Hereditary hemochromatosis (HH)
1/12
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
13 Terms
What is HH
Hereditary Hemochromatosis (HH)
- The most common form of HH is an autosomal recessive disorder of iron overload
- Excessive iron is absorbed in the small intestines (from food)
- Iron then accumulates in many organs/regions such as
o Liver, heart, kidney, pancreas, & joints (+ diabetes)
- Iron causes damage through productive of reactive oxygen species which damage cells
- Diagnosed through measurements of blood iron levels, transferring saturation, or liver biopsy
- Increased serum ferritin and transferrin-iron saturation are indications for mutation testing
What are symptoms of HH
Symptoms:
- Skin pigmentation
- Liver dysfunction
o Hepatocellular carcinoma
- Diabetes
- Cardiomyopathy
- Hypothyroidism
- Hypogonadism
- Arthropathy (joint pain)
What are the genetics behind HH
Genetics of HH:
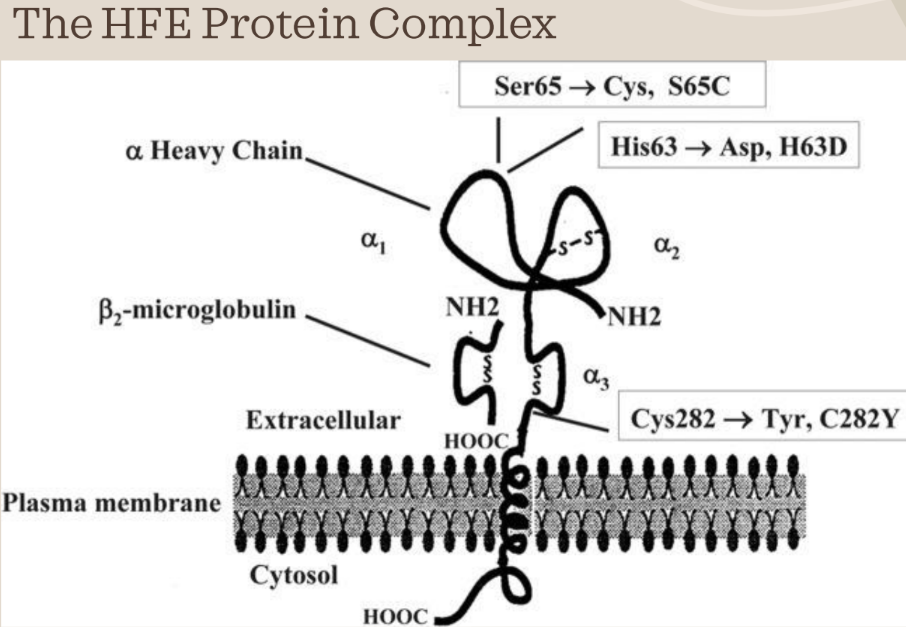
- HFE is found at 6p21.3, which is the short (p) arm of chromosome 6
- HFE has 7 exons through about 10,000 bp of genomic DNA
the mRNA transcript is about 2,700 bp long
- The final protein is translated to 348 amino acids
- The protein is inserted into the plasma membrane with a single transmembrane domain
o Alpha-3 domain spans the membrane
o The extracellular part interacts with beta-2 microglobulin
- The Alpha-1 and alpha-2 domains are extracellular and they interact with transferring receptor
What is the most common mutation for HH
Disease-causing mutations:
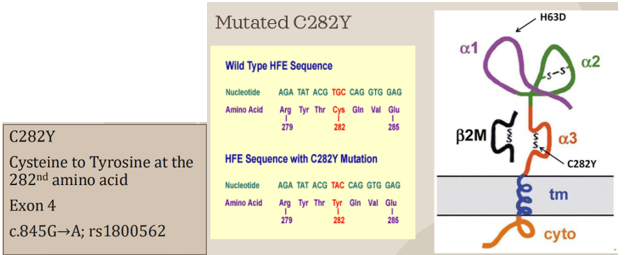
- The most common mutation responsible for HH is the substitution of tyrosine for cysteine at the 282nd amino acid position in the protein sequence (C282Y mutation)
o Found in 10% of Caucasian population
- The cysteine residue at this position is part of a disulfide bond that forms a loop in the alpha-3 domain of the HFE protein
- When cysteine 282 is lost, the disulfide bond cannot be formed and the hfe protein’s alpha-3 domain is no longer able to complex with beta-2-microglobulin, which serves as a stabilization factor
- As a result, the mutated HFE protein is degraded before it has a change to be incorporated into the cell membrane
What are the other (not most common) mutations for HH
Other HFE mutations:
- The second most common HFE mutation is H63D
o There is also S65C mutation
- Both of these mutations affect the alpha-1 binding groove which interacts with the transferrin receptor, but it does not impact the amount of HFE on the cell surface
- Thus, these mutations tend to have a milder phenotype than C282Y, if any
- A small portion of adults with C282Y/H63D or C282Y/S65C compound heterozygosity or H63D homozygosity develop mild iron overload, usually in the presence of concomitant liver disease

What is the cause of hemochromatosis
Causes of Hemochromatosis
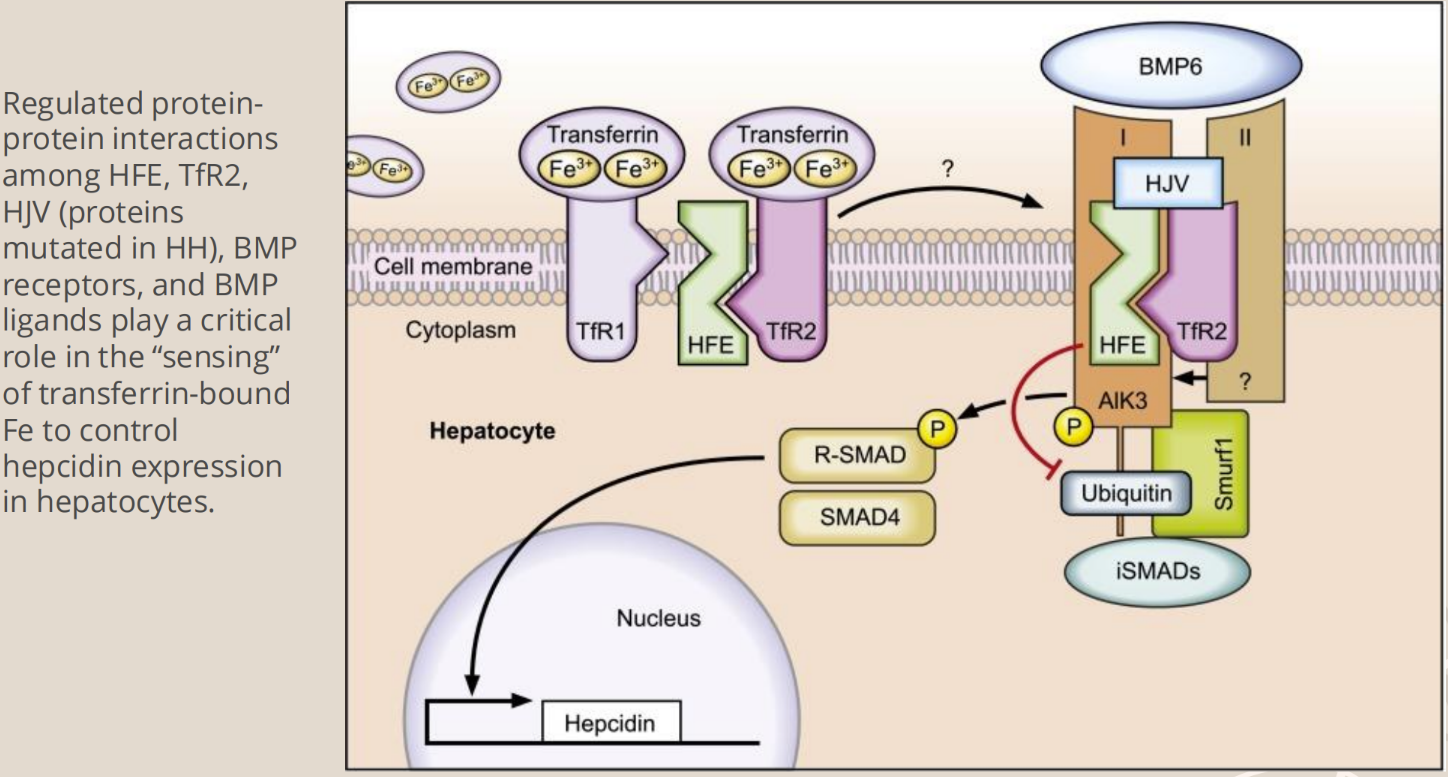
- The Hemochromatosis gene (HFE) = encodes for membrane bound protein that binds with B-microglobulin and transferring on the membrane of cells in the small intestine (and placenta)
o Caused by dysfunction of hemochromatosis type I HFE (HLA-H gene product)
- It is a membrane protein similar to MHC class I-type proteins
- Binds beta-2-microglobulin (B2M) through the alpha-3 transmembrane domain
- The HFE B2M protein complex then binds the transferring receptor (TFRC) through the HFE alpha-1 and alpha-2 domains
o The protein directs iron absorption based on cellular iron loads
- HFE function is required for normal regulation of hepatic synthesis of hepcidin, which is a major controller of iron metabolism
- If HFE is mutated, the gene expression of the hepcidin gene (HAMP) is significantly reduced
o HFE mutation results in decreased hepcidin responsiveness to iron, and thus iron is overloaded in the body
§ Because hepcidin is normally the iron regulator
§ Intestinal cells do not sense iron stores, and iron absorption continues into overload
How is HH diagnosed
Molecular diagnostics of hereditary hemochromatosis
- Any molecular method that can detect a single nucleotide variant can be used to diagnose HH
- Methods include...
o PCR, PCR-RFLP, Sequencing (Sanger, pyrosequencing, NGS), & Hybridization assays
What is used to detect HFE variants
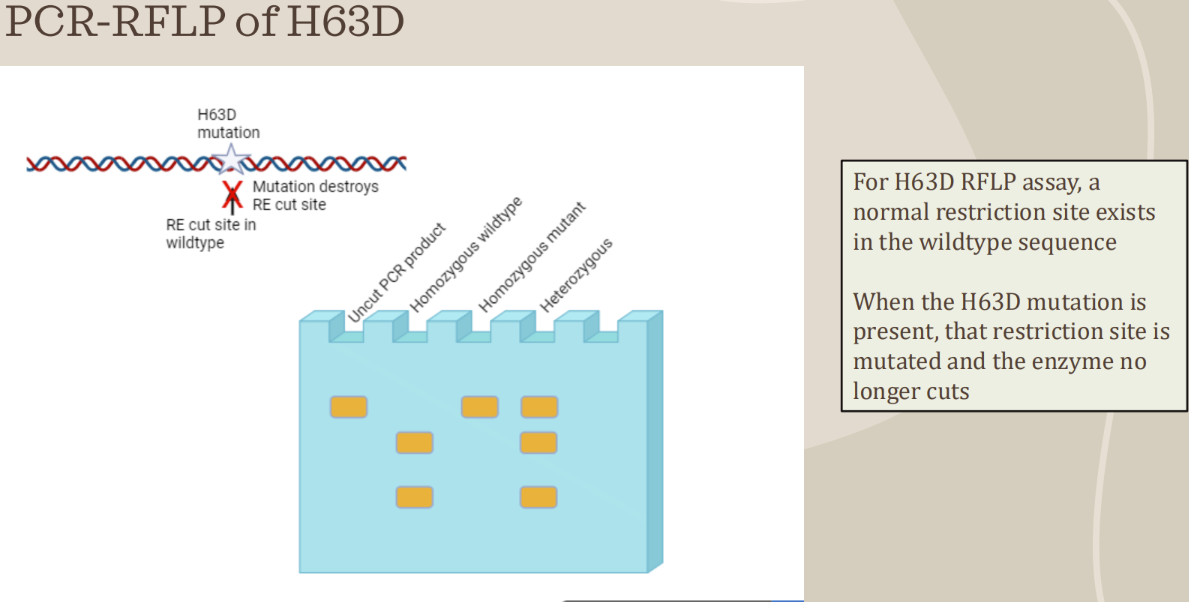
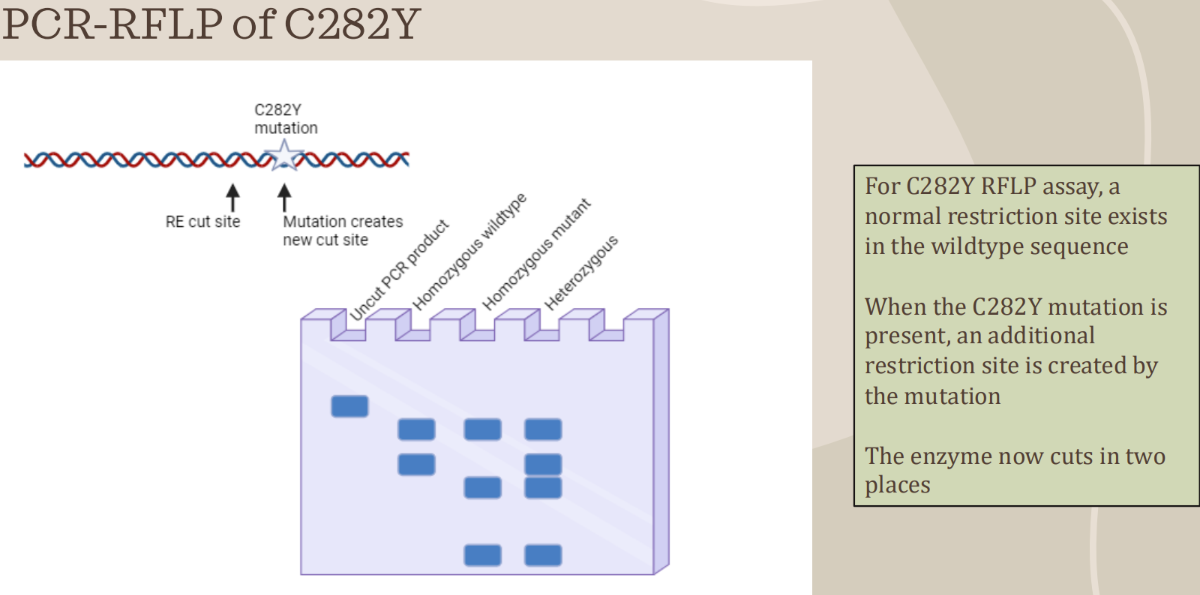
PCR-RFLP to detect HFE variants (C282Y mutation)
- Used to genotype genetic disorders caused by point mutations, insertions, or deletions
- After PCR, amplified products are cut with restriction enzymes at specific locations
- Analyzed results can determine if point mutation is present, and the classification of the individual as heterozygous (carrier) or homozygous (mutant or wild type)
- There are 2 alleles for each gene (potentially different, one from the mother, one from the father)
- Band patterns on a gel will indicate the genotype based on how particular enzymes cut the PCR product
How is HH treated
Treatment:
- Focused on reducing the accumulated iron in the body
- Common and most simple treatment is therapeutic phlebotomy
o Take blood on regular basis
- Iron chelating drugs can also be used to bind free iron
o Deferoxamine
- These treatments prevent further liver damage, reduce cardiomyopathy, returns skin pigmentation to normal, and can improve symptoms of diabetes
What is the disease penetrance of HH
Disease penetrance:
- Depends on age, sex, and clinical symptoms
- Most men with homozygous HH causing mutation do not develop clinical symptoms before the age of 40 years
- Fewer women with homozygous HH causing mutations develop clinical symptoms, if they do, they occur 20 years later than men, due to iron losses during menstruation, gestation, and lactation
Where is the HFE gene found
HFE is found at 6p21.3, which is the short (p) arm of chromosome 6
- HFE has 7 exons through about 10,000 bp of genomic DNA
the mRNA transcript is about 2,700 bp long
What are the demographics of HH
Demographics:
- Determined to affect 1 in 200-300 individuals of European descent
o 1 of every 8 northern European is a carrier of HH
- Affects 1 in 800-1,000 individuals across other ethnicities
- Affects men more frequently than women
o Men are ten times more likely to have disease symptoms (because women bleed every month, remove excess iron)
- Autosomal recessive inheritance pattern

What are evolutionary theories for HH
Evolution of HH:
- Many theories about the prevalence of HH in many populations...
o Did it evolve in Wikings who had iron-poor diets?
o Was it protective against the plague?
o Is there any evidence for or against these theories?
Theories
- Founder effect – mutation inherited from one ancestor and spread through population isolation.
- Nutritional adaptation – mutation favored survival in iron-poor agricultural societies.
- Disease resistance – mutation once offered protection against infections like the plague (later disputed).
- Hitchhiking effect – mutation rose alongside another beneficial gene.
