15 - oxidative phosphorylation
1/15
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
16 Terms
explain the Coupling of the Electron Transport and Oxidative Phosphorylation
is uncoupling a thing?
ATP production linked to exergonic steps of electron transport
Phosphorylation tightly coupled to respiration (due to intact membrane)
Intact membrane
Uncoupling → can be done by some molecules that dissipate the proton gradient therefore ATP cannot be made)
Inhibition
what 2 things drive ATP synthesis
chemical potential (decrease in pH/increase in H)
electrical potential
what is the chemiosmotic hypothesis?
the proton motive force is enough to drive ATP synthesis
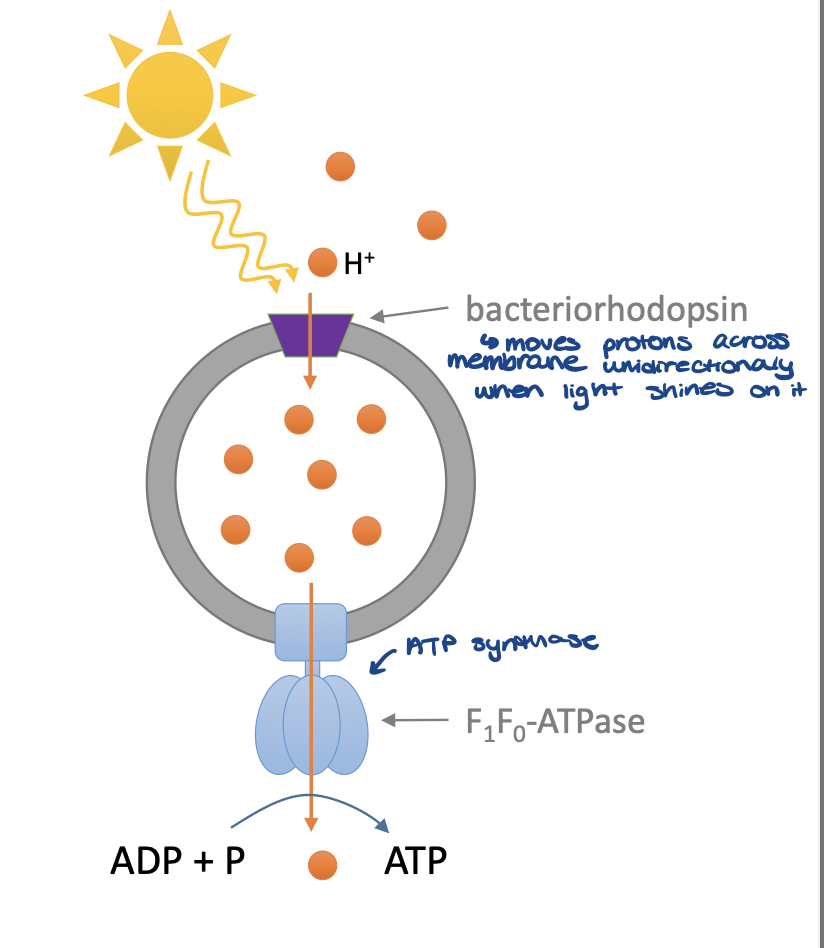
ATP synthase and bacteriorhodopsin
bacteriorhodopsin is a protein on the membrane of a vesicle
when light shines on the bacteriorhodopsin, it moves protons across membrane to build up the H+ concentration in the membrane
the vesicle will then associate with ATPase and cause ATP synthesis as H+ leave the vesicle
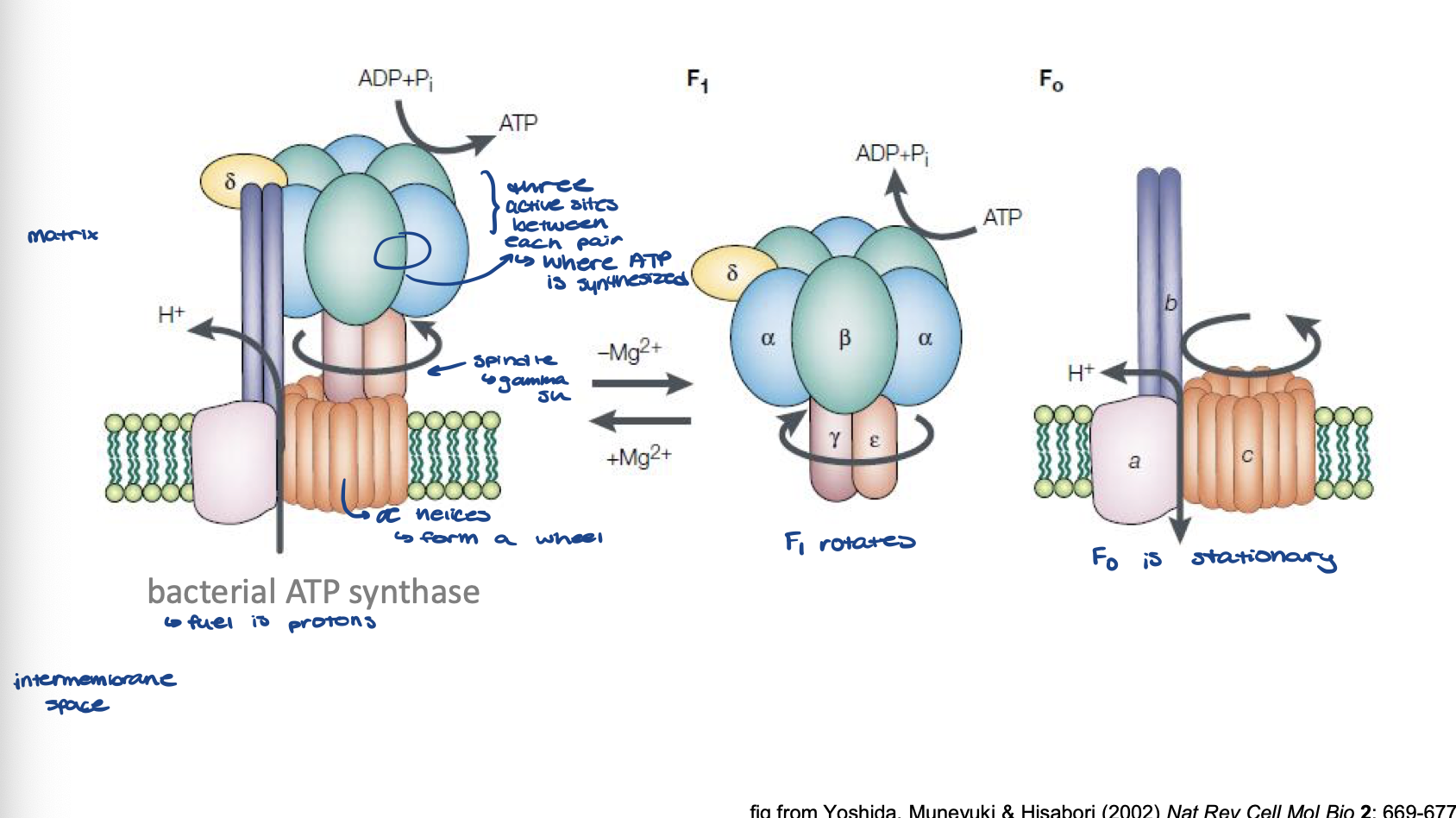
describe the structure of ATP synthase
Fo
stationary
a helices form a wheel in membrane
F1
rotates which is driven by H+ movement
spindle is a gamma subunit that does rotation
at head, 3 active sites btwn each pair of alpha-beta subunits where ATP is synthesized
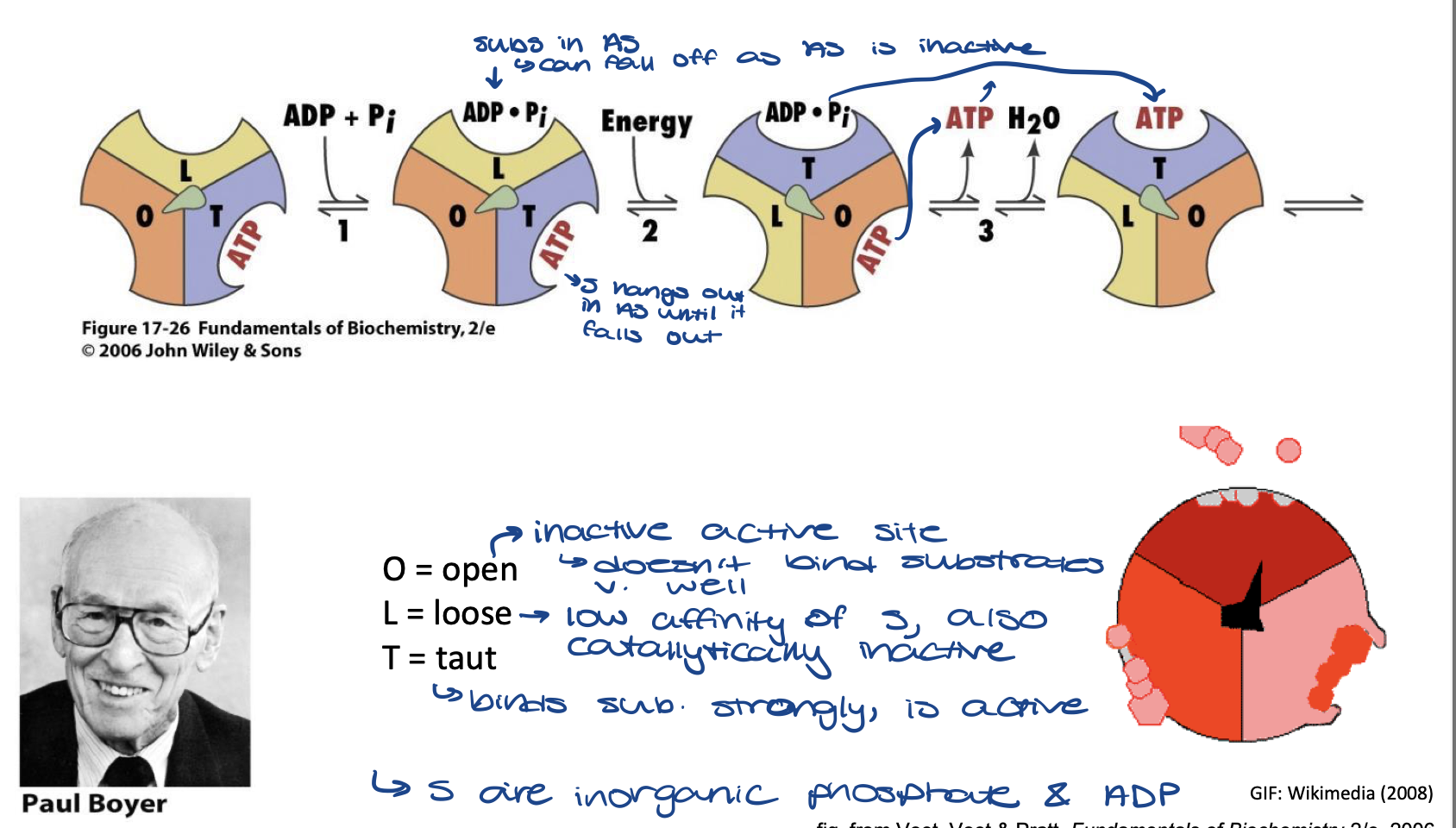
Boyer’s “Binding Change” names of active site
F1 being observed as if it is fixed in place
O = open
inactive active site - doesn't bind substrates (Pi and ADP) very well
L = loose
low affinity of S also catalytically inactive
T = taut
binds substrate strongly, is active
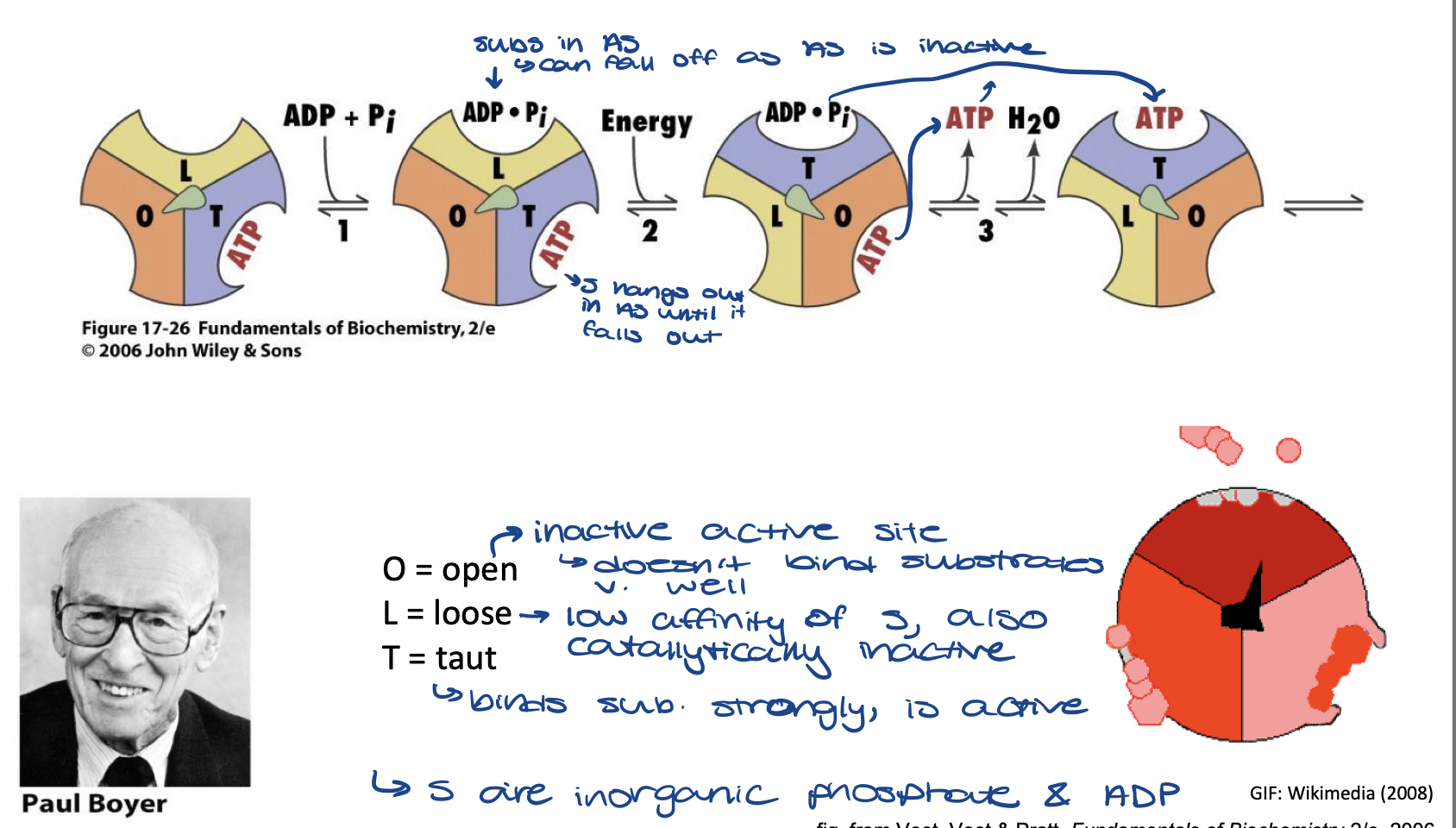
Boyer’s “Binding Change” Mechanism of ATP Synthesis
substrates (ADP and Pi) bind to loose active site. Taut active site still has ATP product from last reaction
energy is added and spindle rotates so the substrates are now in the Taut active site and reaction occurs.
old ATP leaves moves to the open active site and leaves soon, a dehydration reaction occurs
new ATP is in the Taut active site until next reaction occurs and then it will move to the loose active site
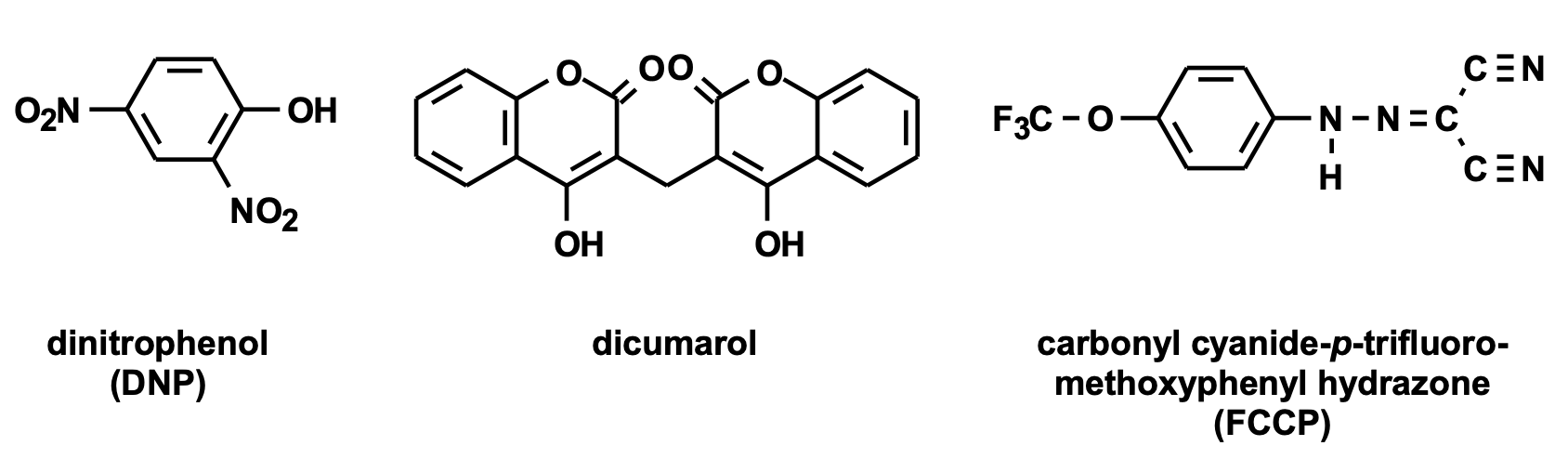
Uncoupling Oxidative Phosphorylation
what happens
what are the characteristics of the molecules that do this?
ETC will still run, but not making ATP so it becomes futile, just using up resources
molecules:
all are hydrophobic (dissolved in membrane)
need weakly acidic or basic FGs:
easily lose H+ at physiological pH
pKa near physiological pH
dissipate gradient via acid-base chemistry
Biological Uncoupling
what is the protein called?
what does it do?
in what organisms is it found?
thermogenin
uncoupling protein
creates heat, found in organisms that need to generate heat
has channel that protons can travel through back to matrix to dissipate H+ gradient which generates heat
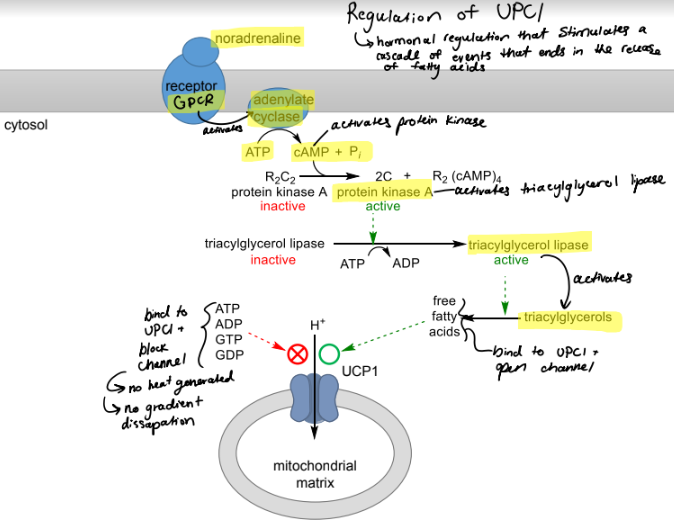
how is thermogenin (UPC1) regulated?
stimulated: hormonal regulation by noradrenaline that stimulates cascade of events that liberates fatty acids to bind UCP1 open channel → allows H+ out and back into matrix, dissipates gradient and generates heat
inhibited: ATP, ADP, GTP, GDP binds to UPC1 and black the channel, no gradient is dissipated
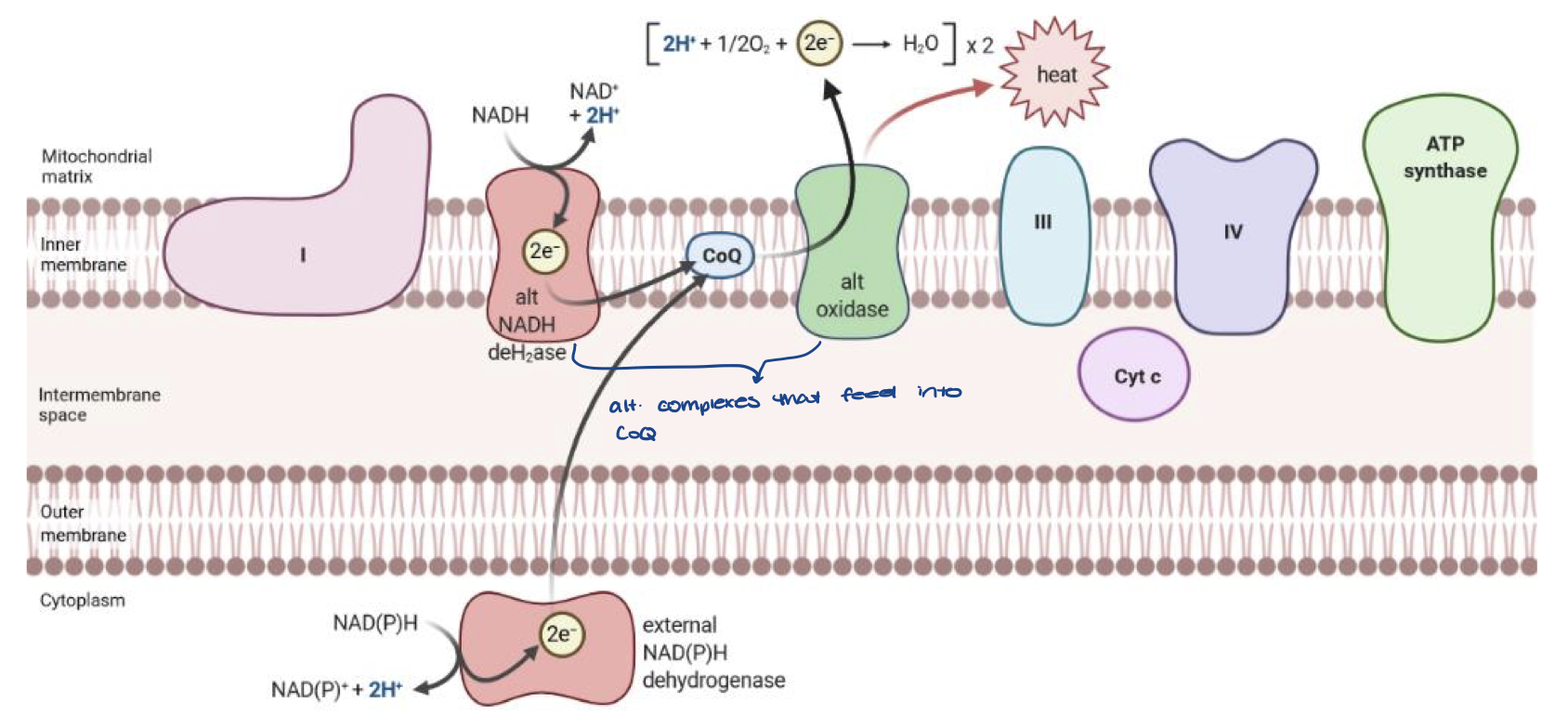
Uncoupling in Plants
oxidase protein transfers H+ and electrons to O2
this takes H+ and electrons out of the ETC so no ATP is made
also generates heat
Transport & Energetics
ATP-ADP Translocase
antiporter
translocates ATP from matrix into IM space and ADP from IM into matrix
energetic cost of 1 H+ to this translocation due to charge diff btwn ATP and ADP
drives reaction by brining in substrate and removing product
Phosphate Translocase
makes up the charge difference between ATP and ADP translocation
symporter of H2PO4- and H+
how many total ATP are made per glucose?
30 or 32
Depends on shuttle system transferring reducing equivalents into the mitochondrion
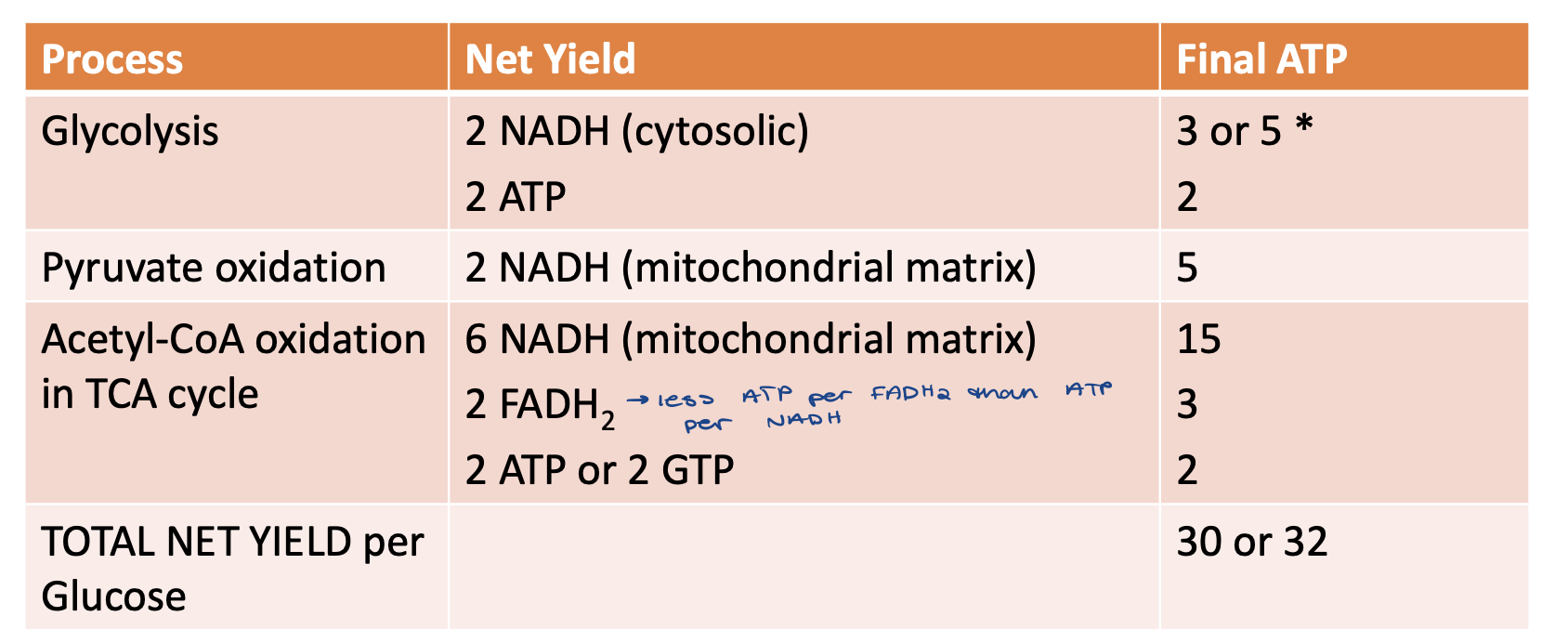
what is the P/O ratio?
P/O ratio
phosphorylation/oxidation
NADH → 5ATP/2NADH = 2.5 ratio
FADH2 → 3ATP/2FADH2 = 1.5 ratio
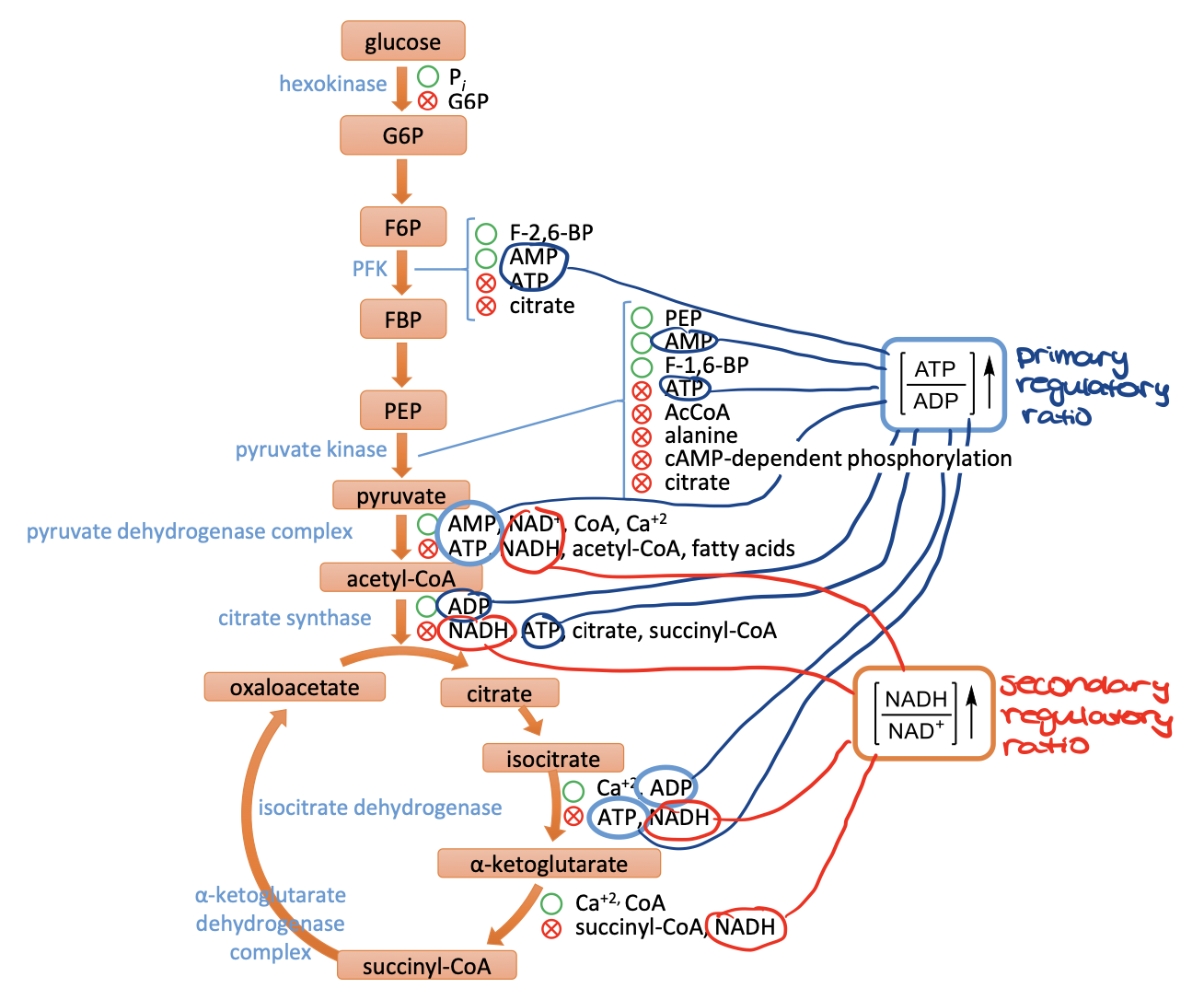
respiratory control
primary regulatory ratio is ATP/ADP
increase the ratio will inhibit ATP making pathways, decreasing will have the opposite effect
secondary regulatory ratio is NADH/NAD+
increase the ratio will inhibit NADH making pathways, decreasing will have the opposite effect
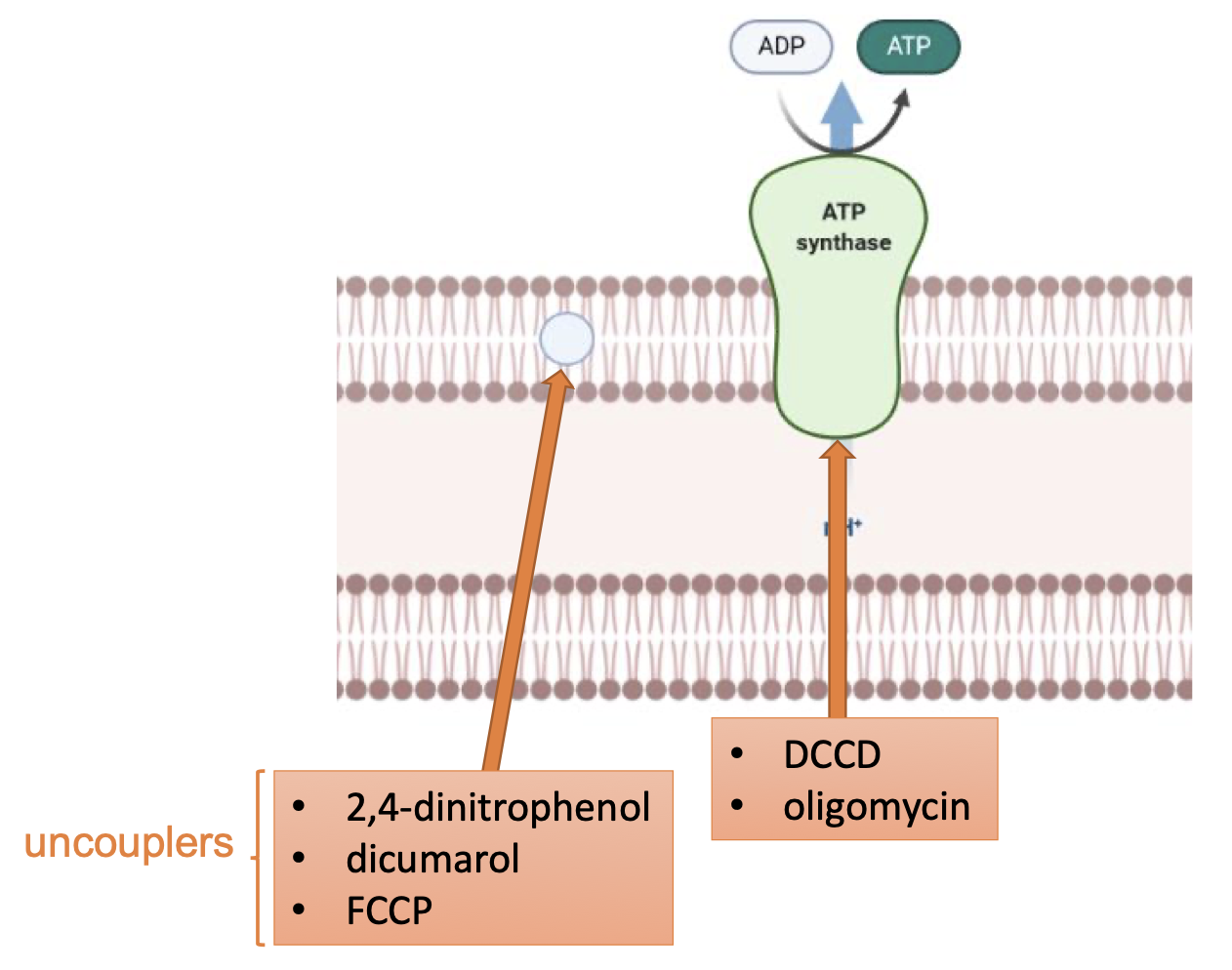
ATP Synthase Inhibitors
2,4-dinitrophenol, dicumarol, FCCP
uncouplers → dissipate the proton gradient to reduce chemiosmotic potential
DCCD, oligomycin
ATP synthase inhibitors → bind to ATP synthase and block it