Transcription and Translation
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
32 Terms
What carries out transcription? What do they lack? What direction does it synthesize? What must it recognize to start
RNA polymerase
do not require primer, lack 3’ to 5’ exonuclease activity
synthesize RNA 5’ to 3’
Must recognize start point for transcription and the template strand of DNA
what are the three factors in why we need transcription?
cell only has one DNA, which is not enough to go around
protect DNA from cytoplasmic environment
regulate the rate of protein synthesis:
number of mRNA copies
life-time of mRNA (degradation)
RNA binding factors (translation)
Compare and contrast prokaryotic and eukaryotic RNA polymerases. What does sigma factors do?
prokaryotic have a single RNA polymerase
transcribes all types of RNA
four subnits: a2bb
sigma factor binds to core enzyme and direct its binding to promotor (sigma 70)
eukaryotic cell have 3 RNA polymerases
I = rRNA
II = mRNA
III = tRNA and 5s rRNA
Different polymerases recognize different type of promotoers
What is a gene and describe its structure
gene = segment of DNA containing information for the expression of RNA
contains:
promoter: binding site for RNA polymerases and transcription factors
promotor-proximal elements and enhancers: binding of nuclear receptors, co-activators, co-repressors
compare and contrast coding and template strnad
template: used by RNA polymerase during transcription; complimentary and ANTI PARALLEL to coding strand and RNA
CODING: analogous to produced RNA
discuss the promotor regions and the concensus sequences that are found to start transcription; What are the additional eukaryotic boxes?
e.coli: TATAAT - Pribnow box
Eukaryotic: TATA(A/T)A- TATA box
additional eukaryotic boxes: CAAT boxes, GC rich sequences, and enhancers
discuss the steps of bacterial transcription. (HINT: What type of factor do they have?) What are DNA topoisomerase? compare/contrast RHO- independent and dependent termination signal
steps of transcription; uses holoenzyme RNA polymerase
recognize and bind to promoter
unwinds and separates DNA strands
sigma factor dissociates from RNA polymerase
RNA polymerase transcribs the DNA
DNA topoisomerase relaxes generated supercoils
Rho-independent: formation of hairpin loop by transcript results in release of RNA polymerase
RHO-dependent: binding of rho-factor causes release of polymerase
what is an operon? what is a cistron? What advantage does the lack of a nucleus provide for prokaryotes?
operon = where many protein-producing gene are linked together and controlled by a single promotor; usually proteins for same metabolic pathway
cistron = region of DNA that encodes for a single polypeptide
no nucleus = coupling of transcription and translation
what is rifampicin?
Used to treat tuberculosis;
inhibits bacterial RNA polymerase and mitochondrial RNA polymerase (higher concentration needed than for bacterial RNA)
what are the five differences of between eukaryotic and bacterial transcription?
occurs in nucleus and in chromatin
more elaborate regulation
more transcription factors and enhancers
nucleosome-modifying enzymes (histone acetyltransferase)
chromatin remodeling complexes
eukaryotes = 3 RNA polymerase
more elaborate mechanism of processing of transcripts
modification of RNA transcript
removal of introns
eukaryotic mRNA has information for only ONE polypeptide
what are GTFs? what are enhancers?
GTFs: general transcription factor
interact with each other and RNA poly II to initiate transcription
enhancers: sequences that initiate transcription
contain response elements: sequences that binds activators
can bind transcription factors
can be upstream or downstream
What are the modifications to the primary transcript of eukaryotic mRNA and its functions
has a 5’ cap (added during transcription)
decreases rate of mRNA degradation
serve as recognition site for binding to ribosome
poly -a - tail at 3’ end
added by poly-a polymerase
increases mRNA stability
has exons and introns
exons = coding sequences
introns are removed by splicing
describe the synthesis of eukaryotic rRNA; Describe its appearence under microscope
by polymerase I
transcribed from a set of tandomly repeated genes
about 1000 copies in genome
rRNA is synthesized as a single large transcript
cleaved into 18s, 28s, and 5.8s rRNAS
multiple Pol I can be on the same gene at a time
christmas tree appearance under electron microscope
forms the ribonucleoprotiein complex= ribosome
how are tRNAS produced?
cleavage of 5’ and 3’ ends
splicing to remove introns
replaces terminal 3’ terminal UU with CCA
multiple modification of bases for specificy of AA
what is Thalassemias?
group of hereditary anemias; most common gene disorder in world
affects synthesis of either alpha or beta chains of hemoglobin
mutation result in a B+ phenotype in TATA box (A → G or A → C in the -28 to -31 region)
reduces accuracy of transcription start point
other mutation futher upstream in promoter region (C→G at -87 and C→T at -88)
Reduces the frequency of Globin Transcription
where do AA attach to at tRNA; how does it recognizes the mRNA codon?
at 3’ end of tRNA, via tRNA’s anticodon
describe the wobble’s hypothesis
explains why multiple codon code for one amino acid
Due to less precise base pairing between the first base of anticodon and 3rd base of codon
what are the four characteristic of the genetic code?
specificity: specific codon = always same AA
universality: exist at the dawn of evolution
redundancy: given AA may have more than one codon
nonoverlapping and commales: Code is read at the same starting spot and always 3 at a time
what are the different types of mutations
point mutations: one base is altered
silent: changed codon = same AA
missense: changed codon = different AA
nonsense: changed codon = become termination codon
Insertion, deletions, and frame-shift mutations
describe the mutation that causes sickle cell anemia and the result
missense mutation: GTG (valine) → GAG (Glu)
in each allele for B-globin
Results: oxygenated molecules are soluble but de-oxygenated = aggregate into insoluble fibers = deforms RBC into spiny or sickle-shaped cells
What are the components required for translation
template
prokaryotic = polycistronic
eukaryotic = monocistronic
ribosome
tRNAS
AA
ATP/GTP
ATP = attach AA to tRNA
GTP = binding of AA-tRNA to A site
ATP and GTP are needed for initiation and termination steps
mRNA
describe the formation of aminoacetyl-tRNA
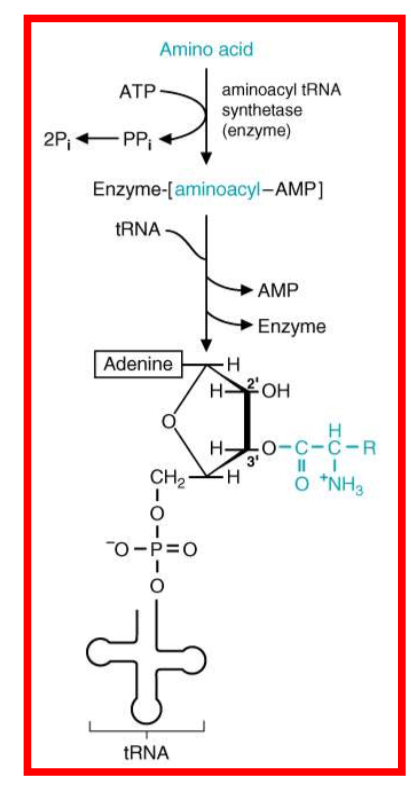
what are the three sites of the ribosome:
A: binds incoming aminoacyl-tRNA and specifies next AA to be added
P: occupied by peptidyltRNA; this tRNA carries the synthesized poly peptide
E: where empty tRNA exits
describe initiation of translation for prokaryotes and eukaryotes
prokaryotes:
16s RNA contain seq. complimentary to Shine-Dalgarno sequence of mRNA
binding of these two sequences on 30s
Eukaryotes:
40s binds to Cap structure of mRNA
What does the initiator tRNA recognize? facilitated by? How is this tRNA different in prokaryotes/mitochondria? Where does the AA attached onto? What is this catalyzed by?
recognizes AUG codon
faciliated by IF-2 in e.coli and several IF in humans
in pro. and mitochondria: initiator tRNA carries N-formylated methionine
AA is attached on carboxyl end of growing polypeptide chain
catalyzed by peptidyltransferase
draw out the steps of translation
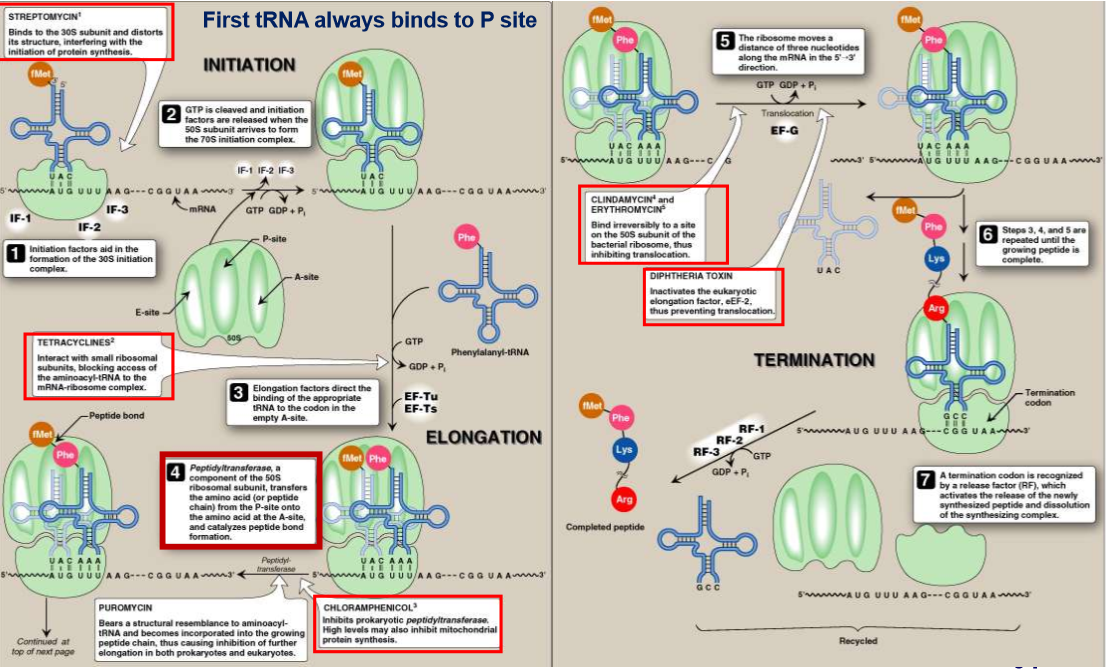
what does streptomycin do?
binds to 30s subunit
interferes with initiation of translation
causes misreading of mRNA
can cause perm. hearing loss
used mainly for treatment of TB
what does tetracycline do?
binds to 30s subunit
inhibits binding of AA-tRNA to A site
what does chloramphenicol do?
interferes with peptidyltransferase of 50s Bacterial subunit
limited use in humans because also affects mitochondria
what does Ricin do?
produced in seeds of castor oil plant
N-glycosylase activity of ricin allows for cleavage of glycosidic bond within a large 60s subunit of eukaryotic ribosome
this depurination = very rapidly and completely inactivates ribosome
list the three regulation of translation
regulatory proteins: stabilize mRNA or can prevent binding of ribosome to mRNA
rare codons: rare codons have fewer tRNA in cell = slows down translation; usually located at beginning of mRNA
small interfering RNA = targets mRNA for degradation
What does clindamycin/erythomycin do? What does diphtheria toxin do?
clindamycin: binds irreversible to 50s subunit → inhibit it
Diphtheria: inactivates the eukaryotic elongation factor (eEF-2) → no translocation