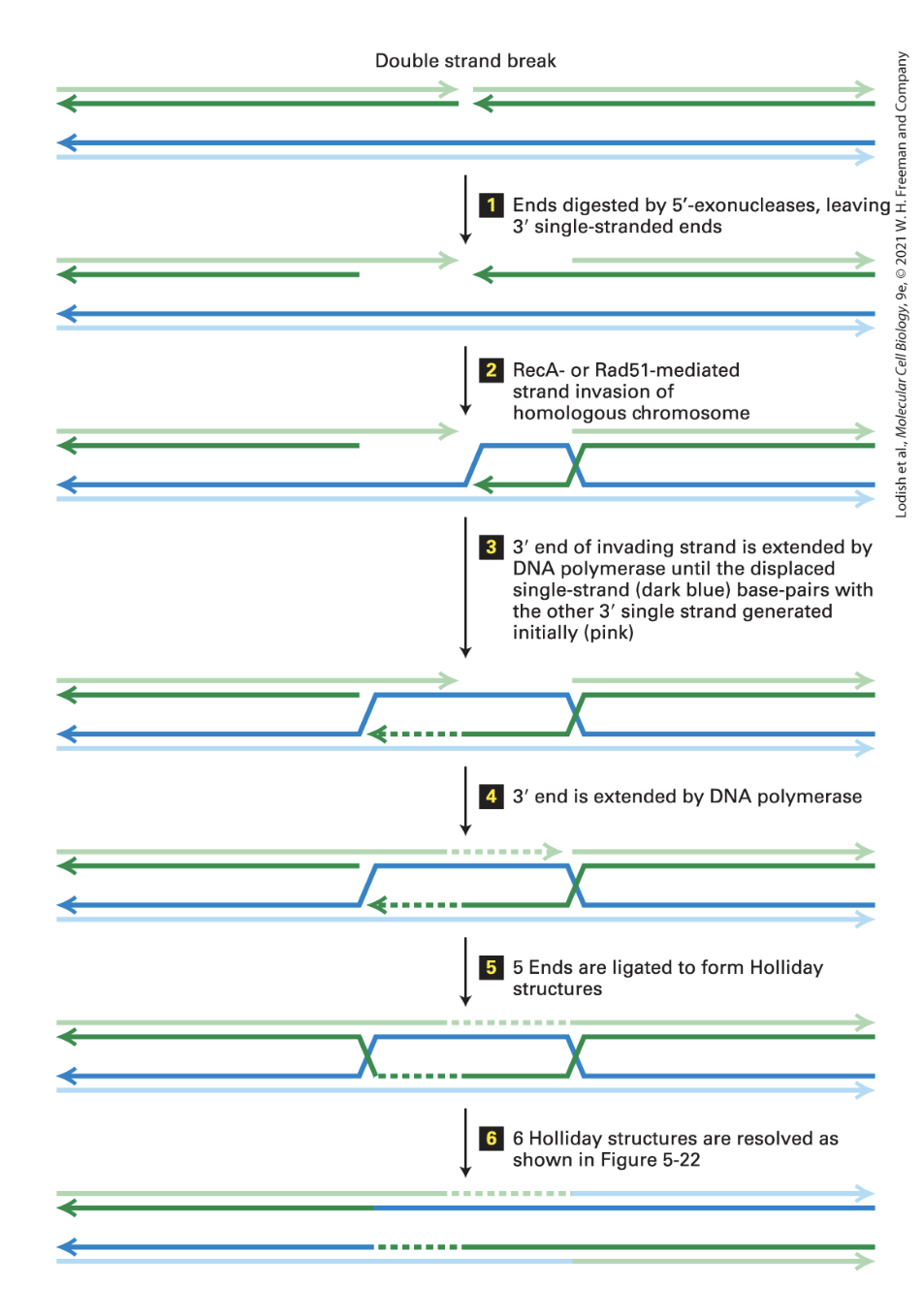
HR double strand break repair when no replication fork and two double strand ends (aka two breaks to repair) (6 steps)
(1) Ends are digested by 5’ exonuclease leaving 3’ single strand ends
(2) RecA- or Rad51- mediated strand invasion of homologous chromosome
RecA wraps around ssDNA and extends it
nb. we don’t yet understand how it searches accross the genome to find complementary sequence
(3) 3’ end of invading strand is extended by the DNA polymerase until the displaced single strand base pairs with the other 3’ single strand generated initially
(4) 3’ end is extended by DNA polymerase
(nb. dotted lines on image represent DNA synthesis by DNA polymerase)
(5) ends are ligated to form Holiday structures
(6)Holiday structures are resolved (break and then ligate)
nb. there are 6 ligations total: 2 at step (4) and 4 at step (5)
What are the two ways holiday junctions can be resolved? what does each lead to?
Both involve rotating the DNA molecules so that they aren’t crossing. then two ways to separate the DNA molecules
1/Original
cutting the original strands
results in gene conversion but no chromosomal crossover
2/Recombinant
cutting other pair of strands
leads to recombinant molecule with chromosomal crossover
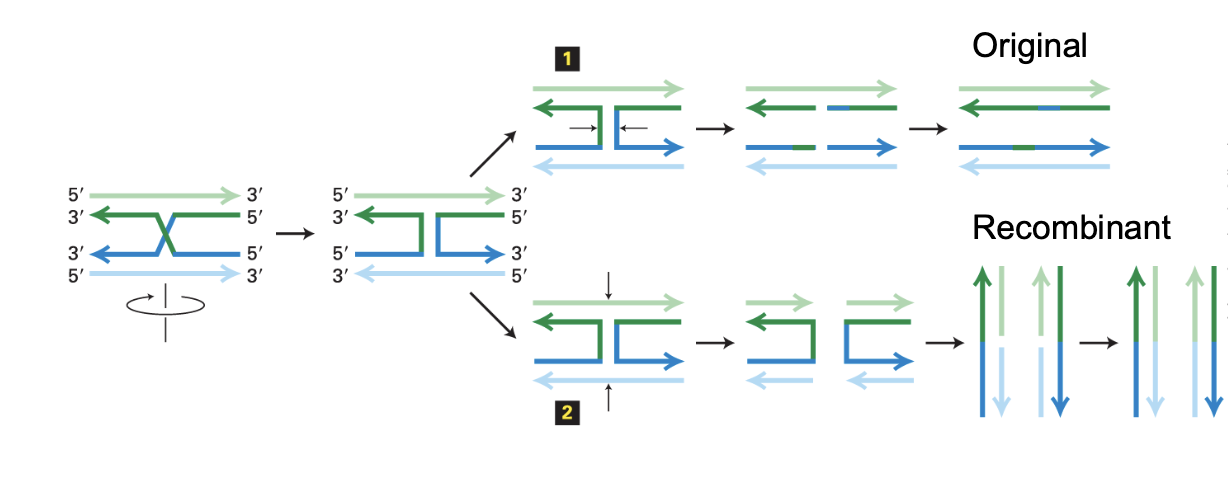
What is the impact of a double strand break during meiosis?
generates genetic diversity !!
during meiosis the DNA recombination occurs from mom and dad to mingle their sequences
the sequences from their chromosomes are mingled producing translocations between homologous chromosomes
Is double strand break bad or good?
generally bad for cells
but can be good for generating genetic diversity during meiosis when done purposefully in very controlled way to separate DNA
whys is Non-Homologous End Joining (NHEJ) less advantageous than HR
is it error prone and can introduce mutations
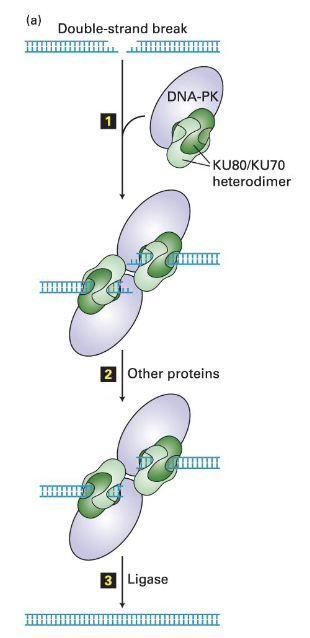
Non-Homologous End Joining (NHEJ) mechanism (3 steps)
(1)
A complex formed by DNA-dependent protein kinase (DNA-PK) and Ku80/Ku70 heterodimer binds to the ends of the double-strand break
(2)
Ends are digested by nucleases or filled in by DNA polymerases
need blunt ends for ligation to occur => exonuclease and DNA polymerase activity is used so that all nucleotides are paired with complementary strand
this results in a loss of information either by the removal or addition of nucleotides
cell chooses this trade off for repair even though mistakes
(3)
Ends are ligated together
chromosomal translocation aka chromosomal rearrangements
An abnormal chromosome region that contains rearranged genetic material, usually from two non-homologous chromosomes
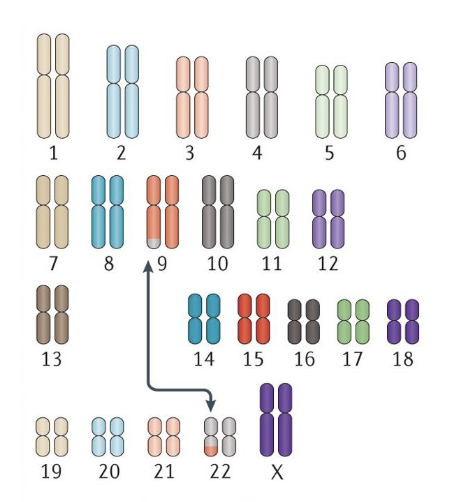
how is NHEJ a source of chromosomal translocations?
Since NHEJ joining of DNA fragments is not driven by sequence similarity, these can result in mistakenly bringing together pieces of two very different chromosomes
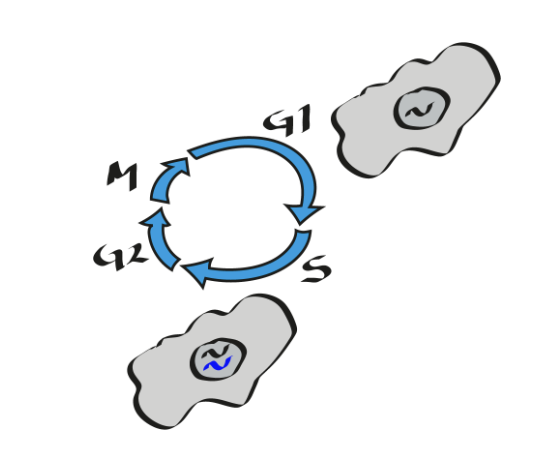
Cells regulate their type of repair according to the cell cycle: which step of the cycle uses what kind of repair?
G1: only one copy of the genome is present
NHEJ repairs DNA breaks
(chromosomal translocation can occur)
S and G2 and M
Homologous REcombination (HR) repairs double strand breaks
S
Translesion Synthesis (TLS)
mismatch repair (MMR)
MMR often corrects TLS erros
Why do many drugs and treatments for cancer like chemotherapy target DNA?
because DNA damage can be repaired efficiently if DNA is not ongoing
(anti-cancer drugs that damage DNA are most effective when cancer cells are rapidly dividing
Cancer cells replicate their DNA continuously, making their DNA-repair mechanisms overworked and less efficient at fixing the additional damage from the drugs. In contrast, healthy, non-replicating cells have time to efficiently repair the drug-induced damage, leading to fewer side effects)
tumors are constantly growing anf dividing
treatments that damage DNA are effectoive against these
Do most cells in our body divide?
No. Only few tissues actively divide.
i.e. in brain muscles cells don’t divide often or at all
What are examples of the few tissues in our body whose cells actively divide?
skin cells
cells in the gastrointestinal tract
blood cells in bone marrow
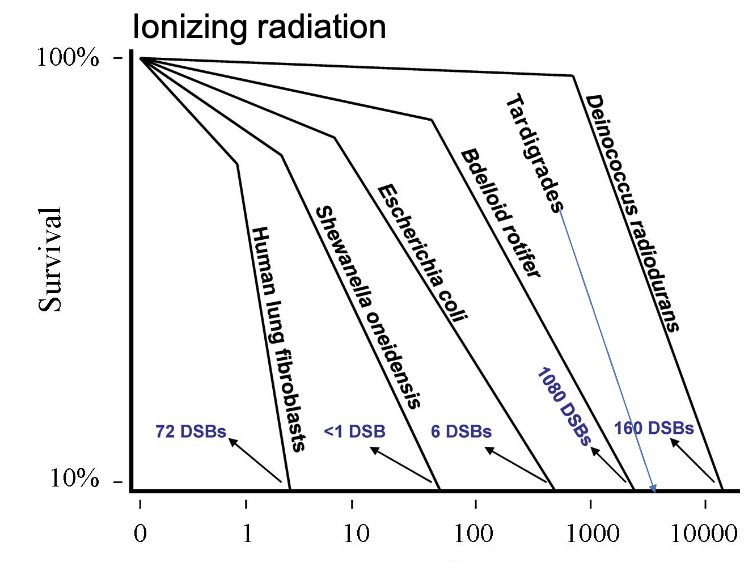
what are two examples of organisms that are (much) better at DNA repair in comparison to humans
tardigrades
most resistant animal
deinococcus
most resistant organism