BIMB10 - Final Exam
1/120
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
121 Terms
What is Mole, Molarity and Molality?
A mole is 6,022 x 10^23, and is a convenient unit to use because of the great numbers of atoms or molecules in a substance.
Molarity is molar concentration. The most common used unit is number of moles per liter (mol/L). Can be calculated through c = n/V, where c is molarity, n is amount of the solute in moles, and V is volume of solution.
Molality is the total moles of a solute contained in a kilogram of a solvent. It is calculated by taking the amount of solute in moles divided by the mass of solvent in kg. Molality = b.
What is Avogadros constant?
Avogadros constant is the number of units in one mole, or 6,022 × 10^23.
What is a limiting reagent?
A limiting reagent is the reactant that gets consumed first in a chemical reaction and therefore limits how much product can be formed. One way of finding the limiting reagent is by calculating the amount of product that can be formed by each reactant, the one that produces less product is limiting.
What is and how do you calculate chemical yield?
Chemical yield is the amount of product created by a reaction in relation to how much reactant was used. To find the chemical yield you take the actual yield divided by the theoretical yield and multiply it by 100 (expressed in percent).
What is concentration, dilution and serial dilution?
Concentration is the mass of the solute divided by the volume of the solution. Expressed in kg/L, but often used molarity units.
Dilution is the process of decreasing the concentration of a solute in a solution, usually by mixing with more solvent.
Serial dilution is a stepwise series of dilutions that are performed to reduce the concentration of a substance in a solution.
What is meant by a primary, secondary, tertiary and quaternary structure of a protein?
Used to describe a protein.
Primary = the amino acid sequence of its polypeptide chain.
Secondary = local spatial arrangement of a polypeptides backbone atoms (no side chains). two most common structural motifs in secondary structures are the alpha helix and the beta pleated sheet, dependent on hydrogen bonds.
Tertiary = the overall three-dimensional arrangement of its polypeptide chain in space (with side chains). This folding dependent on hydrophobic/hydrophilic interactions and other non-covalent bonds.
Quaternary = the overall three-dimensional arrangement of several folded polypeptide chains (subunits) together forming a protein.
What does folding of a protein mean?
Folding of a protein occurs after translation in the ribosome, when the protein is only a long polypeptide chain. Protein folding is dependent on several factors, but the amino acid sequence of the polypeptide chain (primary structure) has all the information needed in order to fold properly. Folding in the secondary structure is dependent on hydrogen bonds between the polypeptide backbone, and folding in the tertiary and quaternary structure is dependent on hydrophobic/hydrophilic interactions, other non-covalent bonds and disulfide bridges. Can fold by itself or with the help of chaperonins.
Explain what a protein domain is.
Protein domains are units of proteins that can fold, function and evolve more or less independently, and each have the characteristics of a globular protein. The polypeptide chain wanders back and forth within a domain. Different domains are connected by short segments of the polypeptide chain.
Where in the cell are proteins modified?
Mostly in the ER, here they are modified and sorted for transport to their eventual destinations within the cell.
Give 5 examples of post-translational modifications of a protein.
N-linked glycosylation, O-linked glycosylation, formation of S-S bridges, hydroxylation, phosphorylation.
What is formation of S-S bridges?
A post-translational modification of a protein. Disulfide bonds (S-S bonds) are formed through the oxidation of sulfhydryl groups (-SH) on two cysteine residues within a protein. Occurs during the folding of the protein when two cysteine residues are brought into proximity. Occurs in the ER, because of the oxidizing environment that promotes formation of S-S bridges. Essential for proper folding and full activity of certain proteins.
What is N-linked glycosylation?
A post-translational modification of a protein. Addition of one or more sugars to a nitrogen atom on Asparagine residues of a protein. Occurs co-translationally and is initiated in the ER. Serine or threonine needs to be two steps away from the glycosylated asparagine in the polypeptide chain. It ensures proper folding of proteins, if they are misfolded they are sent away for destruction.
What is O-linked glycosylation?
A post-translational modification of a protein. Addition of one or more sugars to a hydroxyl group on Serine or Threonine residues of a protein. Occurs in the Golgi. Affect the protein by changing protein stability and regulating protein activity.
What is hydroxylation?
A post-translational modification of a protein. Adds a hydroxyl group (-OH) to the proteins, making hydrophobic molecules hydrophilic. It regulates protein interactions or adds a functional group that can be further modified.
What is phosphorylation?
A post-translational modification of a protein. Reversible phosphorylation of proteins involves addition of a phosphate group on serine, threonine, or tyrosine residues. Several enzymes or signaling proteins are switched 'on' or 'off' by phosphorylation or dephosphorylation. Addition of a phosphate group can convert a previously uncharged pocket of protein into a negatively charged and hydrophilic protein thereby inducing conformational changes in the protein.
Explain what an active site in an enzyme is.
A region on an enzyme that binds to a protein or other substance during a reaction. Each enzyme has an active site to which one or more substrate molecules bind, forming an enzyme-substrate complex. A reaction occurs at the active site, producing an enzyme-product complex. The product is then released, allowing the enzyme to bind further substrate molecules. The active site is made up of amino acid residues that establish temporary bonds with the substrate (binding site) as well as residues that catalyse that substrate's reaction (catalytic site).
Name the main enzyme classes important for the central processes in the cell and shortly their function (11).
Hydrolases - general term for enzymes that catalyze a hydrolytic cleavage reaction.
Nucleases - Break down nucleic acids by hydrolyzing bonds between nucleotides (phosphodiester bonds).
Proteases - Break down proteins by hydrolyzing bonds between amino acids (peptide bonds).
Synthases - Synthesize molecules in anabolic reactions by condensing two smaller molecules together.
Ligases - Join together (ligate) two molecules in an energy-dependent process.
Isomerases - Catalyze the rearrangement of bonds within a single molecule.
Polymerases - Catalyze polymerization reactions such as the synthesis of DNA and RNA.
Kinases - Catalyze the addition of phosphate groups to molecules.
Phosphatases - Catalyze the hydrolytic removal of a phosphate group from a molecule.
ATPases - Hydrolyze ATP.
Oxido-Reductases - General name for enzymes that catalyze reactions in which one molecule is oxidized while the other is reduced.
Explain how protein electrophoresis works.
Called SDS-PAGE. When applying electric charge to a solution containing a protein molecule it will cause the protein to migrate at a rate dependent on its size, net charge, and shape. Proteins are first dissolved in a solution that includes a powerful negatively charged detergent, making them all negative. This also makes them unfold into extended polypeptide chains by binding to their hydrophobic regions. A reducing agent is added to break any disulfide bonds. When voltage is applied, the proteins migrate toward the positive electrode, dependent on size. Smaller proteins travel faster and proteins of the same size travel at the same rate. Result = the mixture of proteins will be fractioned into a series of protein bands arranged in order of molecular weight. provides information about molecular weight and subunit composition.
Explain the use of specific antibodies for immunoblotting (Western blotting).
After fractioning out proteins through SDS-PAGe, a certain protein can be identified by exposing all proteins on the gel to a specific primary antibody that binds to the protein of interest, and then a secondary antibody that recognizes the primary antibody and is labeled with a dye. This is done by first transferring the separated proteins onto a sheet of nylon membrane, and then soaking it in the antibody solutions to reveal the protein of interest. Colors only one specific protein. Can be very useful when assessing the amount of proteins.
Describe principles for protein purification by different chromatography techniques.
Protein purification = separation of proteins.
Affinity chromatography - Molecules separated by their ability to bind to particular small molecules or other macromolecules. An insoluble matrix that is covalently bound to a specific ligand (ex. antibody or enzyme substrate), that will bind a specific protein, all other proteins that do not bind the substrate will be eluted in the column.
Gel filtration/ sizing chromatography - Molecules separated depending on size. Small beads are added that interact with the solved molecule, which slow down the movement of proteins according to size. Smaller molecules travel more slowly, because they enter many pores in the gel, while larger molecules don't enter as many pores and therefore travel faster, and are eluted sooner.
Ion exchange chromatography - Molecules are separated according to charge. Columns are packed with small beads that carry a charge, making proteins fractioned according to arrangement of their surface charges. The affinity between beads and protein depend both on ionic strength and pH, which can both be greatly varied to achieve an effective separation. More highly charged molecules are more tightly bound, travel slower, while less charged molecules bind less and travel faster, eluted sooner.
What are the number of chromosomes in humans and mice?
Humans have 46 chromosomes (23 pairs), while mice have 40 chromosomes (20 pairs).
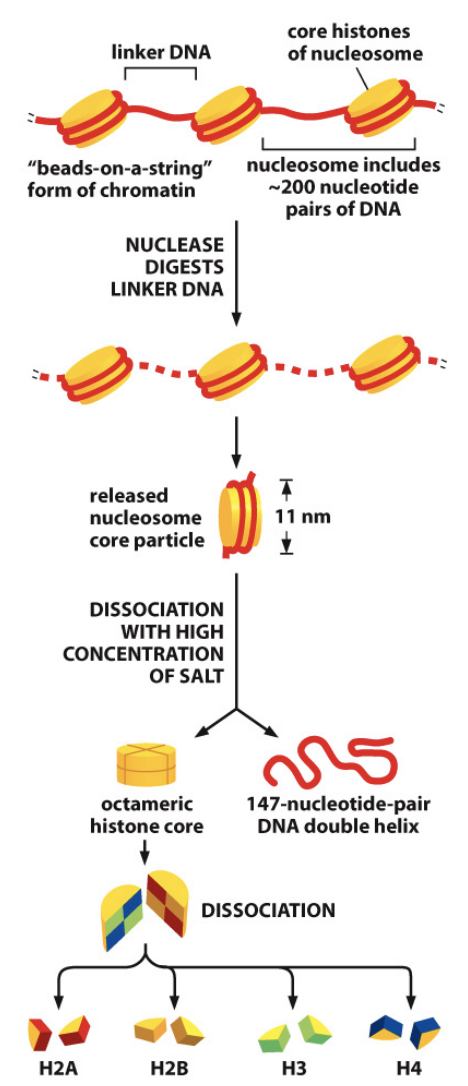
Describe a nucleosome.
String of DNA wrapped around a histone core, the first and most basic level of chromosome packaging. The histone core is made up of eight histone proteins, two molecules each of histones H2A, H2B, H3 and H4. A whole nucleosome includes about 200 pairs of DNA, but only 147 are wrapped around the histone core, with the rest linking the nucleosome to the next. The 147 DNA pairs wrap 1.7 times around the histone core.
What is the difference between mitosis and meiosis?
Mitosis produces two genetically identical “daughter” cells from a single “parent” cell though 1 cell division, whereas meiosis produces 4 cells that are genetically unique from the parent and contain only half as much DNA through 2 cell divisions. Mitosis happens to all cells in the body while meiosis only happens to our sex cells (eggs and sperm).
What are and what happens in the main 4 phases of the cell cycle?
G1, S, G2, M (and G0). G1, S and G2 are collectively called the interphase, while the M-phase is mitosis. G0 is a resting phase (inactive cell cycle) which all cells go into when not duplicating, but duration varies between cell types and differences in environment. In G1 the cells grow and membranes and organelles are synthesized. In S-phase the chromosomes are replicated for DNA synthesis, also histone synthesis for chromosome packaging and centrosome duplication. In G2 the cell grows further and prepares for division. In M-phase cell division occurs in two parts, nuclei division and cytoplasmic division, which both occur in 6 different steps.
Describe the 6 steps of the M-phase in the cell cycle.
Prophase - Chromosomes condense and become visible, spindle fibers emerge from the centrosomes, nuclear envelope breaks down, nucleolus disappears.
Prometaphase - Chromosomes continue to condense, kinetochores appear at the centromeres, mitotic spindle microtubules attach to kinetochores, centrosomes move toward opposite poles.
Metaphase - Mitotic spindle is fully developed, centrosomes are at opposite poles of the cell, chromosomes are lined up at the metaphase plate, each sister chromatid is attached to a spindle fiber originating from opposite poles.
Anaphase - Cohesin proteins binding the sister chromatids together break down, sister chromatids (now called chromosomes) are pulled toward opposite poles, non-kinetochore spindle fibers lengthen which elongates the cell.
Telophase - Chromosomes arrive at opposite poles and begin to decondense, nuclear envelope material surrounds each set of chromosomes, the mitotic spindle breaks down.
Cytokinesis - The cytoplasm is divided into two by a contractile ring of actin and myosin
filaments. This ring pinches the cell in two to create two daughters, each with one nucleus.
What is a cell cycle checkpoint and what are the 3 most common ones?
A control mechanism that prevents entry into next phase until current one is correctly completed. It functions as a binary switch, switching on/off.
The first one is the restriction point, which is in the end of G1 and checks whether the cell is big enough and has made the proper proteins for the synthesis phase, if not it goes into G0.
The second one is G2/M transition, which occurs between G2 and M-phase, and controls if all DNA is replicated correctly and if the environment inside and outside the cell is favorable.
The last one is the metaphase to anaphase transition where it checks if all chromosomes are attached to the spindle and if mitosis is complete before cytokinesis.
Which are the key proteins/enzymes involved in the cell cycle controls?
The cell cycle is driven by kinase proteins called Cdks (cyclin driven kinases). Enzymes control the activity of Cdks, where the most important enzyme is cyclin. Association with cyclin is needed for catalytic activity. When bound, it triggers specific cell-cycle events. Not bound – no Cdk activity. The cyclin also determines specificity of Cdk and participates in the substrate binding (adds specificity). Cdks can both activate and inhibit phosphorylation and act in ubiquitination. The rise and fall of different cyclin levels is what primarily determines cdk activity. However there are also other mechanisms that help control Cdks levels at different stages of the cell cycle, for example:
The Wee1 kinase – inhibits Cdk activity by phosphorylating certain sites on the Cdks where cyclin normally binds in.
The Cdc25 kinase – increases Cdk activity by dephosphorylation of these same sites.
We also have proteins that inactivate the entire Cdk-cyclin complexes, called CIP (Cdk inhibitor proteins). When they bind, the active site structure rearranges. The CIP family can inhibit all Cdks and the INK4 family can only inhibit Cdk 4/6 (active in G1).
Other:
E2F - regulate the genes that encode the passage of proteins required for the S-phase entry.
RB - Retinoblastoma protein, binds to and inhibits E2F, results in gene being deactivated and the cell not passing the restriction point.
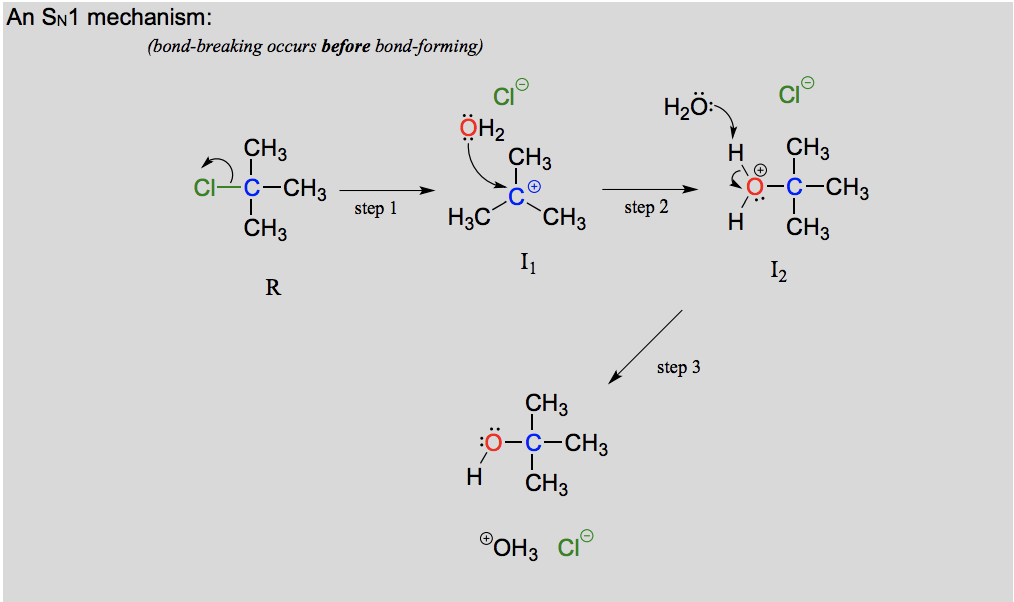
Describe and draw an SN1 mechanism including transition state.
Leaving group leaves first, making a carbocation intermediate before the nucleophile attacks. Actually a three step mechanism with a rapid acid-base step at the end. A unimolecular reaction, the rate determining step is dependent on one molecule (when the leaving group leaves).
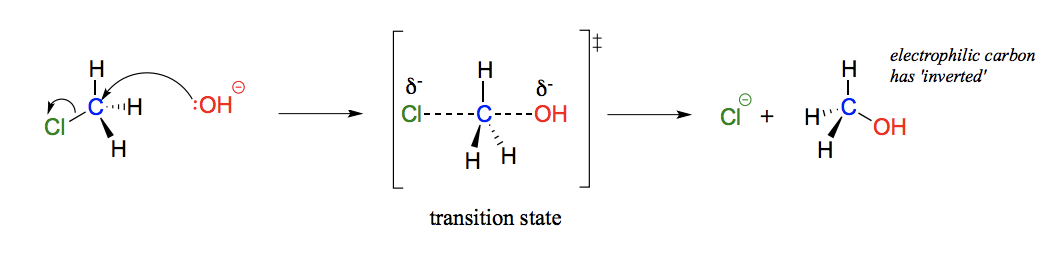
Describe and draw an SN2 mechanism including transition state.
Bond-breaking and bond-forming occur simultaneously, it is concerted (happens in one step). A bimolecular reaction, the rate determining step is dependent on two molecules colliding. Attack occurs on the back side.
What is a nucleophile, electrophile and leaving group?
Nucleophile - electron-rich, nucleus-loving species.
Electrophile - electron-poor species which is attracted to electrons.
Leaving group - the group that leaves in the reaction.
What does the energy diagram of an SN1 reaction look like?
(See picture)
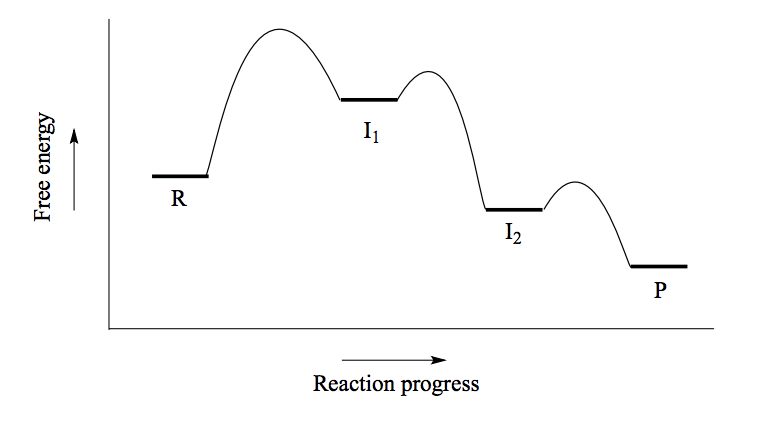
What does the energy diagram of an SN2 reaction look like?
(See picture)
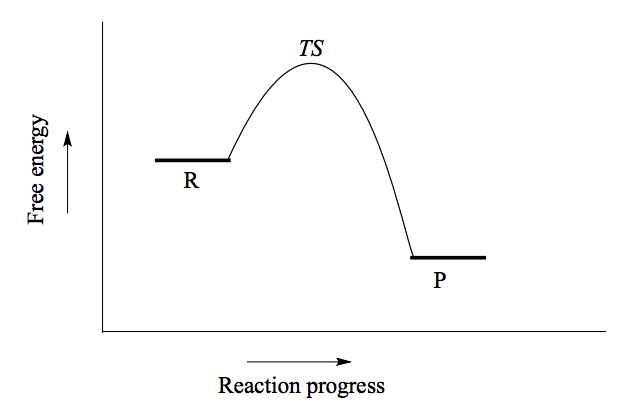
How is stereochemistry affected by SN1 and SN2 reactions (nucleophilic substitution)?
SN2 reactions always invert stereochemistry as it is a backside attack. SN1 reactions can have two products, since the nucleophile can attack from either side of the planar carbocation intermediate, the one with the same stereochemistry and the one with inverse.
Explain the differences in stereochemistry between enzymatic and non-enzymatic SN1 reactions.
Non-enzymatic reactions should result in a racemic product of both stereoisomers of the molecule (if not affected by steric hindrance or other disturbing factors). Enzymatic SN1 reactions usually favour one of the stereoisomers, meaning that the product will almost exclusively contain one stereoisomer. This because the two reactants are bound with specific geometry at the enzymes active site, so that the nucleophile can approach from one side only.
What is hydrolysis vs solvolysis?
Hydrolysis = breaking with water, a water molecule participates in the breaking of a covalent bond.
Solvolysis = more general term, used when a bond is broken by a solvent molecule. Hydrolysis is a form of solvolysis.
How do you know which is a good nucleophile?
When evaluating nucleophilicity, we are speaking in terms of kinetics - how fast does the nucleophile react?
Protonation state - negative charge = stronger nucleophile.
Periodic trends - more nucleophilic to the left, less nucleophilic to the right. In a PROTIC solvent, the stronger nucleophile is at the bottom of the periodic table (by I) and the weaker nucleophile is at the top (by F). In a POLAR APROTIC solvent, the stronger nucleophile is at the top of the periodic table (by F) and the weaker nucleophile is at the bottom (by I). Remember (in biochem), thiols are more nucleophilic than alcohols.
Resonance - more resonance = less nucleophilic (delocalized charges are less reactive).
Steric hindrance - more bulky (more steric hindrance) = less nucleophilic.
Inductive effect - if an electron withdrawing group is bonded (positive), this decreases nucleophilicity since it is taking away charge from the nucleophile, while an electron donating group increases nucleophilicity.
When comparing the rate of nucleophilic substitution reactions, the strength of the nucleophile only matters for SN2 reactions because it is the one where the nucleophile is involved in the rate determining step (it is not in an SN1 reaction).
Why is the nucleophilicity trend different in different solvents?
Protic solvents molecules form strong noncovalent interactions with the electron-rich nucleophile, essentially creating a “solvent cage” of hydrogen bonds, meaning that the nucleophile needs to get past this “cage” in order to be able to attack. Therefore a weaker base (like iodine) interacts weakly with the protons of the solvent and the interactions are more readily disrupted. Also the valence electrons on iodine are far from the nucleus, making its electron cloud polarizable, meaning it can easier escape.
In aprotic solvents, a “solvent cage” is not formed, meaning that it follows the same trends as in basicity. Fluoride is the strongest base and the strongest nucleophile.
What is a protic/ aprotic solvent?
Protic = a solvent that has a hydrogen atom bonded to an oxygen or nitrogen, ex. water, methanol. Relevant in biochemical reactions because water is the solvent.
Aprotic = a solvent that is polar enough to solvate the compounds in the reaction, but is not a hydrogen bond donor, ex. acetone, dimethylsulfoxide (DMSO).
What affects the reaction rate of an SN1 and SN2 reaction?
In an SN1 reaction the rate determining step is when the leaving group leaves, while in an SN2 reaction it is when the two reactants collide. This means that an increase of nucleophiles will affect the reaction rate of an SN2 reaction but not an SN1 reaction.
How do different properties affect the reactivity of an electrophile?
Electrophile is usually a carbon bonded to an electronegative atom.
Steric hindrance - A bulky electrophile will not be able to go through a SN2 reaction because the backside attack is hindered, only SN1 is possible. SN2 reactions occur at methyl, primary and secondary carbon electrophiles, with methyl reacting fastest (least bulky). Tertiary carbon reactions with SN2 do not happen. Steric hindrance on electrophiles do not affect the rate of an SN1 reaction as it is not part of the rate-determining step.
Carbocation stability - this affects the electrophile of an SN1 reaction, depends on the stability of the carbocation intermediate.
What factors affect carbocation stability and how?
Any electron donating group will stabilize a carbocation (EWG destabilizes).
More substituted carbocations are more stable (primary highly unstable, almost not found, tertiary very stable). This is because of hyperconjugation and the fact that alkyl groups are weak electron-donating groups.
Carbonyl groups (double bonded to a substituent) destabilize a carbocation, acts as a EWG, a carbocation further away from a carbonyl group is more stable.
Resonance stabilizes a carbocation, because of delocalization of the charge. This also means that allylic carbocations (carbocation is adjacent to double bond between carbons) are stabilized by resonance.
Hetero atoms (oxygen and nitrogen) can be destabilizing by being electron withdrawing, but are more often stabilizing because they contribute to resonance.
Vinylic carbocations, where the positive charge is on a double bonded carbon, are very unstable.
What does hyperconjugation mean?
When the empty p orbital of a carbocation is stabilized by overlap with a sigma bond on an adjacent carbon.
What are the properties of a good leaving group?
Weaker bases are better leaving groups. I better than F. F better than H.
Give examples of leaving groups in the lab and in the body.
In body - phosphate, water, alcohol, thiol.
Lab - Anything that is too acidic for body, ex acetone or dimethylformamide.
How can you predict if a reaction will proceed by an SN1 or SN2 reaction?
Electrophile - SN1 has a stable carbocation intermediate. SN2 requires unhindered electrophilic center.
Nucleophile - SN2 usually has a more powerful, anionic nucleophile, while SN1 usually has a weak, neutral nucleophile. Based on type of nucleophile, electrophile and solvent.
Solvent - Protic solvents favor SN1, ability to stabilize carbocation intermediates & weaker nucleophile because of solvent cage. Polar aprotic solvents favor SN2 reactions (only in lab).
Factors favoring the SN1 pathway:
Hindered electrophile
Potential for a stable carbocation intermediate
Uncharged nucleophile
Protic solvent such as water
Factors favoring the SN2 pathway.
Unhindered (methyl or primary) electrophile
Powerful, anionic nucleophile
Polar aprotic solvent
How does S-adenosylmethionine (SAM) function as a methyl group donor in a biochemical SN2 reaction?
The sulfide and 2 R groups (basically whole SAM molecule) act as a leaving group, the methyl group becomes the electrophile and a nucleophile on another molecule attacks to form a bond where SAM has donated it´s methyl group.
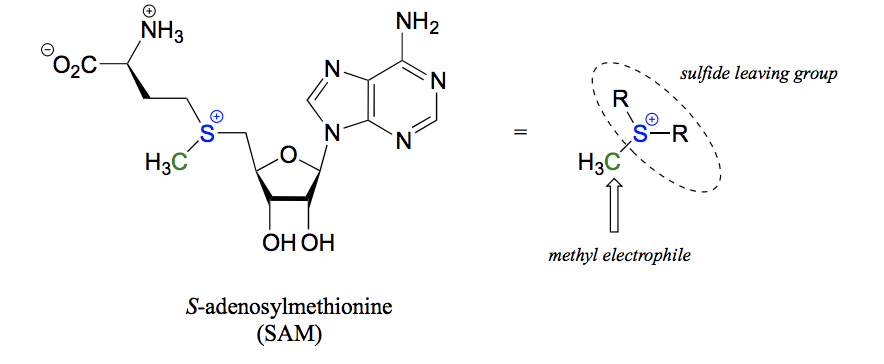
What is a phosphate?
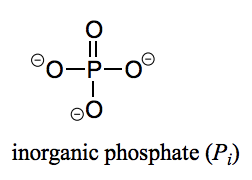
What is a triphosphate?
Three phosphate groups serially bonded.
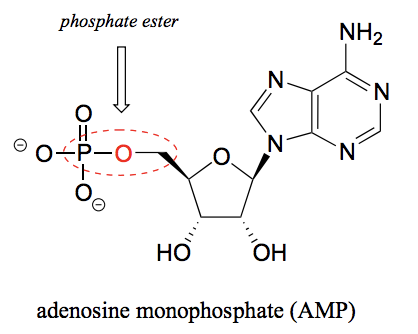
What is a phosphate ester?
The chemical linkage between phosphate and a carbon atom. For example between the ribose and phosphate group in AMP.
What is a pyrophosphate?
Two phosphate groups linked to each other. Inorganic pyrophosphate is a pyrophosphate not bound to anything else (PPi).
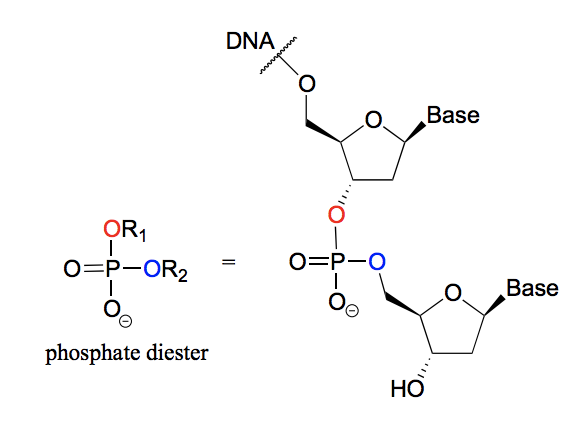
What is a phosphate diester?
A phosphate group with two phosphate ester bonds. For example in DNA, where the nucleotide bases are linked by phosphodiester bonds.
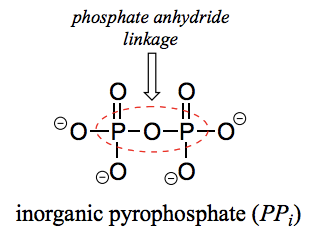
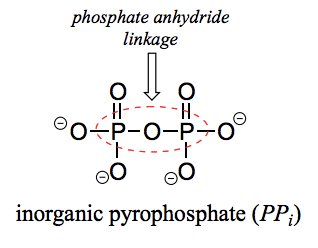
What is a phosphate anhydride?
When two phosphate groups are linked to each other, the linkage itself is called a phosphate anhydride.
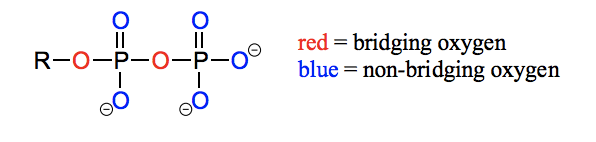
What is diphosphate bridging/ non-bridging oxygen?
Oxygen atoms in phosphate groups are referred to as wither bridging or non-bridging depending on their position. An organic diphosphate has two bridging oxygens (one in the phosphate ester linkage and one in the phosphate anhydride linkage) and five non-bridging oxygens:
What is Pi, PPi, OP, R-OP, R-OPP, R-OAMP, ATP, ADP, AMP?
Pi - Inorganic phosphate (one phosphate group not bound to anything)
PPi - Inorganic pyrophosphate (two phosphate groups not bound to anything)
OP - Organic phosphate (phosphate bound to something)
R-OP - Monophosphate bonded to an R-group.
R-OPP - Diphosphate bonded to an R-group.
R-OAMP - AMP bonded to an R-group.
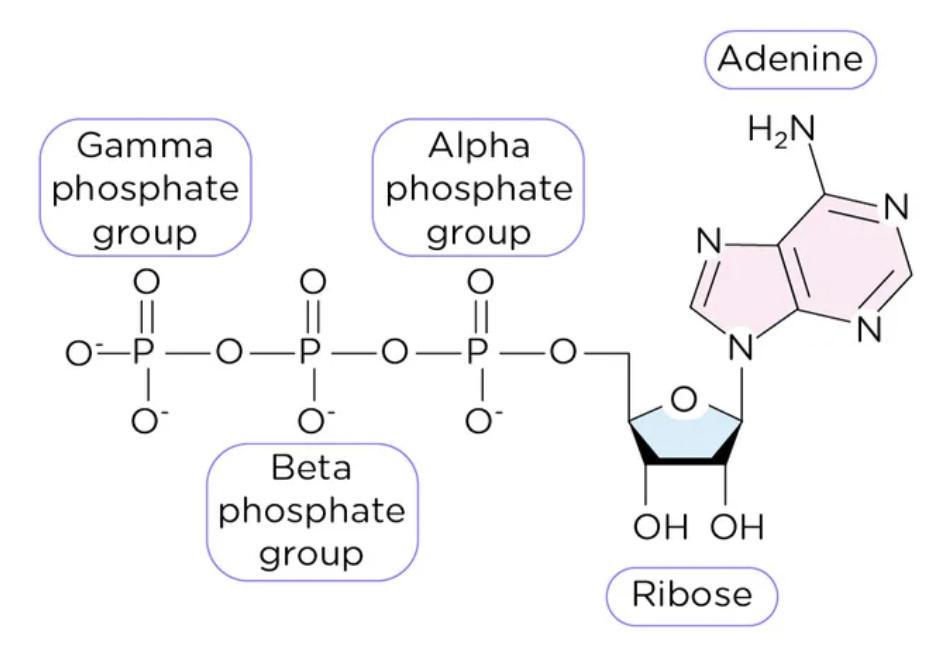
ATP - Adenosine triphosphate = a nitrogenous base (adenine), a ribose sugar, 3 serially bonded phosphate groups.
ADP - Adenosine diphosphate = a nitrogenous base (adenine), a ribose sugar, 2 serially bonded phosphate groups.
AMP - Adenosine monophosphate = a nitrogenous base (adenine), a ribose sugar, 1 bonded phosphate group.
What are the resonance structures of phosphate and why is one minor and one major?
The double bond could be to any of the four oxygens surrounding the phosphate. However, the major resonance contributors are the ones where the double bond is to a non-bridging oxygen, and the minor resonance structure contributors are where the double bond is to a bridging oxygen. This is because in the minor resonance structures, there is an additional separation of charge, and the bridging bonds do not have as much double bond character. It is important to remember however that the negative charges are delocalized over the non-bridging oxygens.
Describe α, β and γ phosphate groups of ATP, as well as ribose and adenosine moieties of the molecule.
How does ATP act as a phosphate group donor?
Can do this by attack on either the gamma, beta, or alpha phosphate, resulting in different products.
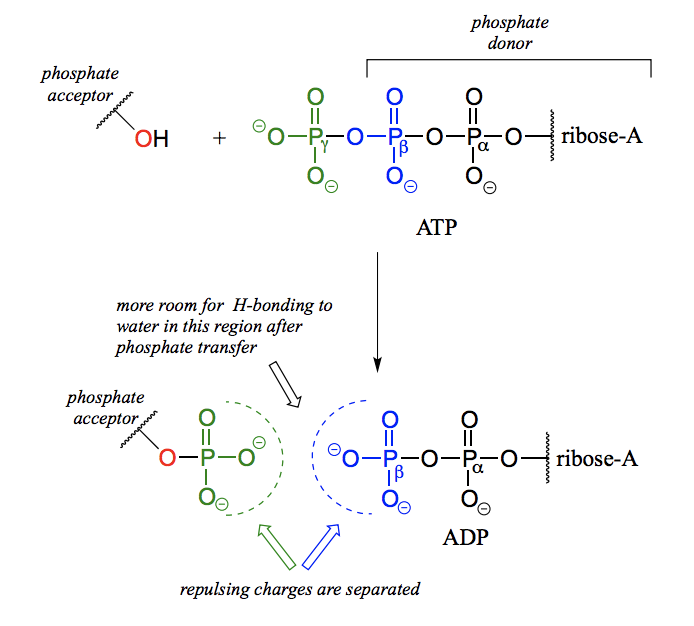
Why are reactions where ATP acts as a phosphate group donor thermodynamically favorable?
Because the phosphate anhydride bonds in ATP are thermodynamically unstable (they contain a lot of chemical energy), and breakage of that bond will mean release of that energy.
Draw the reaction for when ATP acts as a phosphate group donor in the phosphorylation of an alcohol (glucose).
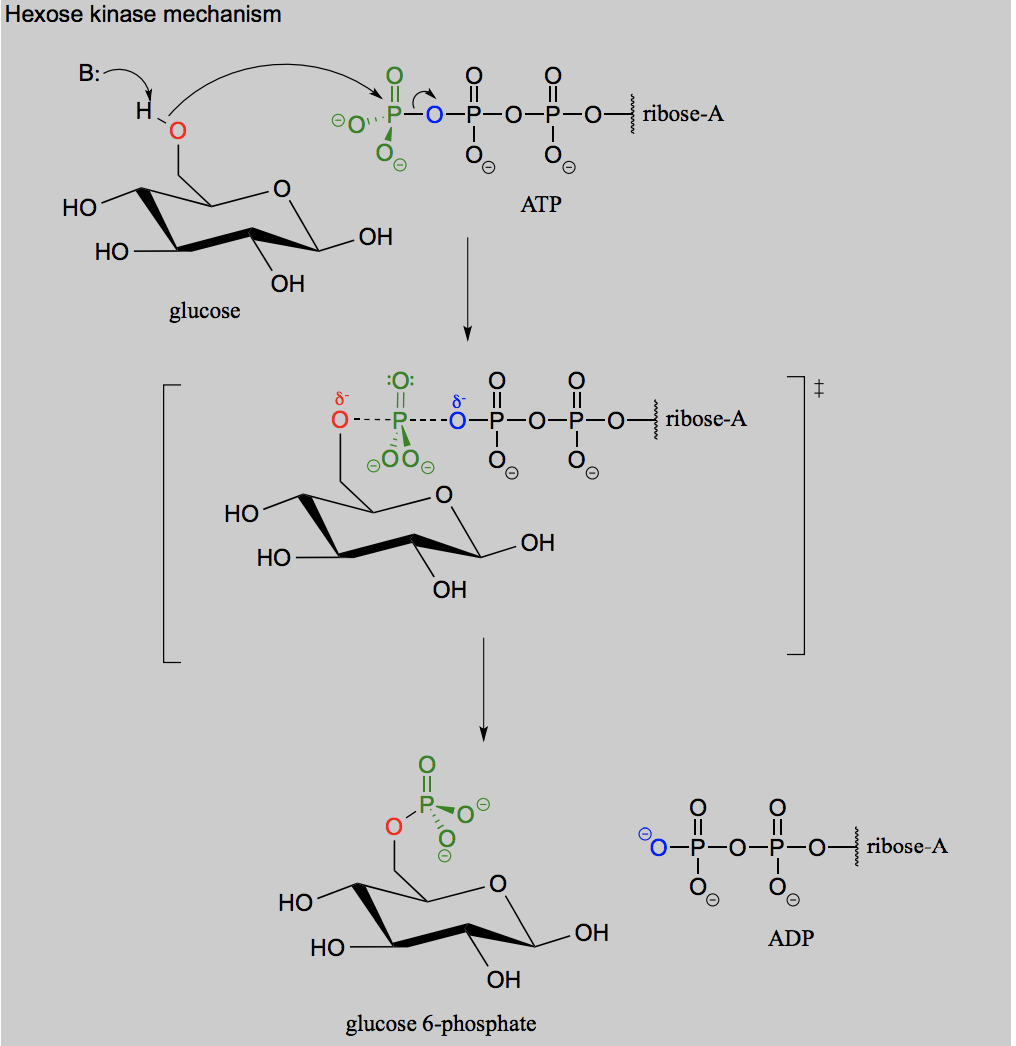
Draw the reaction for hydrolysis of a phosphorylated alcohol.
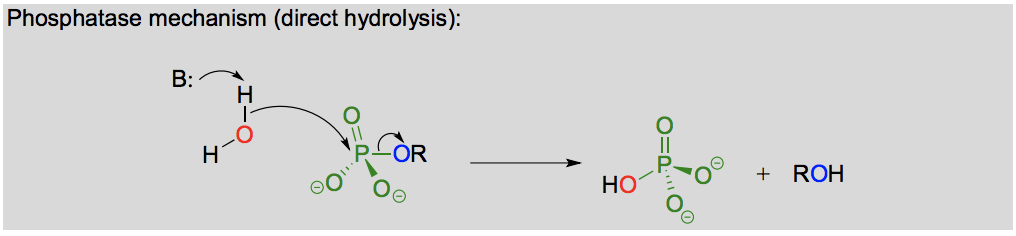
What is the difference between a direct and indirect hydrolysis reaction?
Direct hydrolysis is direct attack on a phosphor to cleave it off, while indirect hydrolysis in when a nucleophilic enzyme group (ex. cysteine) attacks the phosphate first and cleaves it off the substrate, and then the water attacks the phosphate that is bonded to the nucleophilic enzyme group and breaks it off.
Draw the reaction for phosphorylation of carboxylate (conjugate base of carboxylic acid).
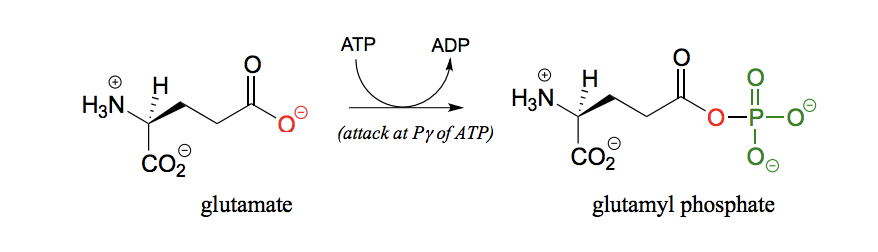
Give examples of phosphate reactions in the construction, repair and degradation of DNA and RNA.
Phosphodiester bonds between nucleotides in DNA and RNA. Nucleases degrade DNA and RNA by hydrolysing these bonds. DNA much more stable than RNA, because the extra -OH group on RNA can hydrolyse itself and degrade the RNA. DNA polymerase uses phosphate reactions to build the backbone of DNA by making phosphodiester bonds between to bases by the -OH group on one and the dNTP on the other. DNA can also be repaired by using ATP to ligate two cleaved off strands of the DNA.
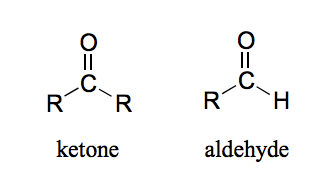
What is an aldehyde and a ketone?
The carbon in a ketone is bonded to two carbons while the carbon in an aldehyde is bonded to one (or two) hydrogens.
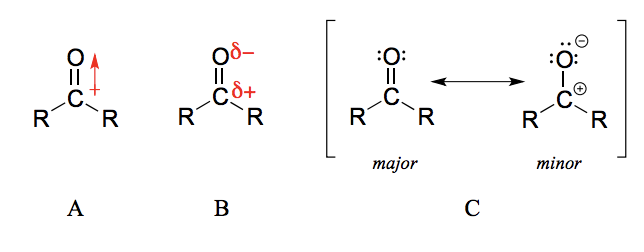
Why is the carbonyl carbon in an aldehyde/ ketone electrophilic?
The carbon-oxygen double bond is polar: oxygen is more electronegative than carbon, so electron density is higher on the oxygen end of the bond and lower on the carbon end.
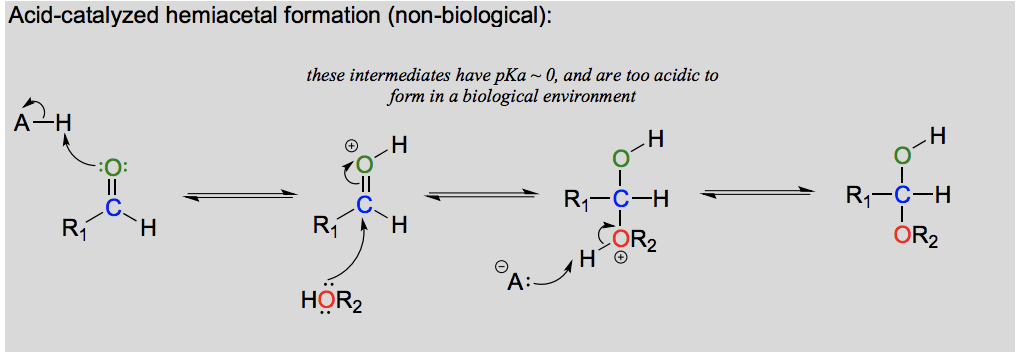
Draw the formation of a hemiacetal from an aldehyde (in the lab)
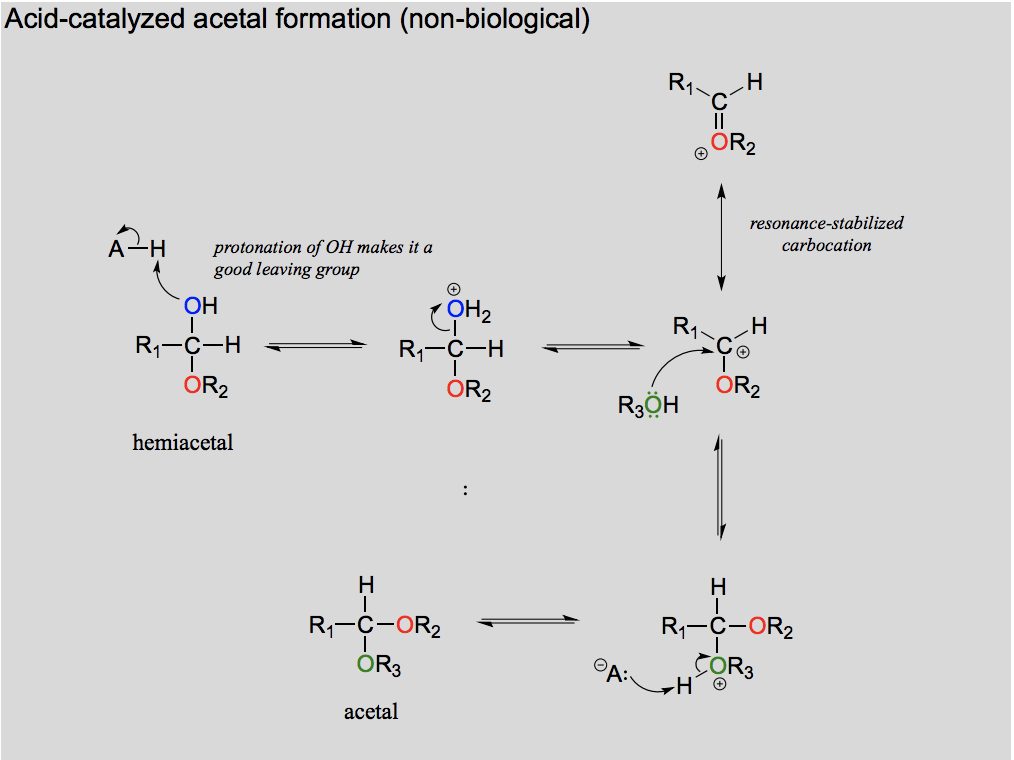
Draw and explain the formation of acetal from a hemiacetal (in the lab).
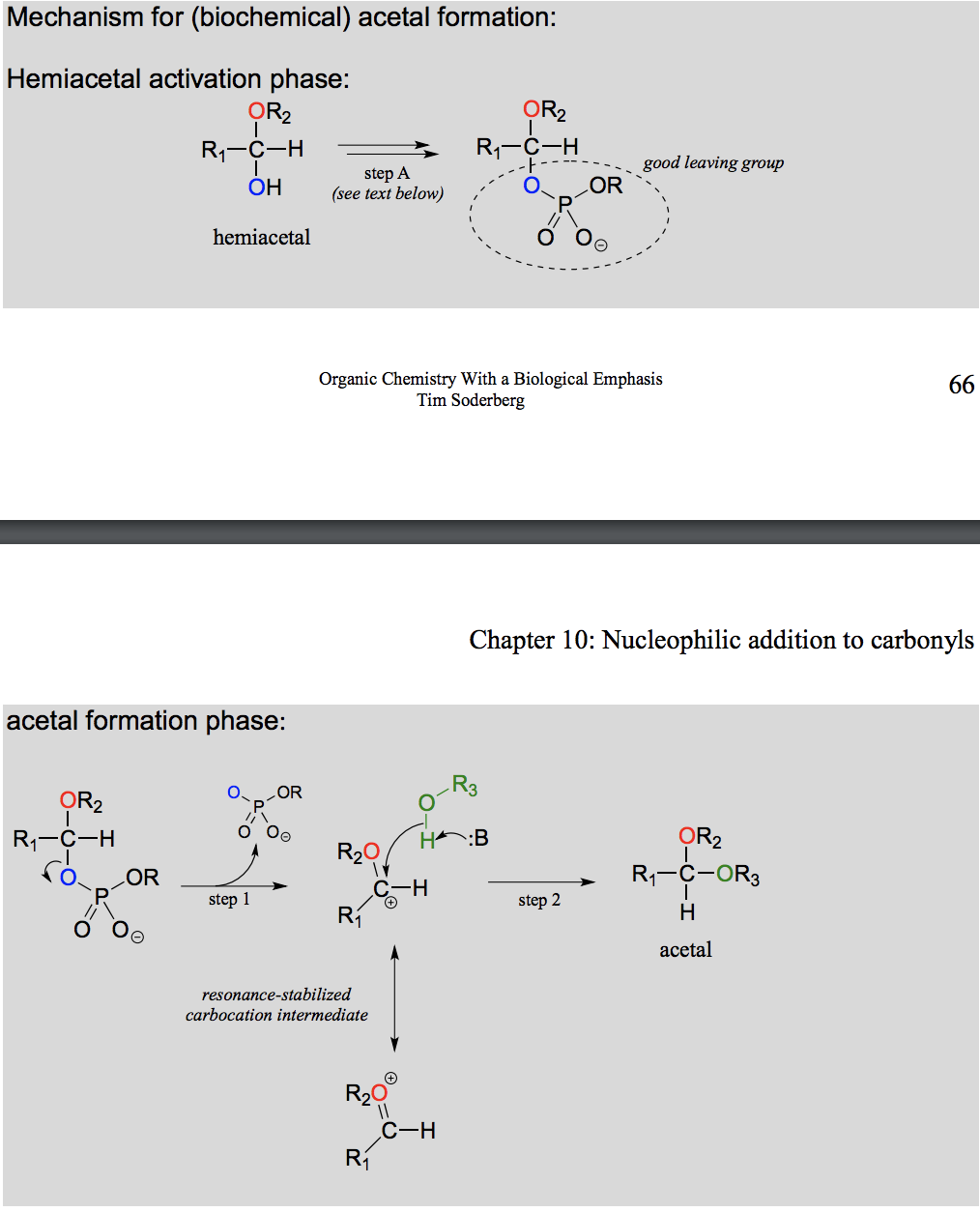
Draw and explain the formation of acetal from a hemiacetal (in the body).
Activation is needed as without it the intermediate is too acidic to be in the body.
What is the difference between a acetal/ ketal formation in the lab and enzymatically in the body?
In the body, the hemiacetal/ hemiketal needs to be activated first, in order to avoid an intermediate that is too acidic for the bodys pH.
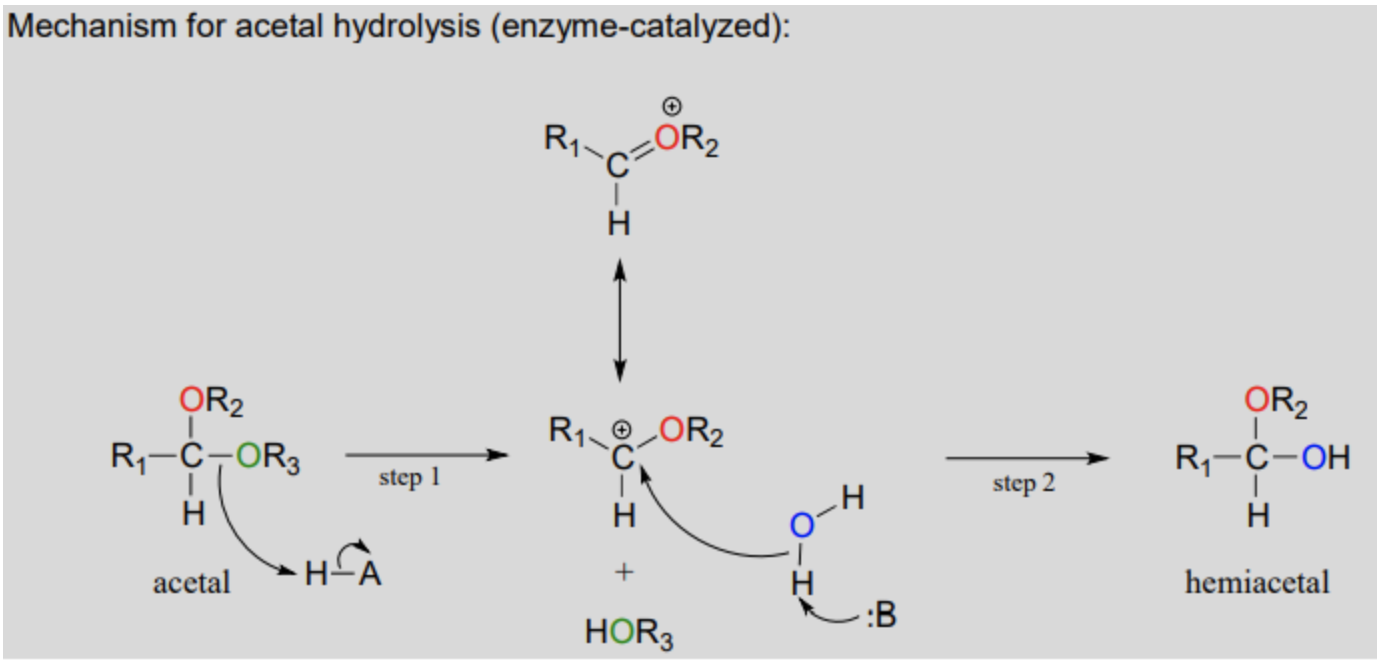
Draw and explain the hydrolysis of an acetal to a hemiacetal.
Draw and explain the formation of an N-glycoside.
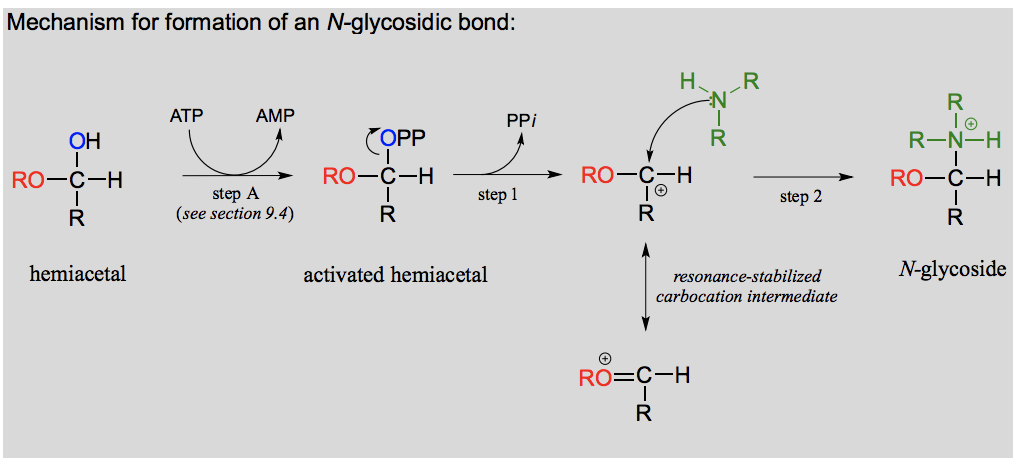
What is an N-glycosidic bond?
If a hemiacetal is attacked not by a second alcohol but by an amine, what results is a kind of ‘mixed acetal’ in which the anomeric carbon is bonded to one oxygen and one nitrogen.
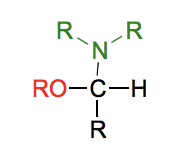
Draw and explain the formation of an imine.
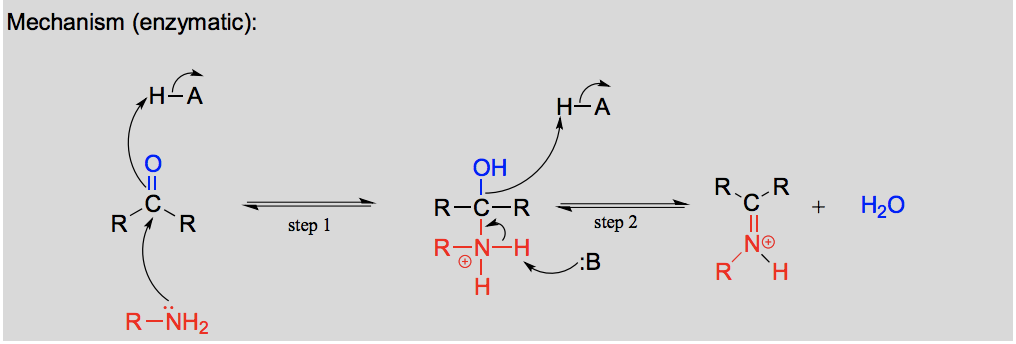
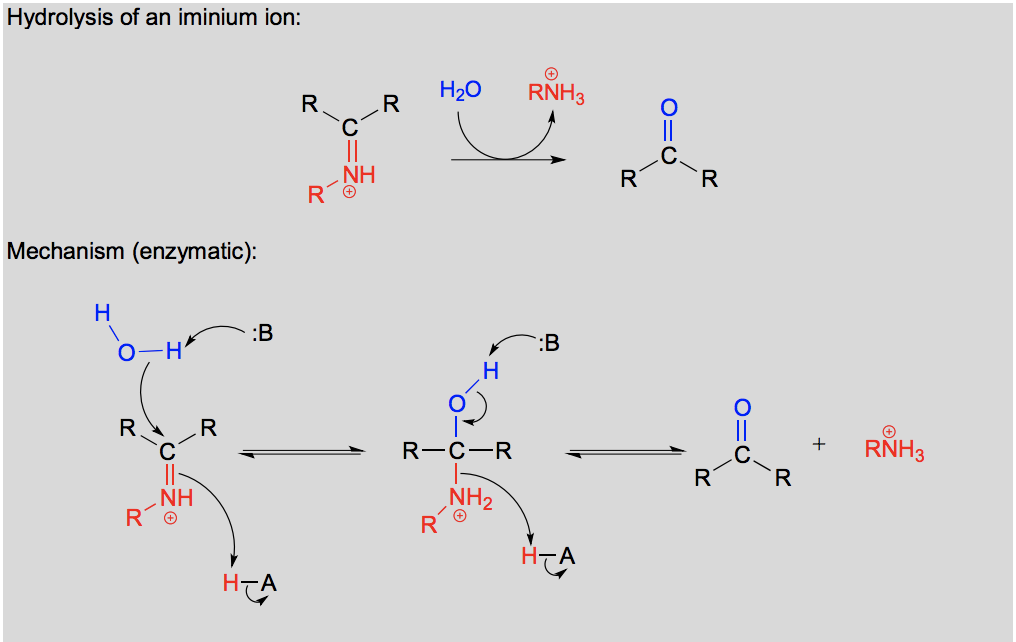
Draw and explain the hydrolysis of an imine.
What is an anomeric carbon?
An anomeric is carbon a carbon in a sugar that is an aldehyde or ketone in the open-chain form and becomes a stereocenter in the cyclic form. The anomeric carbon forms a hemiacetal or hemiketal, in which it is bonded to the ring oxygen and a hydroxyl group.
Explain the balance between open and closed forms of glucose and stereochemical aspects.
Sugars of five or six carbons rapidly interconvert between straight-chain and cyclic forms. This occurs through the formation of intramolecular hemiacetals and hemiketals. Sugar cyclization reactions are not catalyzed by enzymes. The cyclic forms predominate in equilibrium. Nucleophilic attack on a planar carbonyl group can occur at either face of the plane, leading to two different stereochemical outcomes - in this case, to two different diastereomers. These are referred to as the alpha and beta anomers of the sugar. There is almost twice as much of one anomer than the other at equilibrium. This is because six-membered rings exist predominantly in the chair conformation, and that the lower energy chair conformation is that in which unfavorable interactions between substituents are minimized – in most cases, this is the conformation in which larger substituents are in the equatorial position. In the lower-energy chair conformation of the major beta anomer of glucopyranose, all of the hydroxyl groups are in the equatorial position, but in the minor alpha anomer one hydroxyl group is forced into the axial position. As a result, the alpha anomer is higher in energy, and less abundant at equilibrium.
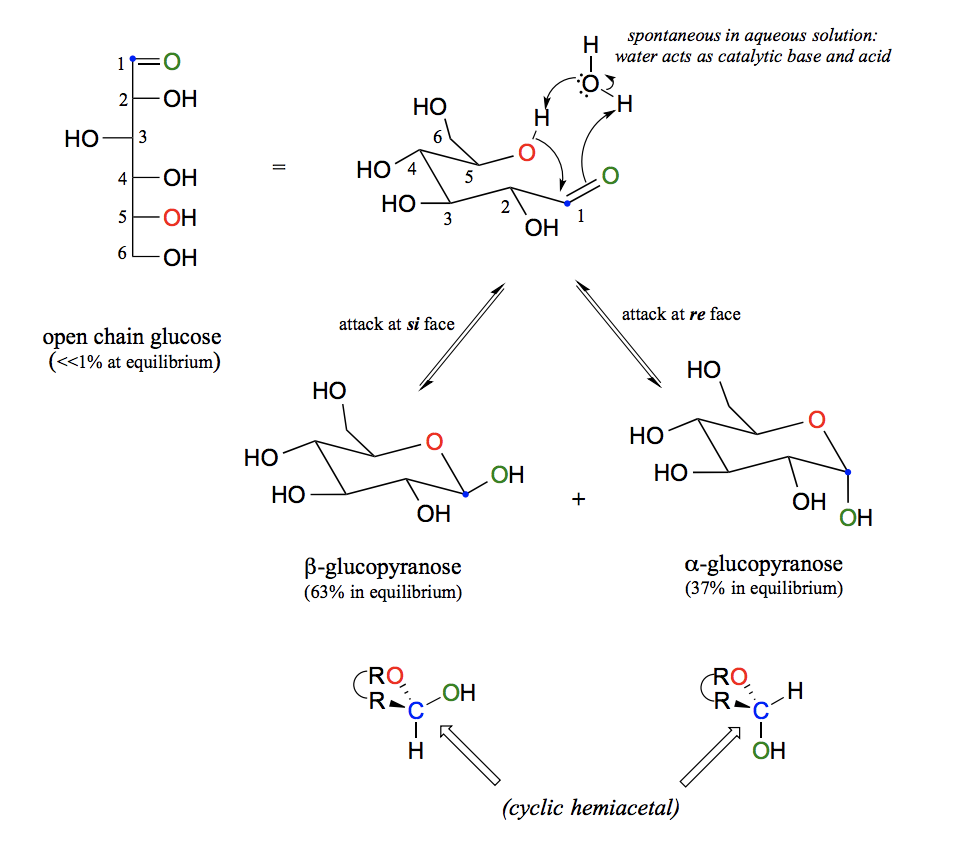
What type of reaction is a glycosidic bond formation?
An acetal/ ketal formation.
Draw the formation of a glycosidic bond.
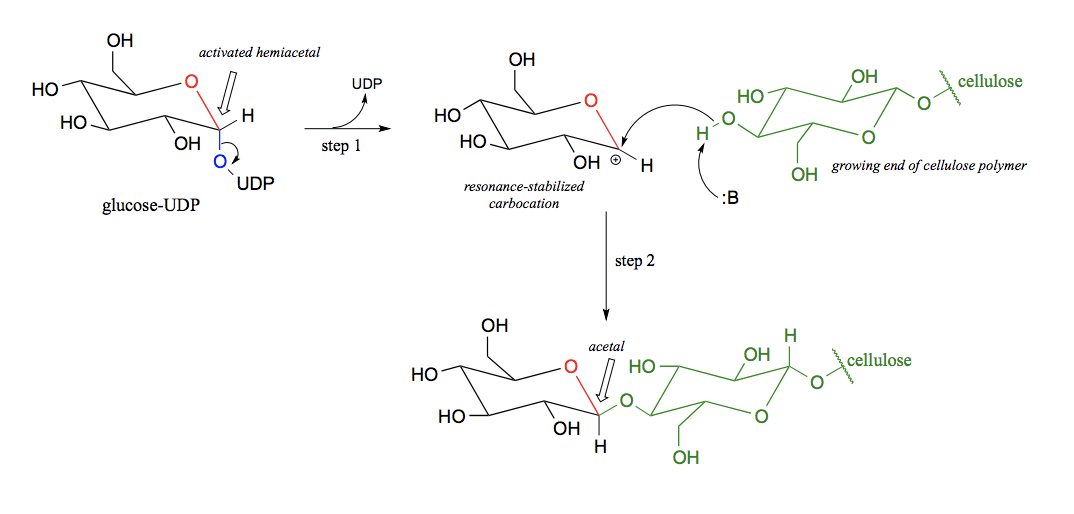
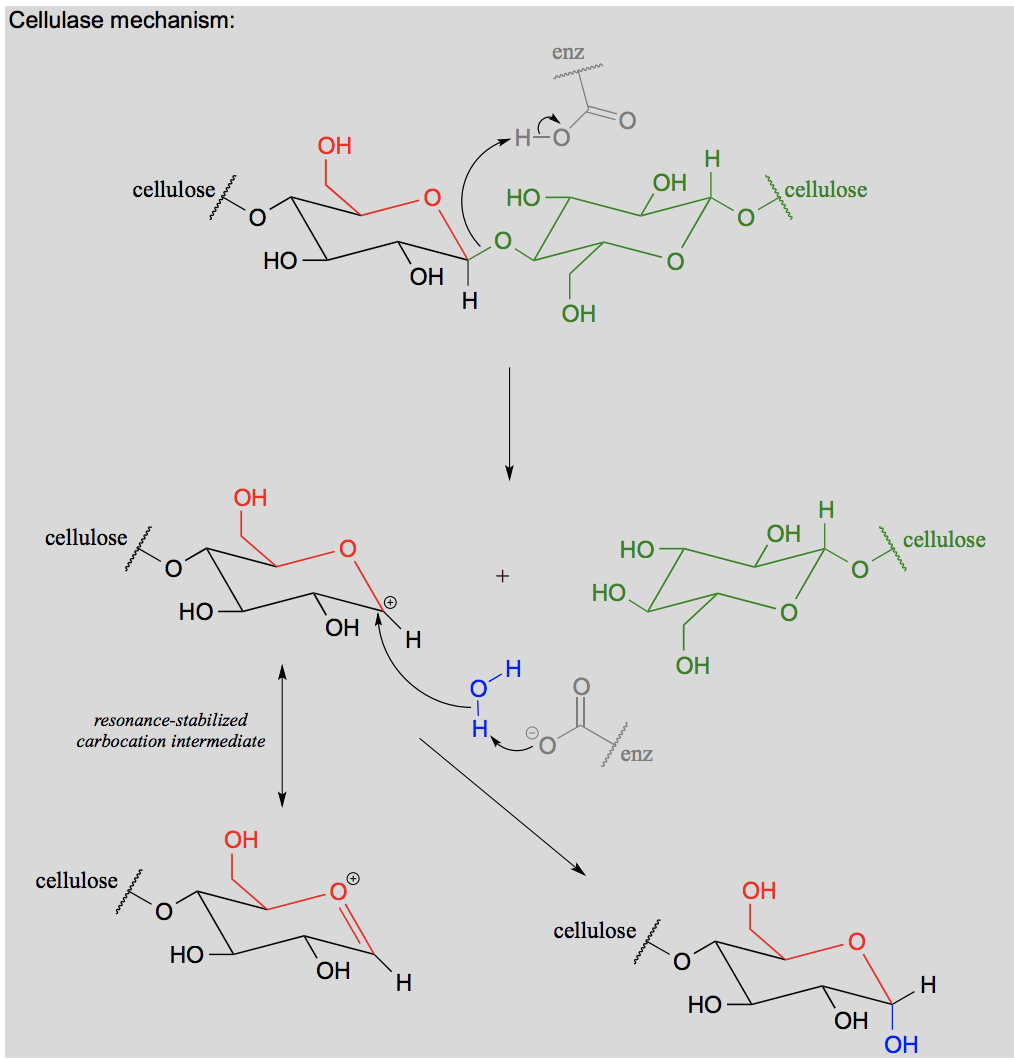
Draw a glycosidic bond hydrolysis.
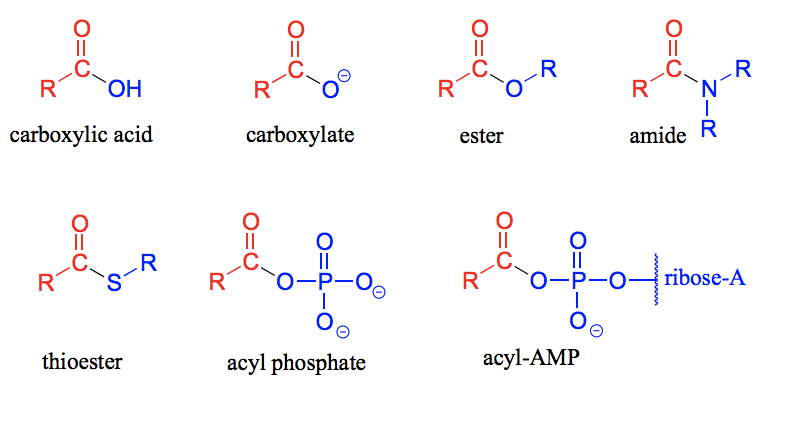
What is a carboxylic acid derivative?
Compounds in which the −OH group of the carboxylic acid is replaced by other functional groups are called carboxylic acid derivatives.
Draw the carboxylic acid derivatives: carboxylic acid, carboxylate, ester, amide, thioester, acyl phosphate and acyl-AMP.
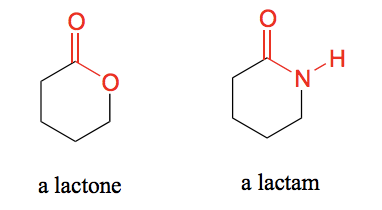
Draw a lactone and a lactam.
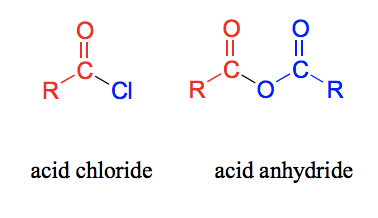
Draw an acid chloride and an acid anhydride.
What is meant by the following terms: acyl, acetyl, formyl, lactone and lactam?
Acyl - The part of a carboxylic acid derivative that is the carbonyl plus the attached alkyl (R) group.
Acetyl - When the R group in a carboxylic acid derivative is a hydrogen.
Formyl - When the R group in a carboxylic acid derivative is a methyl.
Lactone - A cyclic ester.
Lactam - A cyclic amide.
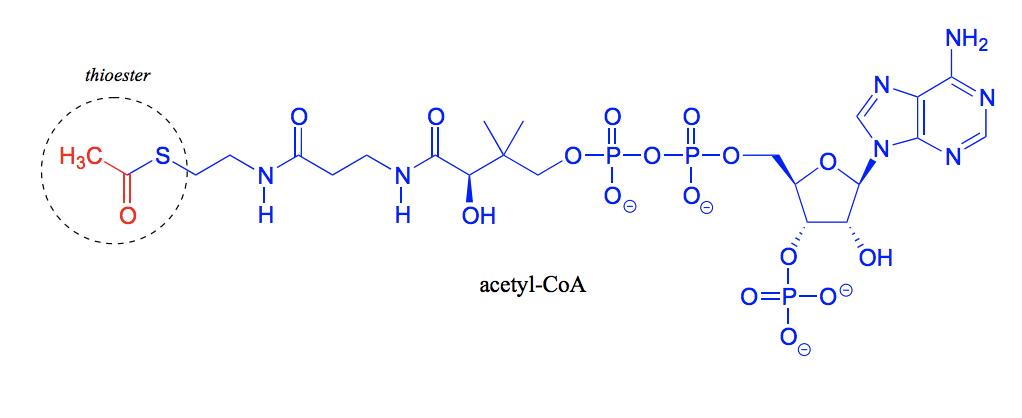
What is this molecule called? And which functional group is most important to point out in this molecule?
Coenzyme A. The thiol group.
What is the difference between a ketone/ aldehyde and a carboxylic acid derivative?
That in a carboxylic acid, the carbon is bonded to a electronegative heteroatom, while a ketone is bonded to a second R group and an aldehyde is bonded to a hydrogen. This means that a carboxylic acid derivative, unlike an aldehyde/ ketone has a potential leaving group (the “acyl X group”). This makes them undergo nucleophilic acyl substitution reactions instead of nucleophilic additions.
How are thioesters formed? And in which process?
In the fatty acid metabolism. With Coenzyme A as the thiol contributing group.
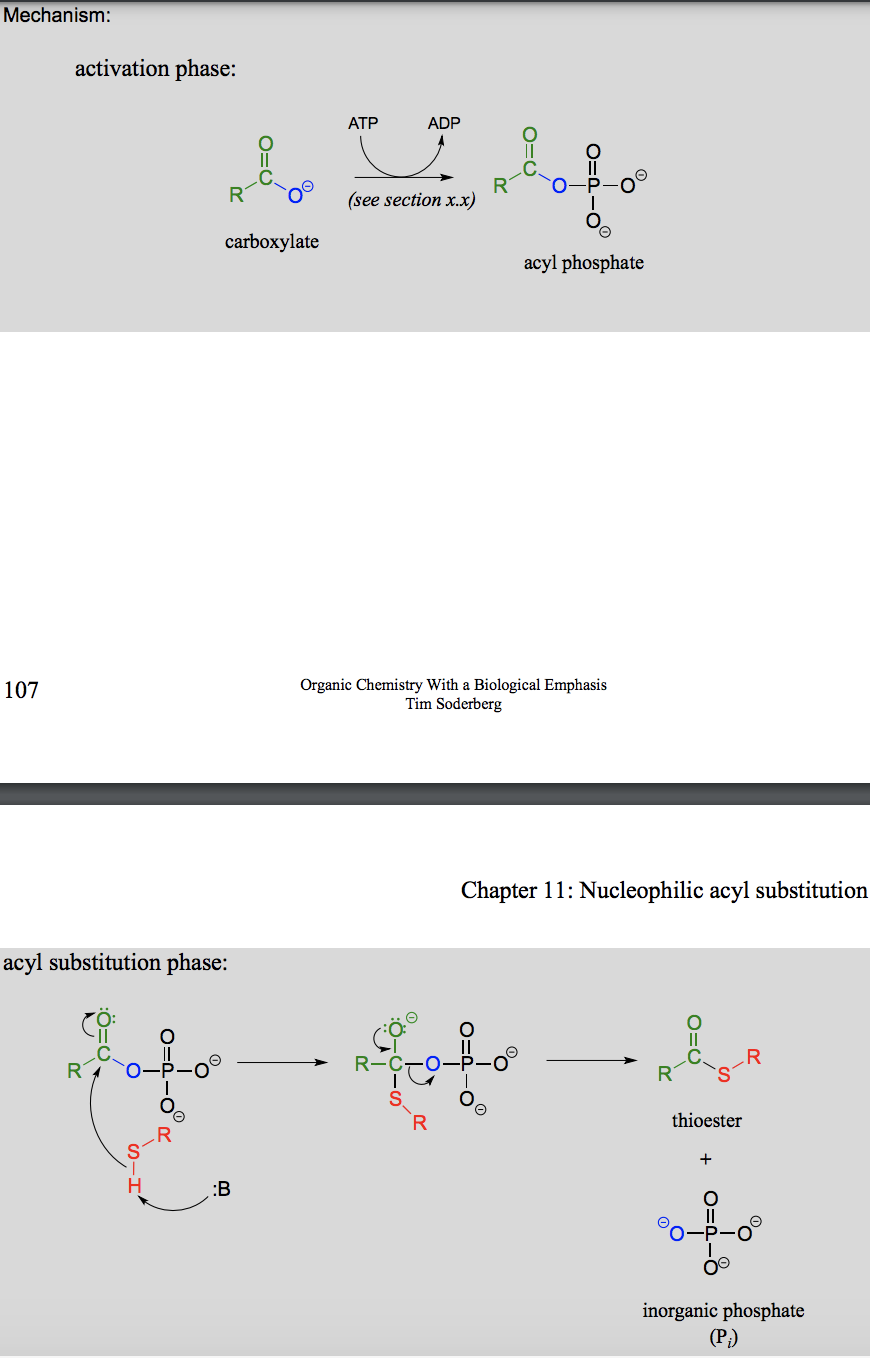
Draw a general mechanism for a nucleophilic acyl substitution reaction.
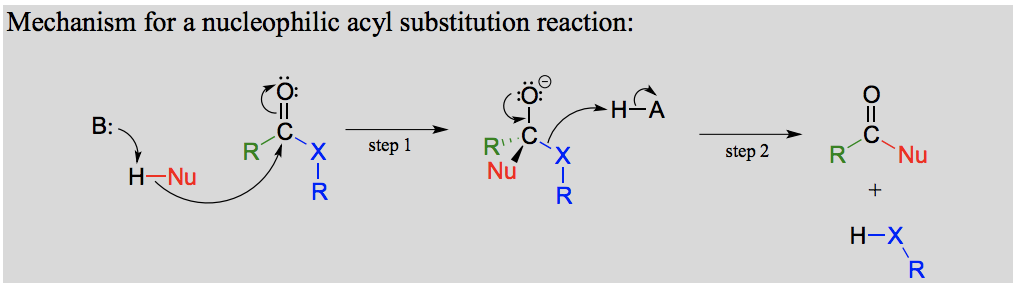
What is an oxyanion hole?
When enzymatic amino acid residues are positioned in the active site so as to provide stabilizing hydrogen bond donating interactions with a negatively charged oxygen.
What is the difference between conditions/ the mechanism in biological systems vs in the lab of a nucleophilic acyl substitution reaction?
Mechanistically, one of the biggest differences between the biological and the lab versions is that the lab reactions usually are run with a strong acid or base as a catalyst, whereas biological reactions are of course taking place at physiological pH. When proposing mechanisms, then, care must be taken to draw intermediates in their reasonable protonation states: for example, a hydronium ion (H3O+H3O+) intermediate is reasonable to propose in an acidic reaction, but a hydroxide (OH−OH−) intermediate is not.
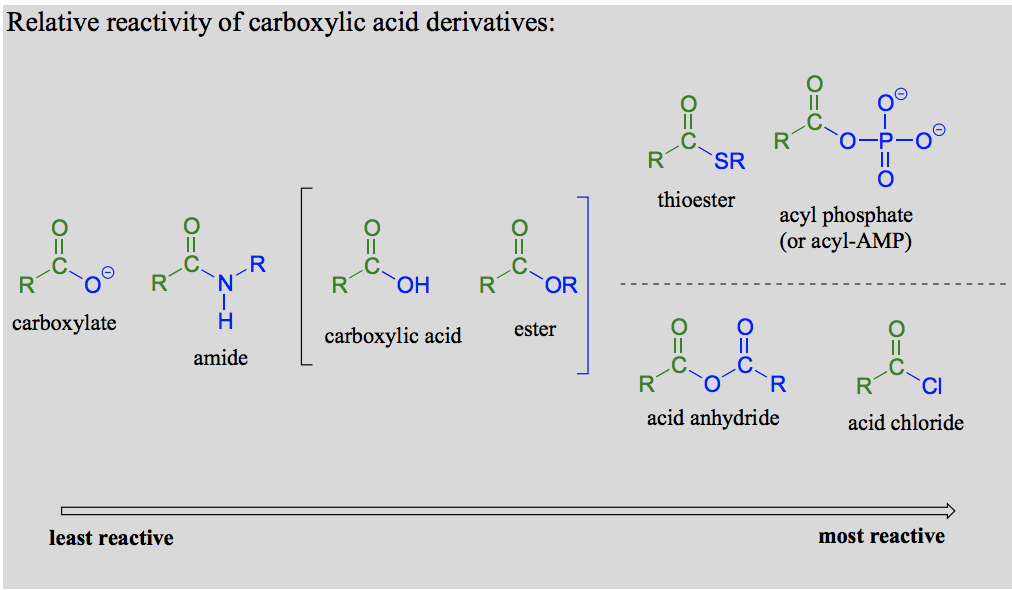
Explain the reactivity trend for carboxylic acid derivatives in biological systems and in the lab.
In carboxylic acid derivatives, the partial positive charge on the carbonyl carbon is stabilized by electron donation from nonbonding electrons on the adjacent heteroatom, which has the effect of decreasing electrophilicity.
Biologically (from highest to lowest): Acyl phosphates, thioesters, carboxylic acids, carboxylic esters, amides, carboxylate groups. (Acyl phosphates and thioesters most relevant).
In the lab: Acid anhydrides and acid chlorides (analogous to thioesters and acyl phosphates in the sense that they too are highly reactive carboxylic acid derivatives).
How is an NHS ester used for modification (tagging) of a protein?
Explain the difference in reactivity between an ester and a thioester.
A thioester is more reactive than an ester because a thiolate (RS-) is a weaker base and better leaving group than an alcoxide (RO- ).
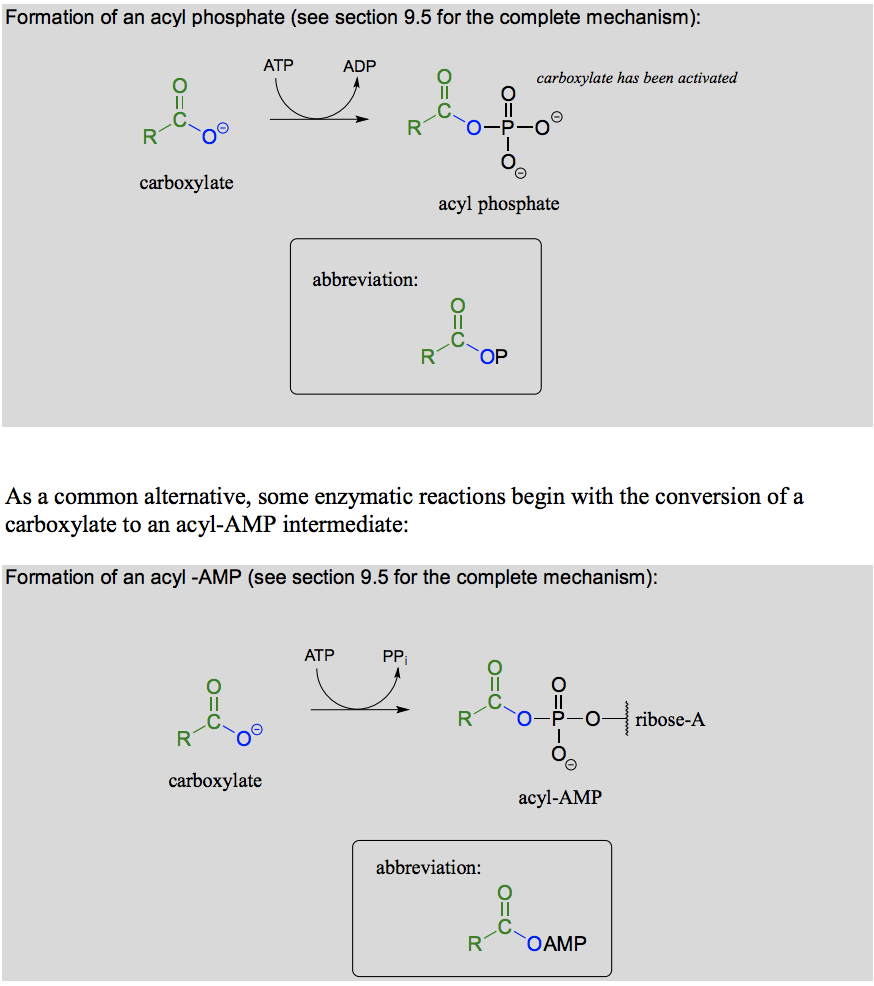
What reaction is this? What is happening mechanistically? What is the energy balance like (endergonic/exergonic) ?
Activation of carboxylic acid and formation of acyl monophosphate or acyl-AMP by ATP dependent phosphorylation. Carboxylate to acylphosphate is endotherm driven by the hydrolysis of 1 ATP molecule
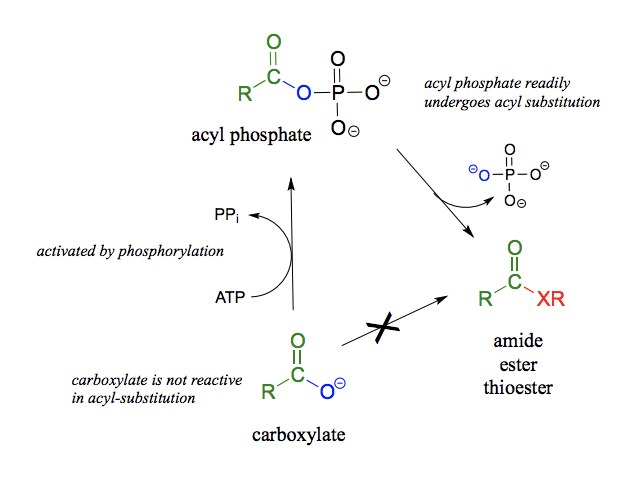
What reaction is this? What is happening mechanistically? What is the energy balance like (endergonic/exergonic) ?
Formation of thioester, ester or amide from acyl phosphate. exothermic in the direction of reaction indicated. Important exception is where there are other benefits such as releasing ring strain.
One such example is penicillin which contains a highly reactive lactam (cyclic amide)
What reaction is this? What is happening mechanistically? What is the energy balance like (endergonic/exergonic) ?
Formation of amide from thioester and ester. exothermic in the direction of reaction indicated. Important exception is where there are other benefits such as releasing ring strain.
One such example is penicillin which contains a highly reactive lactam (cyclic amide)
What reaction is this? What is happening mechanistically? What is the energy balance like (endergonic/exergonic) ?
Hydrolysis of a thioester, a (carboxylic) ester, or amide to carboxylate. exothermic in the direction of reaction indicated. Important exception is where there are other benefits such as releasing ring strain.
One such example is penicillin which contains a highly reactive lactam (cyclic amide)
Draw the mechanism of protein degradation (hydrolysis of amide bonds) via serine proteases.
Serine instead of aspartate.
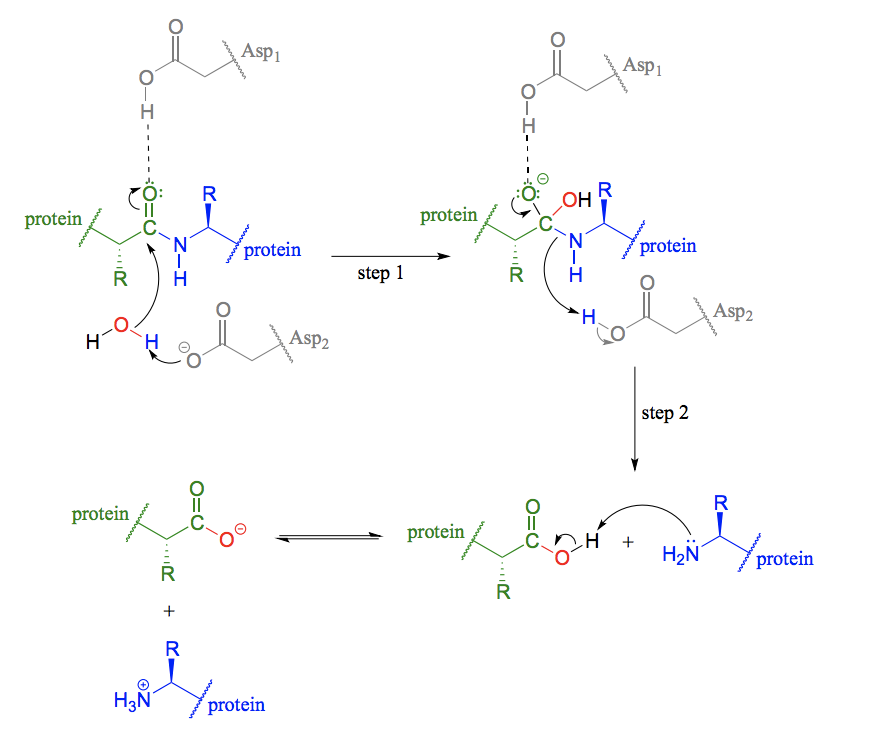
Explain how the oxyanion hole stabilizes the transition state in the active site and the catalytic triad in a protein degradation reaction.
Catalytic triad - a conformation of three amino acids in the active site of some enzymes (common in proteases). An acid-base-nucleophile triad is a common motif.
An oxyanion hole is a pocket in the active site of an enzyme that stabilizes transition state negative charge on a deprotonated oxygen or alkoxide. The pocket typically consists of backbone amides or positively charged residues.
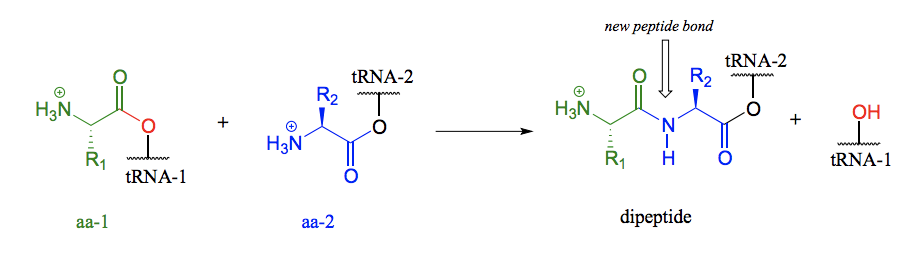
Draw the mechanism of amide formation in the synthesis of protein in the ribosome (focus on the nucleophilic acyl substitution reaction)
The carboxyl group of the first amino acid is first transformed to an acyl-AMP intermediate through a nucleophilic substitution reaction with ATP. In the next step the amino acid is transferred to tRNA. The incoming nucleophile is an alcohol sitting on the tRNA, meaning that we are seeing an esterification: an acyl substitution reaction between the activated carboxylate of the first amino acid and an alcohol on the tRNA. The actual peptide bond-forming reaction occurs when a second amino acid (aa-2) also linked to its own tRNA-2 molecule, is positioned next to the first amino acid on the ribosome. The amino group on aa-2 displaces tRNA1: thus, an ester has been converted to an amide (thermodynamically downhill, so ATP is not required). This process continues on the ribosome, as one amino acid after another is added to the growing protein chain, until a genetically coded signal indicates that the chain is complete, and an ester hydrolysis reaction occurs to stop the growth of the polypeptide chain.