AQA A Level Biology Topic 2
1/115
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
116 Terms
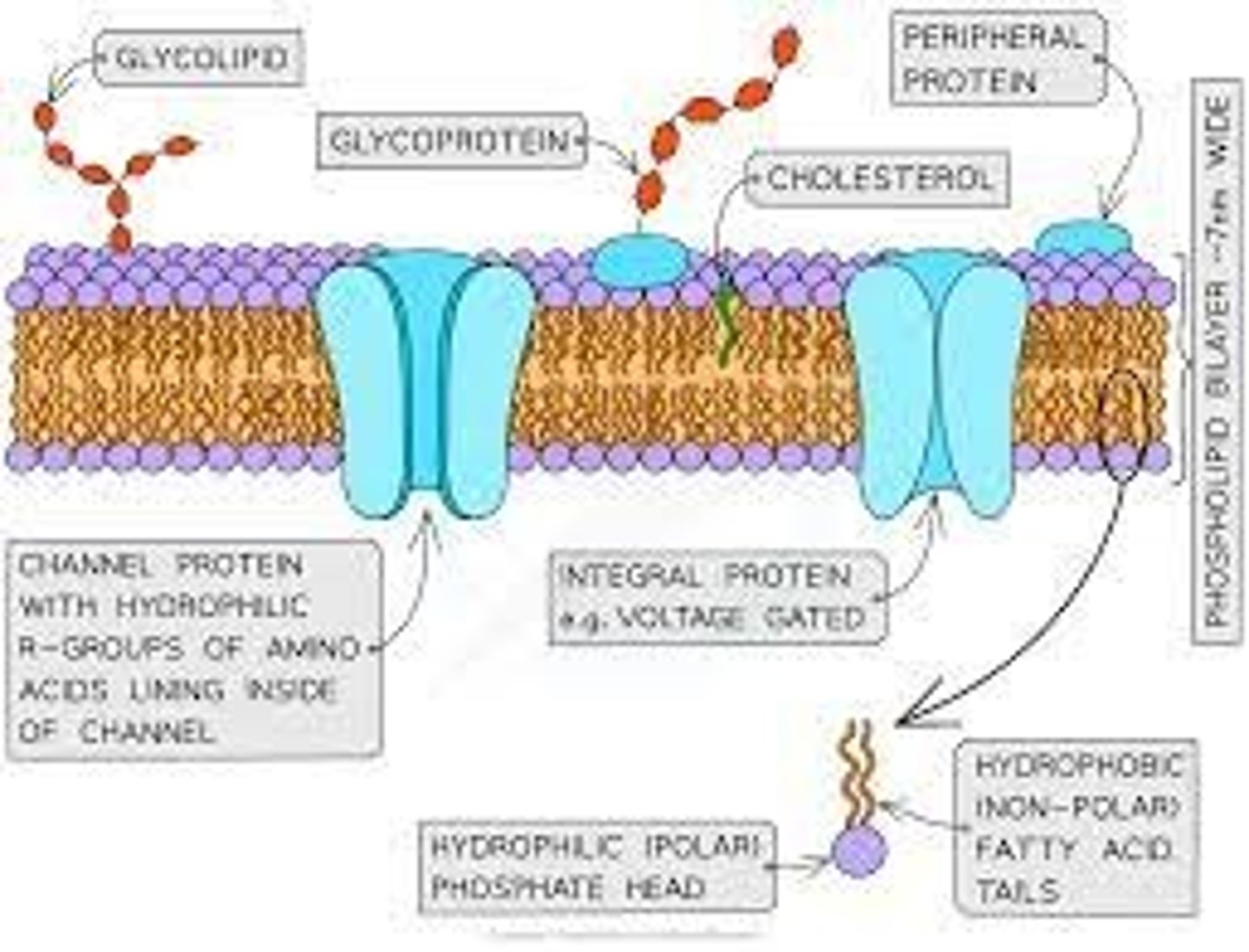
Cell membrane structure
-phospholipid bilayer, with hydrophilic phosphate heads and hydrophobic tails.
- Embedded with proteins
Cell membrane function
● Selectively permeable → enables control of passage of substances in / out of cell
● Molecules / receptors / antigens on surface → allow cell recognition / signalling
Nucleus structure
- Nuclear envelope
- double membrane
-nuclear pores
- nucleoplasm
- nucleolus
- protein/histone-bounds, linear DNA
-chromatin
-chromosome
Nucleus function
● Holds / stores genetic information which codes for polypeptides (proteins)
● Site of DNA replication
● Site of transcription (part of protein synthesis), producing mRNA
● Nucleolus makes ribosomes / rRNA
Mitochondria structure
Double membrane with inner membrane folded into cristae. 70S small ribosomes in matrix. Small circular DNA. Enzymes in matrix
Mitochondria function
● Site of aerobic respiration
● To produce ATP for energy release
● Eg. for protein synthesis / vesicle movement / active transport
Chloroplast structure
- Double membrane
- Stroma:
- Thylakoid membrane
- Small/70S ribosomes
- Circular DNA
-starch granules/lipid droplets
- Lamella
- Grana
Chloroplast function
● Absorbs light energy for photosynthesis
● To produce organic substances eg. carbohydrates / lipids
Organisms containing chloroplasts
Plants and Algae
Golgi Apparatus structure
Golgi apparatus = flattened membrane sacs
Golgi vesicle = small membrane sac
Golgi apparatus function
Golgi
apparatus
● Modifies protein, eg. adds carbohydrates to produce glycoproteins
● Modifies lipids, eg. adds carbohydrates to make glycolipids
● Packages proteins / lipids into Golgi vesicles
● Produces lysosomes (a type of Golgi vesicle)
Golgi
vesicles
● Transports proteins / lipids to their required destination
● Eg. moves to and fuses with cell-surface membrane
Lysosome structure
Type of golgi vesicle containing hydrolytic enzymes
has membrane
Lysosome function
● Release hydrolytic enzymes (lysozymes)
● To break down / hydrolyse pathogens or worn-out cell components
RER function
● Ribosomes on surface synthesise proteins
● Proteins processed / folded / transported inside rER
● Proteins packaged into vesicles for transport eg. to Golgi apparatus
SER function
● Synthesises and processes lipids
● Eg. cholesterol and steroid hormones
Ribosome structure
● Made of ribosomal RNA and protein (two subunits)
● Not a membrane-bound organelle
Ribosome function
Site of translation in protein synthesis
RER structure
system of membranes with bound ribosomes
SER structure
system of membranes with no bound ribosomes
function of cell wall
● Provides mechanical strength to cell
● So prevents cell changing shape or bursting under pressure due to osmosis
Cell Wall structure
found in plant, fungal and bacterial cells.
● Composed mainly of cellulose (a polysaccharide) in plants / algae
● Composed of chitin (a nitrogen-containing polysaccharide) in fungi
Cell vacuole structure
fluid filled (cell sap). surrounded by a single membrane (tonoplast)
Describe the function of the cell vacuole in plants
● Maintains turgor pressure in cell (stopping plant wilting)
● Contains cell sap → stores sugars, amino acids, pigments and any waste chemicals
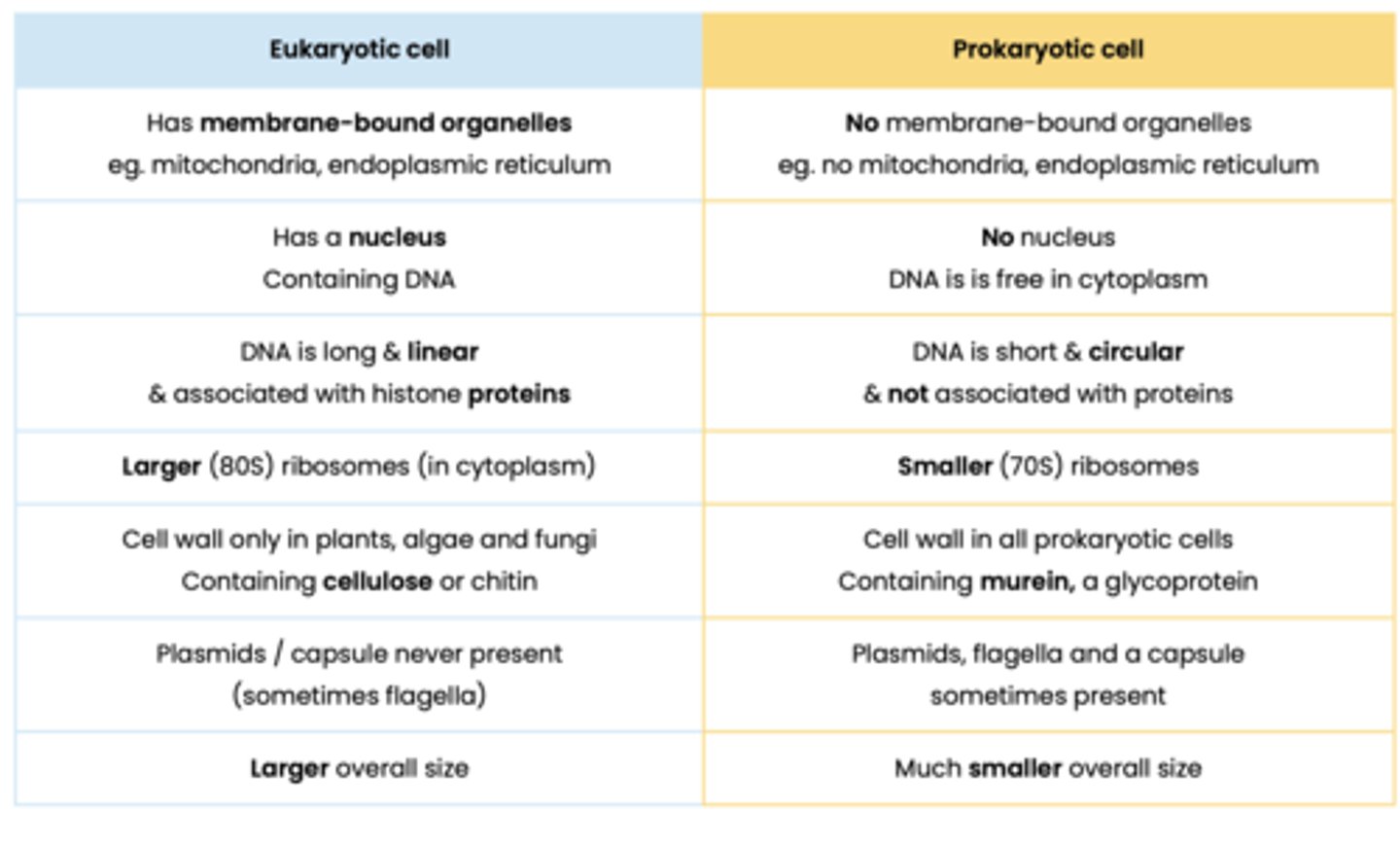
Contrast prokaryotic and eukaryotic cells
- Prokaryotic cells are smaller
- Prokaryotic have no membrane bound organelles
- Prokaryotes have smaller 70S ribsomes
- Prokaryotes have no nucleus
- Prokaryotes have circular DNA that is not associated with histones
- Prokaryotic cell wall made of murein instead of cellulose/chitin
Describe how eukaryotic cells are organised in complex multicellular
organisms
Tissue - Group of specialised cells with a similar structure working together
to perform a specific function, often with the same origin
Organ - Aggregations of tissues performing specific functions
Organ system - Group of organs working together to perform specific functions
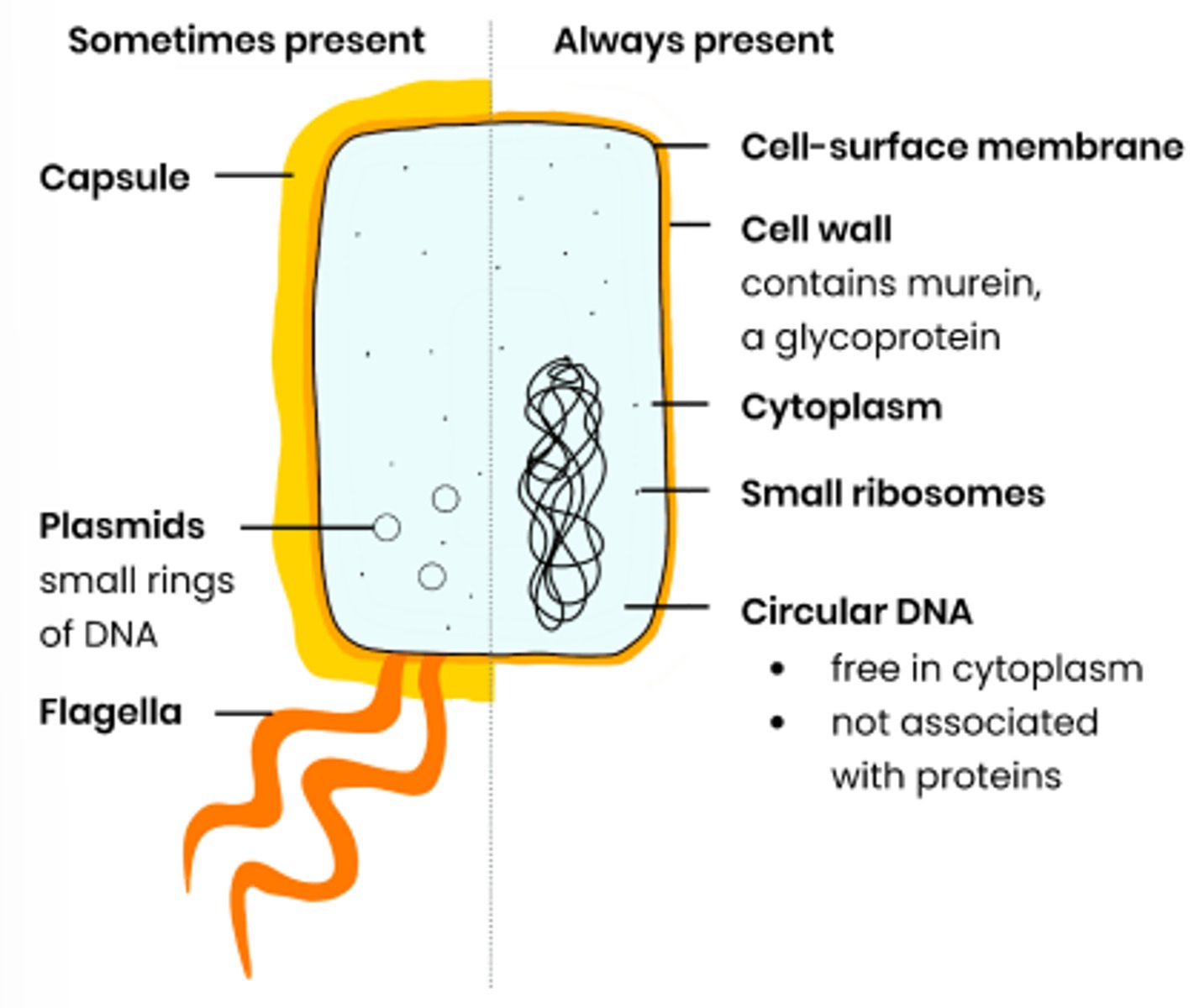
What are the distinguishing features of prokaryotic cells
● Cytoplasm lacking membrane-bound organelles
● So genetic material not enclosed in a nucleus
Describe the general structure of prokaryotic cells
Protein carriers
binds with a molecule e.g. glucose, which causes a change in shape of the protein.
this change in shape enables the molecule to be released to the other side of the membrane
Protein channels
Tubes filled with water enabling water soluble ions to pass through the membrane
channel proteins only open in the presence of certain ions when they bind to the protein
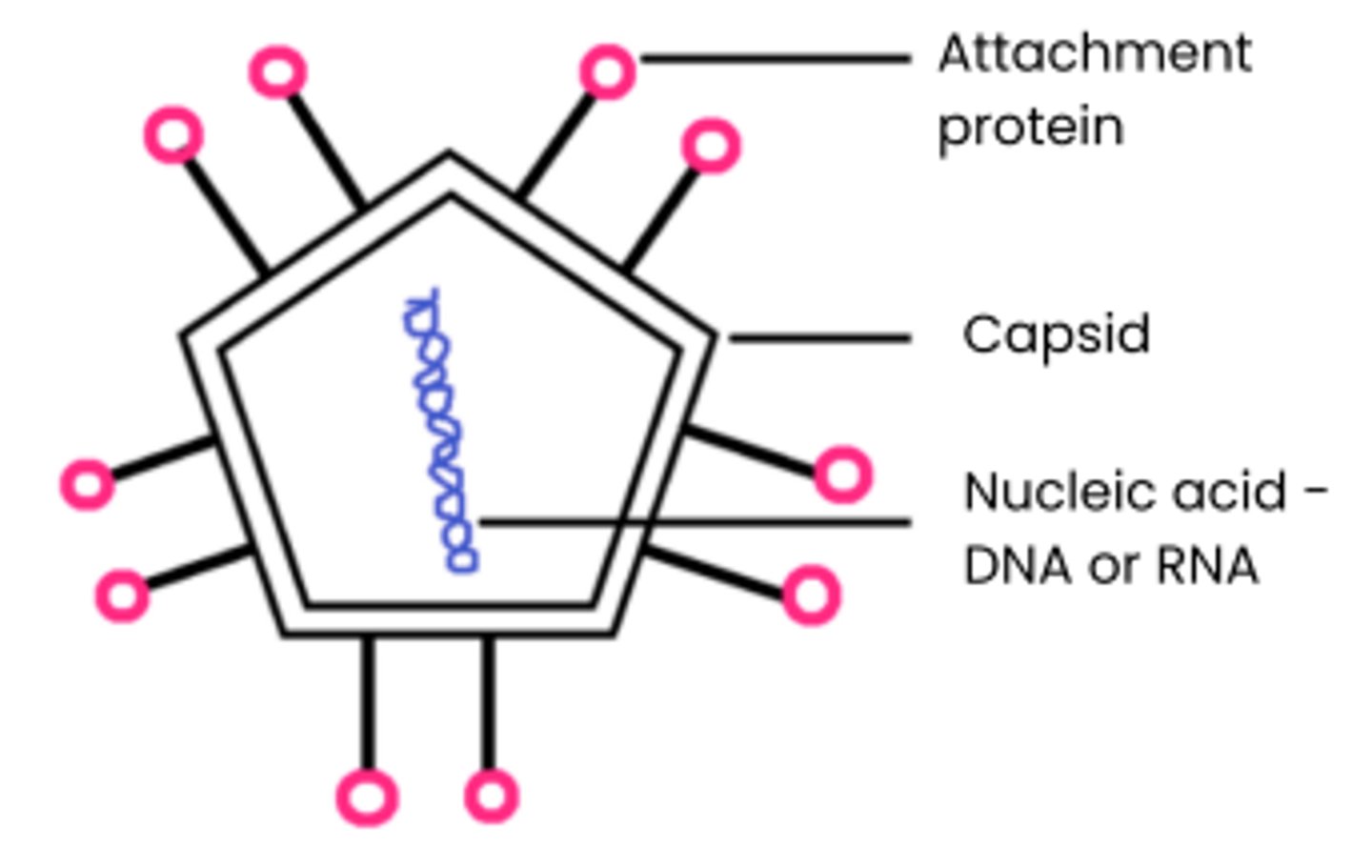
Explain why viruses are described as acellular and non-living
● Acellular - not made of cells, no cell membrane / cytoplasm / organelles
● Non-living - have no metabolism, cannot independently move / respire / replicate / excrete
Describe the general structure of a virus particle
1. Nucleic acids surrounded by a capsid
(protein coat)
2. Attachment proteins allow attachment
to specific host cells
3. No cytoplasm, ribosomes, cell wall, cell-surface membrane etc.
4. Some also surrounded by a lipid envelope eg. HIV
3 types of microscopes
- Optical (light) microscopes
- Scanning electron microscope (SEM)
- Transmission electron microscope (TEM)
Magnification
how many times larger the image is compared to the real object.
Equation: magnification = image size/ actual size
Resolution
The minimum distance between 2 objects in which they can still be viewed as separate.
determined by wavelength of light (for optical microscopes) or electrons (for electron microscopes)
Optical microscopes
- Beam of light used to create image.
- glass lens used for focusing
- 2D coloured image of a cross section
Evaluate optical microscopes
- Poorer resolution as long wavelength of light- so small organelles are not visible
- Lower magnification
- Can view living samples
- Simple staining method
- Vacuum not required
Transmission electron microscopes
- Beam of electrons pass through the sample used to create an image
- Focused using electromagnets
- 2D, black & white image
- can see internal ultrastructure of cell
- structures absorb electrons and appear dark
Evaluation of TEMs
- Highest resolving power
- high magnification
- extremely thin specimens required
- complex staining method - specimen must be dead.
- vaccum required
Scanning electron microscopes
- Beam of electrons pass across sample used to create image
- focused using electromagnets
- 3D, black and white image produced
- electrons scattered across specimen producing image
Evaluation of SEMs
- High resolving power
- high magnification
-thick specimens usable
- complex staining method
-specimen must be dead
- vaccum required
Why calibrate eyepiece graticule?
- Calibration of the eyepiece is required each time the objective lens is changed
- calibrate to work out the distance between each division at that magnification
Describe how to convert between different units
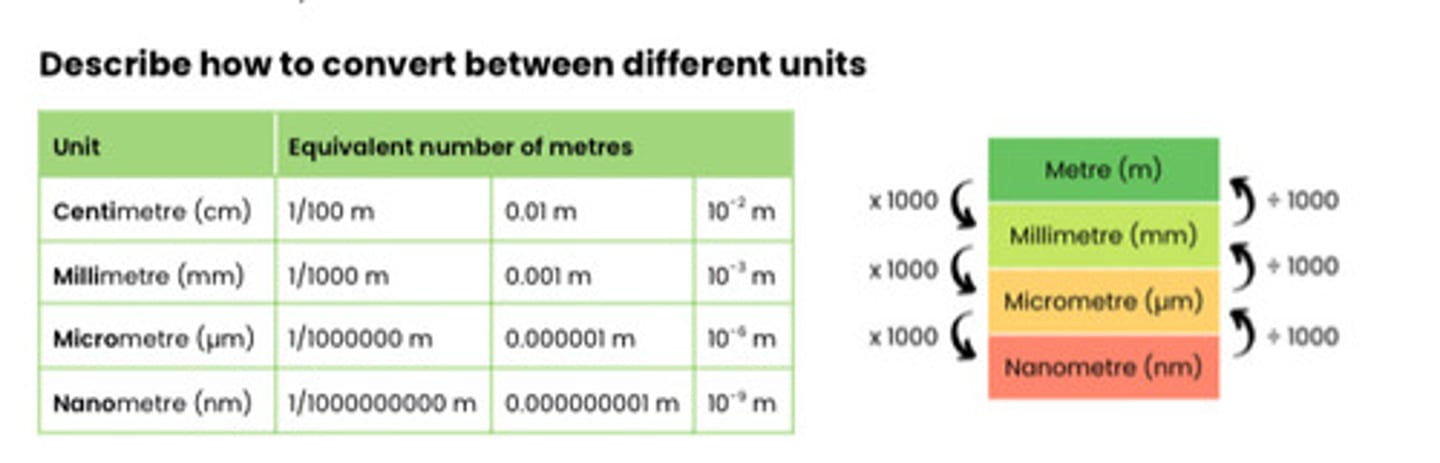
Describe how the size of an object viewed with an optical microscope can be measured
1. Line up (scale of) eyepiece graticule with (scale of) stage micrometre
2. Calibrate eyepiece graticule - use stage micrometre to calculate size of divisions on eyepiece graticule
3. Take micrometre away and use graticule to measure how many divisions make up the object
4. Calculate size of object by multiplying number of divisions by size of division
5. Recalibrate eyepiece graticule at different magnifications
Purpose of cell fractionation
Break open cells & remove cell debris so organelles can be studied
Describe and explain the principles of cell fractionation and
ultracentrifugation as used to separate cell components
1. Homogenise tissue /
use a blender
● Disrupts the cell membrane, breaking open cells to release
contents / organelles
2. Place in a cold,
isotonic, buffered
solution
● Cold to reduce enzyme activity
○ So organelles not broken down / damaged
● Isotonic so water doesn't move in or out of organelles by osmosis
○ So they don't burst
● Buffered to keep pH constant
○ So enzymes don't denature
3. Filter homogenate ● Remove large, unwanted debris eg. whole cells, connective tissue
4. Ultracentrifugation -
separates organelles
in order of density /
mass
● Centrifuge homogenate in a tube at a low speed
● Remove pellet of heaviest organelle and respin supernatant
at a higher speed
● Repeat at increasing speeds until separated out, each time the
pellet is made of lighter organelles (nuclei → chloroplasts /
mitochondria → lysosomes → ER → ribosomes)
Binary Fission
- Involves circular DNA & plasmids replicating
- cytokinesis creates two daughter nuclei
- each daughter cell has one copy of circular DNA and a variable number of plasmids
Cell Cycle
- Interphase (G1, S, G2)
- Nuclear division - mitosis or meiosis
- Cytokinesis
Interphase
- longest stage in the cell cycle
- when DNA replicates (S phase) and the organelles duplicate while cell grows ( G1 & G2 phase)
- DNA replicates and appears as 2 sister chromatids held by centromere
Mitosis
- one round of cell division
- two diploid, genetically identical daughter cells
- growth and repair (e.g. clonal expansion)
- includes prophase, metaphase, anaphase and telophase
Prophase
- chromosomes condense and become visible
- nuclear envelope disintegrates
- in animals - centrioles separate and spindle fibres structure forms
Metaphase
- Chromosomes line up at the equator of the cell
- spindle fibres released from poles attach to centromere and chromatid
Anaphase
- spindle fibre contracts (suing ATP) to pull chromatids, centromere first, towards opposite poles of cell
- centromere divides in two
Telophase
● Chromosomes uncoil, becoming longer / thinner
● Nuclear envelopes reform = 2 nuclei
● Spindle fibres / centrioles break down
Mitotic index
- used to determine proportion of cells undergoing mitosis
- calculated as a percentage or decimal
mitotic index= the number of cells in mitosis / the total number of cells x 100
Describe how tumours and cancers form
● Mutations in DNA / genes controlling mitosis can lead to uncontrolled cell division
● Tumour formed if this results in mass of abnormal cells
○ Malignant tumour = cancerous, can spread (metastasis)
○ Benign tumour = non-cancerous
Suggest how cancer treatments control rate of cell division
● Some disrupt spindle fibre activity / formation
○ So chromosomes can't attach to spindle by their centromere
○ So chromatids can't be separated to opposite poles (no anaphase)
○ So prevents / slows mitosis
● Some prevent DNA replication during interphase
○ So can't make 2 copies of each chromosome (chromatids)
○ So prevents / slows mitosis
Fluid mosaic model
● Molecules free to move laterally in phospholipid bilayer
● Many components - phospholipids, proteins,
glycoproteins and glycolipids
Explain the arrangement of phospholipids in a cell membrane
- phospholipids align as a bilayer
- hydrophilic heads are attracted to water
- hydrophobic tails repelled by water
Explain the role of cholesterol (sometimes present) in cell membranes
- present in eukaryotic organisms restrict lateral movement of the membranes
- adds rigidity to membrane - restricts movement
Selectively permeable membrane
- molecules must have specific properties to pass through plasma membrane
- lipid soluble (hormones e.g. oestrogen)
- very small molecules
- non-polar molecules (oxygen)
Suggest how cell membranes are adapted for other functions
● Phospholipid bilayer is fluid → membrane can bend for vesicle formation / phagocytosis
● Glycoproteins / glycolipids act as receptors / antigens → involved in cell signalling / recognition
Simple diffusion
● Lipid-soluble (non-polar) or very small substances eg. O2
, steroid hormones
● Move from an area of higher concentration to an area of lower conc., down a conc. gradient
● Across phospholipid bilayer
● Passive - doesn't require energy from ATP / respiration (only kinetic energy of substances)
Facilitated diffusion
● Water-soluble / polar / charged (or slightly larger) substances eg. glucose, amino acids
● Move down a concentration gradient
● Through specific channel / carrier proteins
● Passive - doesn't require energy from ATP / respiration (only kinetic energy of substances)
Osmosis
● Water diffuses / moves
● From an area of high to low water potential (ψ) / down a water potential gradient
● Through a partially permeable membrane
● Passive - doesn't require energy from ATP / respiration (only kinetic energy of substances)
Water potential
- the pressure created by water molecules
- measured in kPa
- pure water has a water potential of 0 kPa
- the more negative the water potential, the more solute must be dissolved
Hypertonic solution
- when the water potential of a solution is more negative than the cell
- water moves out of the cell by osmosis
- both animal and plant cells will shrink and shrivel
Hypotonic solution
- when the water potential of a solution is more positive (closer to zero) than the cell
- water moves into the cell by osmosis
- animal cells will lyse (burst)
- plant cells will become turgid
Isotonic
- when the water potential of the surrounding solution is the same as the water potential inside the cell
- no net movement in water
- cells would remain the same mass
Active transport
- the movement of ions and molecules from an area of lower concentration to an area of higher concentration using ATP and carrier proteins
Describe the role of carrier proteins and the importance of the hydrolysis of
ATP in active transport
1. Complementary substance binds to specific carrier protein
2. ATP binds, hydrolysed into ADP + Pi, releasing energy
3. Carrier protein changes shape, releasing substance on side
of higher concentration
4. Pi released → protein returns to original shape
Co-transport
● Two different substances bind to and move simultaneously via a
co-transporter protein (type of carrier protein)
● Movement of one substance against its concentration gradient is often coupled with the movement of another down its concentration gradient
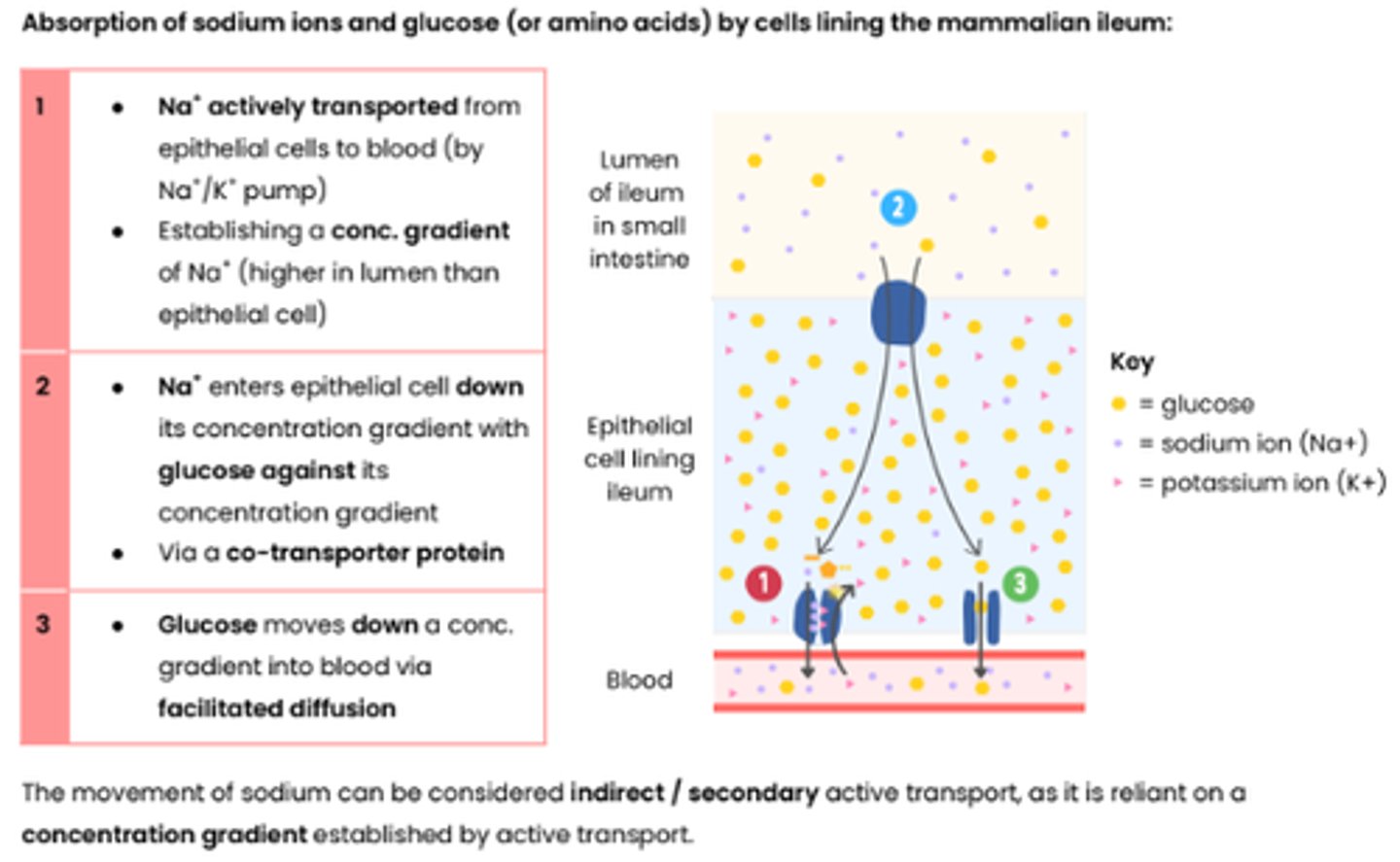
Describe an example that illustrates co-transport
1 ● Na+ actively transported from
epithelial cells to blood (by Na+/K+ pump)
● Establishing a conc. gradient of Na+ (higher in lumen than epithelial cell)
2 ● Na+ enters epithelial cell down its concentration gradient with glucose against its
concentration gradient
● Via a co-transporter protein
3 ● Glucose moves down a conc. gradient into blood via facilitated diffusion
Describe how surface area, number of channel or carrier proteins and
differences in gradients of concentration or water potential affect the rate of movement across cell membranes
● Increasing surface area of membrane increases rate of movement
● Increasing number of channel / carrier proteins increases rate of facilitated diffusion / active transport
● Increasing concentration gradient increases rate of simple diffusion
● Increasing concentration gradient increases rate of facilitated diffusion
○ Until number of channel / carrier proteins becomes a limiting factor as all in use / saturated
● Increasing water potential gradient increases rate of osmosis
Explain the adaptations of some specialised cells in relation to the rate of transport across their internal and external membranes
● Cell membrane folded eg. microvilli in ileum → increase in surface area
● More protein channels / carriers → for facilitated diffusion (or active transport - carrier proteins only)
● Large number of mitochondria → make more ATP by aerobic respiration for active transport
Molecules lymphocytes identify
- Pathogens (bacteria, fungi, viruses)
- Cells from other organisms of same species (transplants)
- Abnormal body cells (tumour cells)
- Toxins (released from bacteria)
Antigens
- proteins on the cell-surface membrane
- trigger an immune response when detected by lymphocytes
How are cells identified by the immune system?
● Each type of cell has specific molecules on its surface (cell-surface membrane / cell wall) that identify it
● Often proteins → have a specific tertiary structure (or glycoproteins / glycolipids)
Antigenic variability
● Antigens on pathogens change shape / tertiary structure due to gene mutations (creating new strains)
● So no longer immune (from vaccine or prior infection)
○ B memory cell receptors cannot bind to / recognise changed antigen on secondary exposure
○ Specific antibodies not complementary / cannot bind to changed antigen
Physical barriers
anatomical barriers to pathogens
- skin
- stomach acid
- mucus
- tears
Describe phagocytosis of pathogens (non-specific immune response)
1 Phagocyte attracted by chemicals / recognises (foreign) antigens on pathogen
2 Phagocyte engulfs pathogen by surrounding it with its cell membrane
3 Pathogen contained in vesicle / phagosome in cytoplasm of phagocyte
4 Lysosome fuses with phagosome and releases lysozymes (hydrolytic enzymes)
5 Lysozymes hydrolyse / digest pathogen
Phagocytosis leads to presentation of antigens
Describe the response of T lymphocytes to a foreign antigen (the cellular
response)
Specific helper T cells with complementary receptors (on cell surface) bind to antigen on antigen-presenting cell → activated and divide by mitosis to form clones which stimulate:
● Cytotoxic T cells → kill infected cells / tumour cells (by producing perforin - binds to cell surface membrane of target cell and pierce holes)
● Specific B cells (humoral response - see below)
● Phagocytes → engulf pathogens by phagocytosis
Antigen presenting cells
- Any cell that presents a non-self antigen on their surface
- infected body cells
- macrophage after phagocytosis
- cells of transplanted organ
- cancer cells
Cytotoxic T cells
- destroy abnormal/ infected cells by releasing perforin
- so that any substances can enter or leave the cell and this causes cell death
B lymphocytes
- made in the bone marrow and mature in the bone marrow
- involved in humoral immune response
- involved antibodies
Describe the response of B lymphocytes to a foreign antigen (the humoral
response)
1. Clonal selection:
○ Specific B lymphocyte with complementary receptor (antibody on cell surface) binds to antigen
○ This is then stimulated by helper T cells (which releases cytokines)
○ So divides (rapidly) by mitosis to form clones
2. Some differentiate into B plasma cells → secrete large amounts of (monoclonal) antibody
3. Some differentiate into B memory cells → remain in blood for secondary immune response
B memory cells
- derived from B lymphocytes
- remember specific antibody for particular antigen
- will rapidly divide by mitosis and differentiate in plasma cells upon secondary encounter
- resulting in large numbers of antibodies rapidly
Antibodies
● Quaternary structure proteins (4 polypeptide chains)
● Secreted by B lymphocytes eg. plasma cells in response to specific antigens
● Bind specifically to antigens forming antigen-antibody complexes
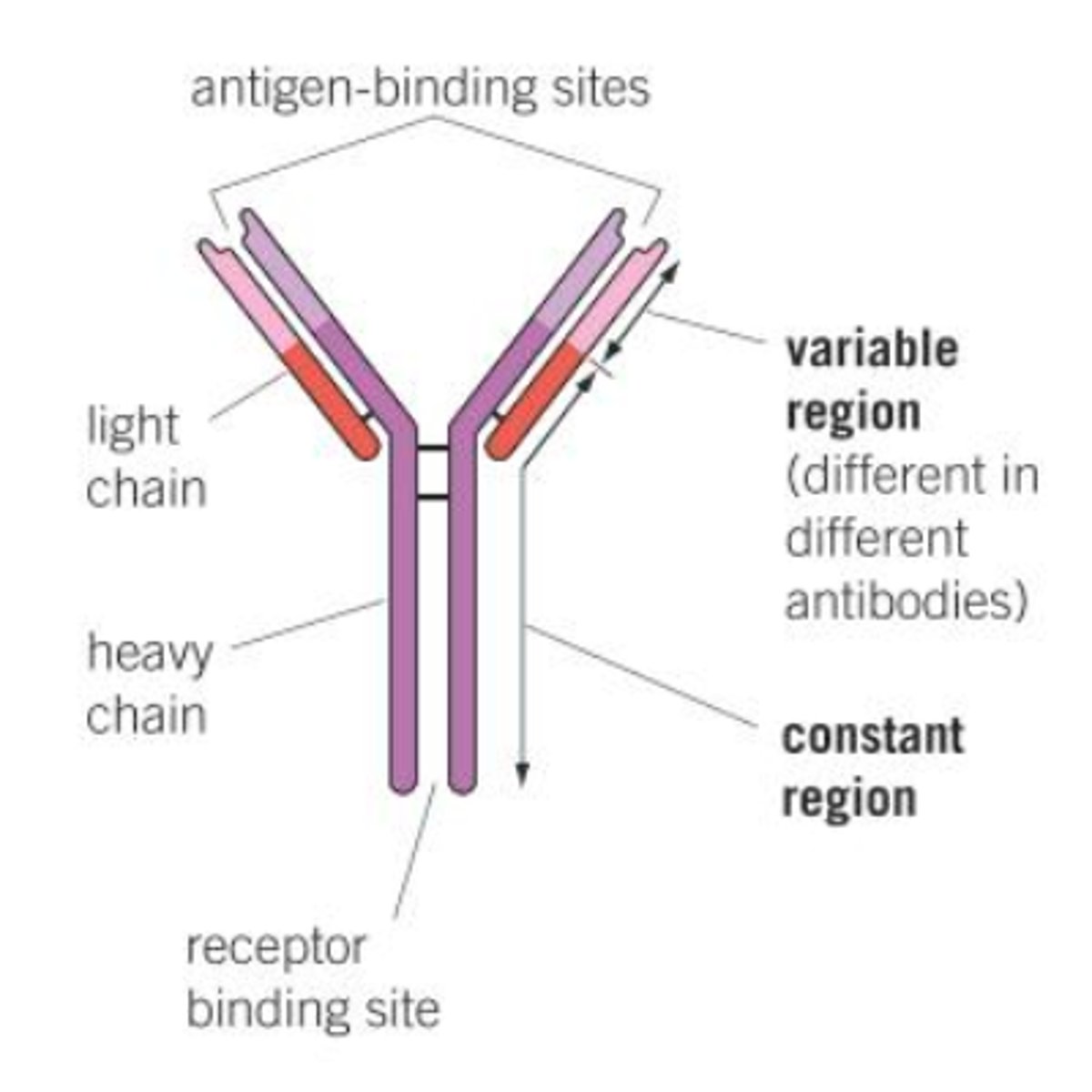
Antibody structure
- 2 light chains and 2 heavy chains
- constant region
- variable region
- has disulfide bridges
Explain how antibodies lead to the destruction of pathogens
● Antibodies bind to antigens on pathogens forming an antigen-antibody complex
○ Specific tertiary structure so binding site / variable region binds to complementary antigen
● Each antibody binds to 2 pathogens at a time causing agglutination (clumping) of pathogens
● Antibodies attract phagocytes
● Phagocytes bind to the antibodies and phagocytose many pathogens at once
Passive immunity
- antibodies introduced into body
- plasma and memory cells not made as no interaction with antigen
- short-term immunity
- fast acting
Active immunity
- immunity created by own immune system - antibodies made
- exposure to antigen
- plasma and memory cells made
- long term immunity
- slower acting
Natural active immunity
- after direct contact with pathogen through infection
- body creates antibodies and memory cells
Artificial active immunity
- creation of antibodies and memory cells following introduction of an attenuated pathogen or antigens
- vaccination
Vaccinations
- small amounts of dead or attenuated pathogens injected
- humoral response activated
- memory cells are able to divide rapidly into plasma cells when re-infected
Explain how vaccines provide protection to individuals against disease
1. Specific B lymphocyte with complementary receptor binds to antigen
2. Specific T helper cell binds to antigen-presenting cell and stimulates B cell
3. B lymphocyte divides by mitosis to form clones
4. Some differentiate into B plasma cells which release antibodies
5. Some differentiate into B memory cells
6. On secondary exposure to antigen, B memory cells rapidly divide by mitosis to produce B plasma cells
7. These release antibodies faster and at a higher concentration
Primary vs Secondary response
- Primary = first exposure to the pathogen
- longer time for plasma cell secretion & memory cell production
- for the secondary response, memory cells divide rapidly into plasma cells
- so a large number of antibodies made rapidly upon reinfection
Herd immunity
- when the majority of the population is vaccinated so pathogen is not transmitted and spread easily
- provides protection for those without vaccine
Monoclonal antibodies
- a single type of antibody that can be isolated and cloned
- antibodies that are identical - from one type of B lymphocyte
- complementary to only one antigen
Uses of monoclonal antibodies
- Medical treatment - targeting drugs by attaching antibody complementary to tumour cell antigen
- medical diagnosis - pregnancy tests
Pregnancy test
- ELISA test which uses 3 monoclonal antibodies and enzymes to test for hCG