SCSC 304 Exam 2
1/73
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
74 Terms
Heritability
Degree to which variability of a quantitative trait is transmitted to progeny
P (phenotype) =
G (genotype) + E (environment)
Vp=
VG + VE + VGE
VG
VA + VD + VI
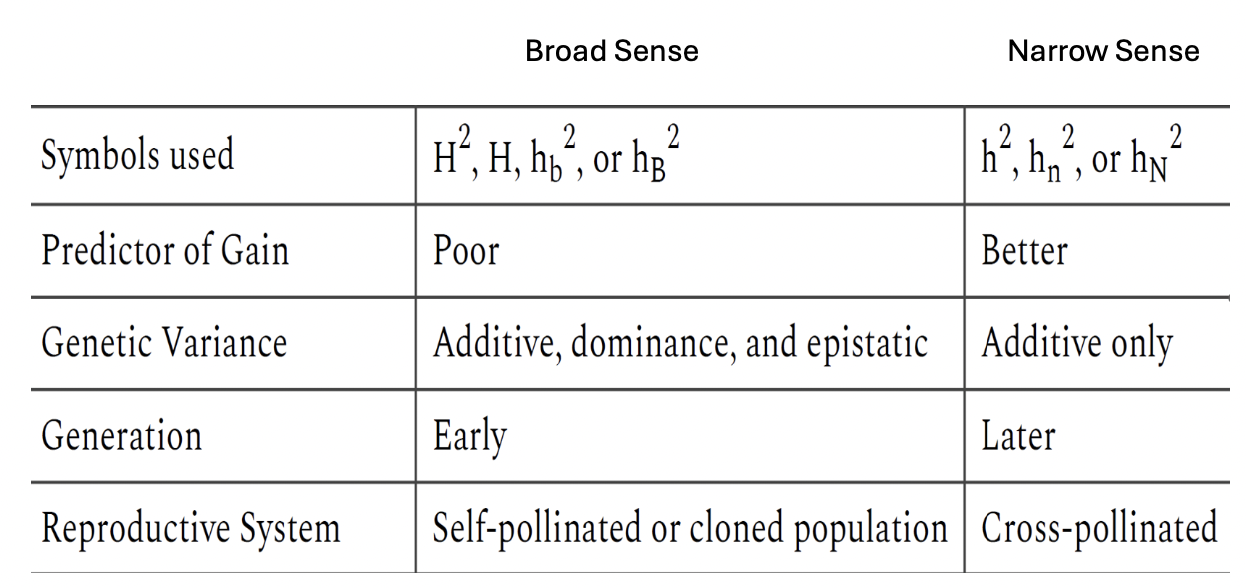
Broad Sense Heritability (H)
VG / VP
Narrow Sense Heritability (h2)
VA / VP X 100
Heritability from Variance Components
H = σ2g / ( σ2g + σ2gy + σ2gl + σ2gyl + σ2e)
σ2g = total genetic variance
σ2gy = genotype * year variance
σ2gl = genotype * location variance
σ2gyl = genotype year location
σ2e = experimental error
5 Heritability from Parent Assumptions?
Trait has diploid mendelian inheritance
Population from which the parents are taken are in random mating equilibirum
No linkage
Parents are non-inbred
No environmental relationship between the performance of the offspring and parent
Parent-Offspring Regression
h2= b * 100
b = Σ ( x – x ) ( y – y ) / Σ ( x – x )2
x = parent value
y = progeny value
Parent-Progeny Regression
y = a + bx
y = response or dependent variable
x = predictor or independent variable
a = y-intercept of line
b = regression coefficient, slope of line
Broad Sense vs Narrow Sense Heritability Table
Uses of Heritability in Selection
determine the relative importance of genetic effects which could be transferred from parent to offspring
determine which selection method would be most useful to improve the character
predict gain from seletion
if additive gene action is only important gene action, than an h2 estimate is needed
Selection Intensity and Genetic Advance
Gs = (i) (√VP) (h2)
Gs= predicted genetic advance
i= constant based on selection intensity in standard deviation units
√VP= square root of phenotypic variance
Gain per Year
Gs = [( c ) (i) (√Vp) (h2)] / y
y = # of years required to complete breeding cycle
if selection is based on one parent, c = 0.5
if two parents, c = 1.0
Complex Inheritance
Cannot be predicted by Mendelian ratios
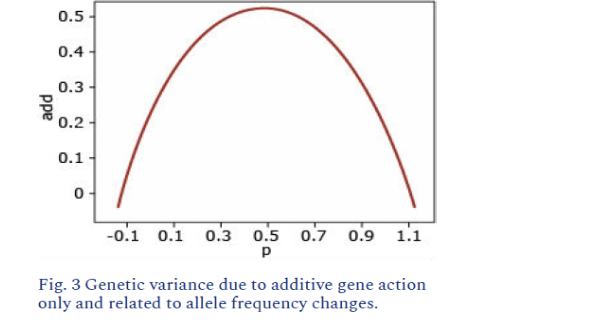
Additive Gene Action
each gene enhances trait expression
aabb=0, Aabb=1, AAbb=2, AABb=3, AABB=4
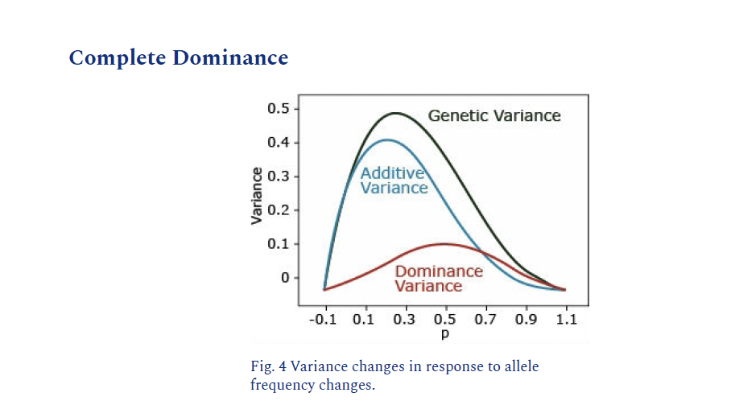
Dominance Gene Action
Heterozygote is more like one parent or the other
aa=0,Aa=2, AA=2
Epistatic Effects
Nonallelic interactions- individually genes have no effect, but together they do
AAbb=0, aaBB= 0, A_B_=4
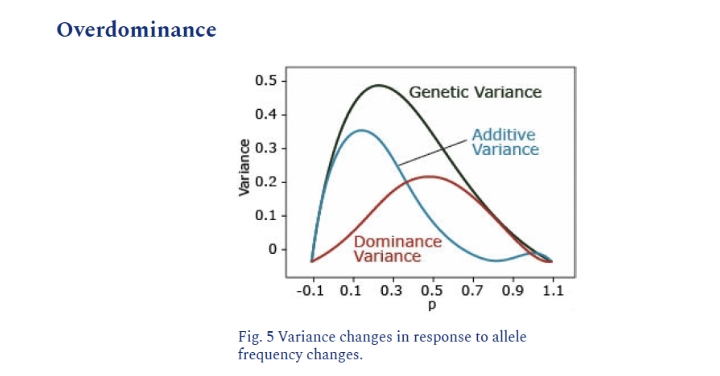
Overdominance
Each allele contributes, but together they exceed the sum of individual allele contribution
aa=1, AA=1, Aa=3
Gene Interactions (Epistasis)
Complementary
Modifying
Inhibiting
Masking
Duplicate
Additive
Pleiotropic genes
Complementary Action
Two non-allelic genes required to produce a single effect
AB= resistant; Ab, aB, ab = susceptible
Modifying Action
One gene produces an effect only in the presence of a second gene at another locus
In corn, Pr =purple aleurone color in the presence of dominant R.
But expresses no effect in the absence of R.
PrR=purple aleurone
prR=red aleurone
Prr and prr = colorless aleurone
Inhibiting Action
One gene inhibits action of another gene
In corn, the dominant gene R for red kernel color does not produce an effect in the presence of a dominant “inhibitor” gene I.
Ri= red
RI, rI, and ri = white
Masking Action
One gene may hide the effect of a second gene when both are present
In oats, a dominant gene Y produces yellow seed coat color
A dominant gene B produces a black seed coat.
The gene Y will have no visible effect in the presence of B because the black seed coat color will mask the yellow color.
• BY, By = black
• bY=yellow
• by=white
Duplicate Action
Either of the two genes may produce a similar effect, or the same effect is produced by both of them together
Common shephards purse has a triangular-shaped seed capsule produced from either of the dominant genes C or D , or by both together, CD.
With both recessive, the seed capsule has an ovoid shape.
Cd, cD, CD = triangular shape seed capsule.
cd= ovoid shape seed capsule
Additive Effect
Two genes may produce the same effect, but the effects are additive if both genes are present
In barley, either A or B will produce medium-length awns, while the two dominant genes together produce long awns.
The recessive genes produce awnless plants.
Ab, aB= medium length awns
AB= long awns
ab= awnless
Pleiotropic Effects
Single gene having more than one effect- simultaneous influence over size, shape, color, or function of several organs
In barley, the “uzu” gene in the recessive state may shorten stem and rachis internodes, reduce seed size, and produce an erect coleoptile leaf.
Uz= normal appearance
uz= semi-dwarf, dense spike, short awns, small seeds, short erect flag leaf.
Uses of Molecular Markers
selection of parents
mapping
marker assisted selection
cultivar identification
genetic diversity studies
4 Important Applications of Molecular Markers in Plant Breeding?
Screening single traits
Speeding up breeding programs
Germplasm evaluation
Cultivar identification and protection
Types of Molecular Markers
Isozymes- codominant
RFLPs- codominant
RAPDs- dominant
SSRs- codominant
AFLPs- dominant
SNPs- codominant
Advantages & Disadvantages of Commonly Used DNA Markers
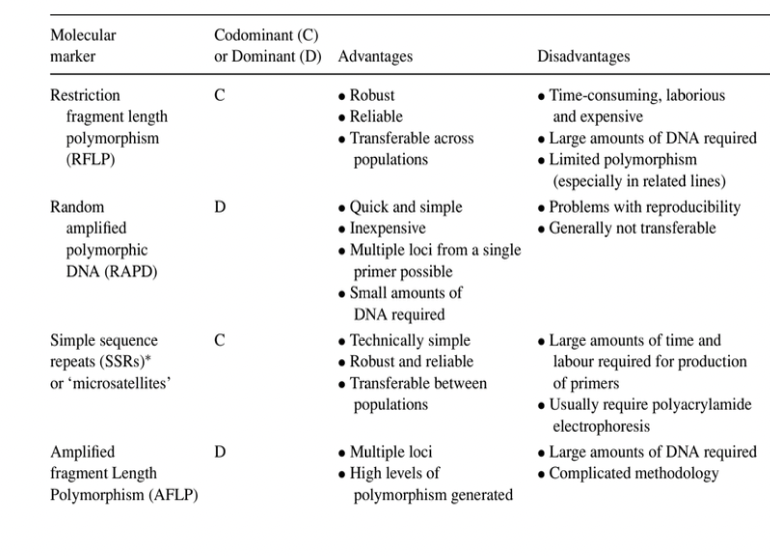
Simple Sequence Repeats (SSRs)
microsattellites- short, repetitive sequences common in eukaryotes
random repeat of 2-5 nucleotides
PCR bases
Single Nucleotide Polymorphisms (SNPs)
PCR bases
Single base pair site in genome that is unique per individual
often linked to genes
Requirements for Mapping QTLs
Trait phenotypes
Polymorphic markers
Genetic structure of populations
Single Marker Analysis
Compare trait means of different classes for each marker locus individually in the form of a single factor ANOVA
tests the association between a single marker and a trait
Single Marker Analysis Drawbacks
cannot accurately estimate QTLs effect
Doesn’t indicate on which side of the marker the QTL is located
Interval Mapping
QTL lies between two linked marker loci
Analyzed by statistical software (MapMarker/QTL)
Interval Mapping Drawbacks
can only map a single QTL location
many genes throughout genome affect quantitative traits
Functional Marker (FM)
derived from genomic region of trait-controlling gene
directly linked to a plant phenotypic trait
enable efficient characterization of germplasm without recombination
can be used to select for complex traits
100% predictive of a phenotype
Association Mapping (AM)
identifies polymorphisms near or within a gene of interest that controls the phenotypic differences within genotype
Linkage Disequilibrium (LD)
certain alleles are more likely to be found together on the same chromosome than expected if they were inherited independently
can be maintained for several generations between loci that are genetically linked, which enables marker assisted selection (MAS)
Random DNA Markers (RDMs)
most common markers for indirect selection
usually located near gene of interest, but don’t always lead to a selection of the desired phenotype
Genome Wide Association Studies (GWAS)
covers entire genome to identify regions associated with phenotypic traits
pinpoints which parts of genetic code are associated with traits
Methods of Transforming Plants
Agrobacterium
Particle bombardment
Electroporation
Promoters
drive DNA expression by determining the level of transcription of a selectable coding sequence
Constitutive Promoters
Promote frequent transcription and constantly “turned on.”
A common promoter is from the cauliflower mosaic virus called CaMV 35S
Tissue-Specific and Developmentally Regulated Promoters
Genes are only expressed in certain tissues or at certain times.
Inducible Promoters
Expression in response to injury, chemical, temperature stress, etc.
Agrobacterium
invades a plant’s DNA, performs a coup de’tat, and makes the plant form a gall for the benefit of the pathogen
Particle Bombardment
considered direct gene transfer
gene gun
not very efficient
plants with larger cells usually more successful
Electroporation
Callus culture, or explants such as immature embryos or protoplasts, is placed in a cuvette.
It is inserted into an electroporator.
This procedure widens the pores of the protoplast membrane by
means of electrical impulses.
The widened pores allow foreign DNA to enter and become integrated with nuclear DNA
GMO
the insertion of DNA (gene) into a species where the gene is not normally found or expressed
Non-GMO
the insertion of DNA (gene) that is found to exist naturally in the same species (taxa) or closely related and often cross-compatible species. (e.g. CRISPR)
CRISPR
Bacteria capture pieces of viral DNA when they are infected
These pieces are inserted into the bacteria’s DNA
These segments are called CRISPR arrays
Scientists have repurposed this system to edit DNA in living organisms
Mutations
source of all variation in plants
most are recessive
chromosomal- rearrangement, loss, or duplication of chromosome segments
loss or duplication of entire chromosomes
Tissue often treated to induce mutations
seed (most often)
vegetative tissue (rhizomes, stolons, bulbs, corms)
pollen
callus(tissue culture)
frame-shift mutations
changes reading frame
usually results in complete silencing or malfunction of gene
Germplasm
genetic source material used by plant breeders to develop new cultivars
Sterility
failure to complete fertilization and obtain seed as a result of defective pollen or ovules, or other aberrations
Flower parts may not exist or function
pollen may not be compatible with stigma, etc
Embryo may not be able to survive
Resulting hybrid progeny may be infertile
Incompatibility
results from the failure to obtain fertilization and seed formation after self-pollination, usually due to failure of the pollen tube to penetrate the stigma or due to reduced growth of the pollen tube in the style
encourages cross-pollination
discourages inbreeding
Male Sterility
failure to form functional anthers or pollen
flowers can be cross-pollinated
Genetic male sterility
nuclear genes inhibit normal development of anthers and stamens
Male sterility genes
recessive(ms)- inhibits male floral function
Uses of male sterility in breeding programs
eliminates emasculations for producing hybrid progeny
increases natural cross-pollination in normally self-pollinated crops
Cytoplasmic Male Sterility (CMS)
controlled by factors in cytoplasm, but influenced by genes in nucleus
flowers have non-functional male parts
result of nuclear chromosomes in foreign cytoplasm
How CMS Works
• Fertility restoring genes, located in the chromosome, can make cytoplasm operative.
• Parent with the sterile cytoplasm used as the female plant
• Parent with fertility restoring gene used as the male parent
Utility of CMS
production of hybrid seed in many crops
Chemically Induced Male Sterility
flexible system that kills pollen or prevents its formations
drawback is getting complete pollen sterility is often difficult
Apomixis
seeds formed without union of sperm and egg
Obligate apomixis
plants reproduce only by apomixis
facultative apomixis
sexual and asexual embryos may coexist in the same ovule or same plant
Chasmogamous flowers
when flowers open male and female floral parts at same time
promotes cross-pollination
Cleistogamous flowers
flowers are closed when male and female floral parts mature
promotes self-pollination
Dichogamous flowers
male and female floral parts mature at different times- either within a flower or in different flowers on the same plant
promotes cross-pollination