MICR2000 - All Modules
1/242
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
243 Terms
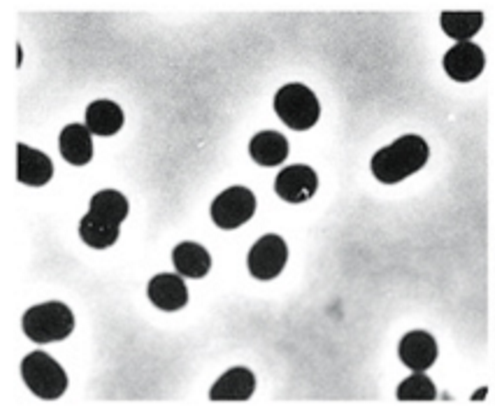
What is this
Coccus (cocci) - sphere
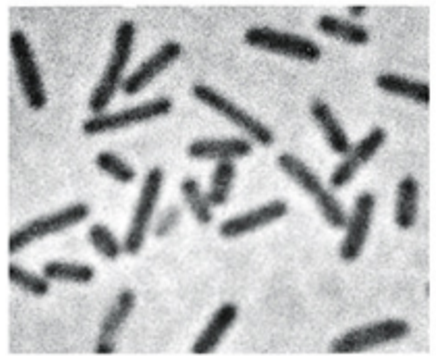
What is this
Rod shaped
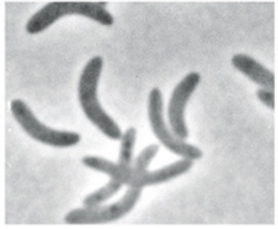
What is this
Spirillum (spirilla)

What is this
Spirochete
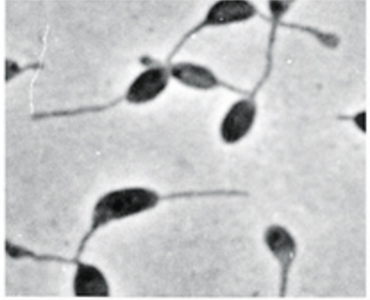
What is this
Budding and appendaged
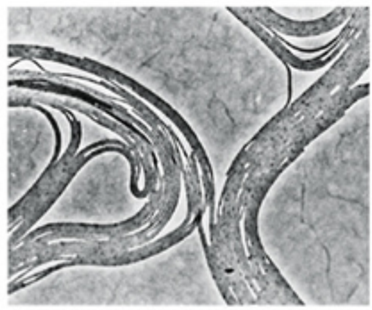
What is this
Filamentous
Robert Hooke
First description of microbes
fruiting structures of molds
Antoni Van Leeuwenhoek
First to describe bacteria
Louis Pasteur
Showed microbes responsible for fermentation
Disproved theory of spontaneous generation (microbes appear from thin air)
Koch’s postulates
used to establish causal relationship btwn microbe and disease
The suspected pathogen must be present in all cases of disease and absent from healthy animals
Suspected pathogen must be grown on pure cultures
Cells from a pure culture must cause disease in a healthy animal
The suspected pathogen must be reisolated and shown to be the same as the originals
Cytoplasmic membrane structure and function
Phospholipid bilayer with embedded proteins
Functions as highly selectively permeable barrier
Bacterial vs Archaeal Cytoplasmic Membranes
both phospholipids
Bacteria
fatty acids joined to glycerol through ester linkages
Archaea
isoprene units joined to glycerol via ether linkages
Gram +ve vs Gram -ve cell walls
Both
peptidoglycan - rigidity
composed of G + M
peptide crosslinking provides strength in X and Y direction
Gram +ve
90% peptidoglycan
peptidoglycan above cytoplasmic membrane
crosslinking via peptide interbridge
teichoic acids (embedded) and lipoteichoic (teichoic acids covalently bonded to lipids) acids in cell wall
Gram -ve
10% peptidoglycan
peptidoglycan sandwiched btwn outer and inter membranes
crosslinking via NH2 group of DAP and COOH of D-alanine link
Periplasm
gel-like compartment containing proteins btwn outer and cytoplasmic (inner) membranes
Lipopolysaccharides embedded in outer membrane
Lipid A (endotoxin) buried in membrane - highly toxic
core polysaccharide and lipid A conserved
O-antigen (sticking out) varies
Gram staining
Process
add crystal violet
crystal violet makes cells purple but will wash out unless u cross link with iodine
add iodine
iodine only cross links with crystal violet in peptidoglycan - only g+ve have peptidoglycan exposed
wash with alcohol
washes off crystal violet on gram -ve cells
counterstain with saffranin
stains g-ve pink
Acid fast bacteria
E.g., Mycobacterium
Waxy lipid mycolic acid makes cell impermeable to gram stain components
Able to be stained red using acid fast stain
Cell walls of Archaea
S layer
interlocking protein and glycoprotein arranged in paracrystalline surface structure
no peptidoglycan or outer membrane
Some archaea contain pseudomurein - similar to peptidoglycan
Capsules of bacteria
Polysaccharide layers
assist in attachment and biofilm formation
aid in evading immune system
resist desiccation
Fimbriae
Filamentous bacterial appendages involved in adhesion
Different fimbriae can be expressed to allow adhesion in different environments
Flagella (arrangements, structure, biosynthesis)
3 arrangements
peritrichous
spread evenly around cell
polar
one flagella
lophotrichous
multiple flagella concentrated on one end
Structure
filament
composed of flagellin
hook
connects filament to motor
mot proteins
motor
anchored to cytoplasmic membrane and cell wall
fli proteins
motor switch
reverses rotation of flagella in response to intracellular signals
Biosynthesis
requires many genes
flagellin synthesised in cytoplasm, moved through filament to build (filament core is hollow)
Types of flagella movement
Polar
move forwards and backwards
can reorient to move forwards/backwards in different heading
Peritrichous or lophotrichous
can bundle flagella to move like polar cells
tumble and run
flagella pushed apart to tumble
bundled to run
Gliding motility
Flagella independent motility - very slow
slime
shoot slime for propulsion
type IV pili
attach to surface and twitch
specific proteins
membrane fluidity - proteins move along cell membrane and push against a surface
Microbial taxes
Chemotaxis
moving up a gradient of attractant chemicals
increased tumble and run
chemoreceptors
measured using capillary tube assay if run and tumble behaviour not observed)
things attracted or repelled by tube
Cell inclusions
Aggregate of a molecule
lipid or glycogen storage for extra energy
accumulate inorganic compounds
magnetosomes
magnetite, orient bacteria in magnetic field
Gas vesicles
spindle shaped gas filled protein structrues
buoyancy
optimise position in water column
Endospores
Dormant bacteria resistant to extreme conditions
ideal for dispersion in harsh environments e.g., wind, water, gut
only in some g+ve
Germinate to produce mature spore (sporulation)
Structure
exosporium = outermost protein coat
spore coats = more layers made of peptidoglycan or carbohydrates
cortex = peptidoglycan
core = cellular components surroudned by core wall
Generation time/doubling time
Interval for formation of two cells (from one cell)
Batch culture
Closed system microbial culture of fixed volume
Phases of growth curve
Lag phase
period before growth begins at maximal rate
cells adapted to stationary phase have to adapt to fresh medium (adapt for growth) or adapt to different medium (e.g., minimal) by producing new enzymes
Exponential phase
doubling/generation time (g) constant
N = N0×2^n
n = number of generations
t = incubation time (time)
g=t/n
mean growth rate constant (k) = n/t
Stationary phase
no net increase in cell number
something limiting growth - nutrient or inhibitory products
adaptation to stationary phase - specific genes
RpoS (RNA polymerase sigma factor)
directs RNA polymerase to transcribe stationary phase adaptation genes
Death phase
cells die
associated with lysis
Total cell count
counting cells in square and extrapolating
rapid but imprecise, hard to see small cells, can’t distinguish live from dead
Viable cell count
serial dilutions until little enough cells to count, then use dilution factor
Turbidimetric measurements
measures turbidity - how much light is scatterede by density of culture
optical density (OD)
Primary vs secondary metabolites in microbial growth
Primary
made during exponential growth
Secondary
produced during stationary phase
not essential for growth
overproduced
Biofilms
community of bacterial cells enclosed in self produced matrix (polysaccharide, protein, extracellular DNA) and adhered to surface
Model of biofilm development
Reversible attachment - bacteria contact surface with flagella
Irreversible attachment - fimbriae/pili attach to surface, adhesins
Microcolony formations
matrix of polysaccharide, protein, DNA surrounds colony and holds it together
quorum sensing
bacteria communicate with homoserine lactone (HSL)
cells start communicating when quorum achieved (minimum conc of HSL for comms)
regulate gene expression, control cell density, change behaviours
Metabolic interactions
Dispersal of bacteria
Why do bacteria form biofilms?
Self defense (physical forces, phagocytosis, antibiotics)
Colonise favourable niches (remain attached to nutrient rich areas)
Enable bacteria to live together (quorum sensing, genetic exchange)
Survival strategy when nutrients limited
Why are biofilms antibiotic resistant?
Slow penetration of antibiotic
Inner layers have more time to adapt to antibiotic
Cells in nutrient low zones inactive and thus antibiotics may not act on them - persister cells are in dormancy
Antimicrobial depletion due to adsorption
Sterilisation
killing or removal of all viable organisms within a growth medium
Inhibition
Effectively limiting microbial growth
Decontamination
Treatment of an object to make it safe to handle
Disinfection
Directly targets removal of all pathogens, not necessarily all microbes
Microbial growth control
Heat sterilisation
endospores can survive heat that would kill vegetative cells
decimal reduction time = time req for 10 fold reduction in viability
thermal death time = time to kill all cells at given temp
autoclave - uses steam under pressure
Pasteurisation
precise heat to reduce microbial load, controls pathogens not killing all
Radiation sterilisation
ionising radiation can also produce reactive molecular species
Filter sterilisation
filter organisms out
Antimicrobial agent classification
Bacteriostatic
stops growth of bacteria but doesn’t kill them
Bacteriocidal
kills but doesn’t lyse cells - viable cell count decreases
Bacteriolytic
kills and lyses bacteria
Measuring antimicrobial activity
Minimum Inhibitory Concentration
smallest amount of agent req to inhibit growth of microbe
Disk diffusion assay
Microbial community vs population
Community = multiple interacting populations
Population = one species
Classes of antimicrobials
Synthetic
growth factor analogues - man made
Semi synthetic
microbially produced and chemically modified
How are drugs that block the synthesis of folic acid able to maintain
selective toxicity?
humans acquire folic acid from diet while bacteria synthesis it themselves
Beta lactam antibiotics
Beta lactam ring
incl penicillins
inhibit cell wall synthesis by binding to penicillin binding proteins (PBPs) and preventing cross linking
cell wall synthesis continues but weakened
Penicillin-PBP complex stimulates release of autolysins, which degrade the cell wall (normally used for cell division)
AMR mechanisms
resistant organisms can:
lack structure antibiotics inhibit
impermeable to antibiotic
inactivate antibiotics
modify target of antibiotic
resistant biochemical pathways (take up folic acid instead of secreting)
efflux of antibiotic
Vancomycin resistance
Vancomycin = last line antibiotic
Resistant S. aureus replaces components of cell wall with different ones still recognised by PBPs for cross linking but no by vancomycin
Conjugation
Conjugative R plasmid
DNA transfer from one cell to another
Attach two cells via pilus
transfer one strand of plasmid to recipient cell
synthesis of complementary strands in both cells
cells separate
Can carry AMR genes
Transposable elements
DNA sequences that can move positions within or between DNA strands
Two types:
Insertion sequences (IS)
smallest, encode transposase (endonuclease and integrase activities)
this cuts it out and puts it in somewhere else
IS does not encode any other genes
flanked by inverted repeat (IR) that is recognised by transposase
Transposons
contain transposase and IR as well as non transposition related genes - often AMR
often contain integrons
contain promotor and attachment site for gene cassettes (free floating DNA)
cassettes encoding gene followed by integrase specific recombination site - 59 base element recognised by integrase
integrase integrates cassettes to attachment site and promoter tests what they do
cassettes excised and transferred btwn integrons
Transposition methods
Conservative - cut and paste
transposase cuts target DNA (staggered nick)
IS integrates
Replicative - copy and paste
TnpA gene transcribed to make transposase
transposase binds to IR and initiates transposition
cuts donor plasmid at ends and makes staggered nicks on target plasmid
ligation of transposon to target ends
3’ ends replicate through DNA polymerase, replicating the carried genes of the transposon
this forms a cointegrate - a single molecule of DNA between the donor and target plasmid (looks like 8)
contains 2x copies of transferred DNA
Resolvase binds to ‘res’ regions on transposon
cuts and recombines forming 2x plasmids with the transposon
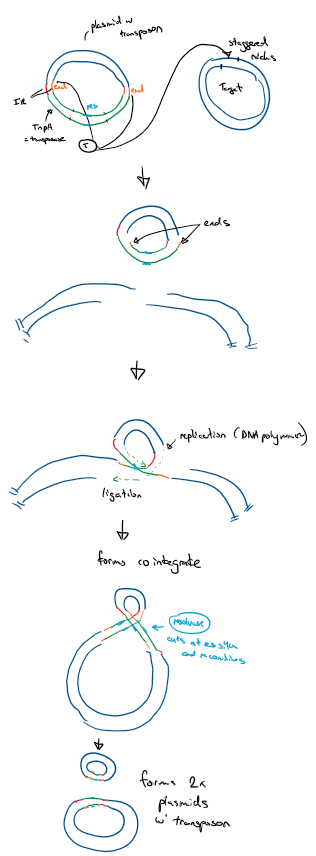
How to stop spread of AMR
stop inappropriate use of antibiotics to reduce selective pressure
remove ineffective antibiotics from use
monitoring, isolation and treatment programs to prevent establishment and spread of multiple resistant pathogens
Normal flora
microbes that live on and in our body without causing infection under healthy conditions
balance btwn enough for microbes to survive and not enough to cause infection
pathogens can be transient members of normal flora
Virulence factors
Bacterial product or strategy contributing to virulence or pathogenicity
colonisation of host
evade immune system
damage host
6 categories
motility
motile bacteria can target host cells in dynamic environments like mucosal environments
ability to contact host cells
adhere to host cells and resist physical removal
pili/fimbriae and adhesins
invade host cells
invasins allow penetration
inside the cell access to nuritents, hide from immune system, divide and multiply
resist phagocytosis and complement
capsules hard for macrophages to attach and engulf, biofilms
evade immune defenses
phase variation of surface
vary surface structures to evade detection
capsules can resemble human tissue
don’t evade - KILL
endotoxin
lipid A in gram -ve outer membrane
released when bacteria attacked (membrane breached) and can be secreted
exotoxin
soluble excreted toxins
toxin genes spread on plasmids
cytotoxins kill or inhibit cells
neurotoxins interfere with nerves
enterotoxins affect epithelial cells of GI tract
requires production of antitoxins
ability to compete for nutrients
compete with host tissue and normal flora for limited nutrients
Staphylococcus aureus virulence factors
g+ve cocci
adhesins - adhere to host cells
secrete exotoxins that kill host cells
secrete enzymes that deteriorate red blood cells and immune system enzymes
neutralise hydrogen peroxide from macrophages - resist phagocytosis
protein a - evade immune system
capsule - resist phagocytosis
coagulase - slow down immune system through blood clot
Helicobacter pylori and virulence factors
Host adapted pathogen that colonises human stomach and duodenum - inhabits mucosal layer )noninvasive) so not cleared by immune response (persistent infection), can be treated by antibiotics
symptomatic or asymptomatic infection - virulent strains have cag a pathogenicity island
Virulence factors:
Urease
bacteria imports urea from gastric juice (through porin to periplasm, UreI to cytoplasm)
urease in cytoplasm catalyses urea → ammonia reaction
ammonia makes gastric acid more basic allowing H pylori to survive
Flagella
motility
lophotrichous arrangement
move in mucosal lining
Adhesins
BabA and SabA allow adherence to gastric epithelium
Mucinase
degrades gastric mucus locally for easier motility
CagA - cagA pathogenicity island confers high virulence
CagA protein and type IV secretory system transcribed and translated
injected into host cells via type IV secretory system (syringe) to release pro-inflammatory cytokines
increases acid which wears away mucous
Testing for H. pylori
Rapid urease test
pH test for urease catalysing ammonia production
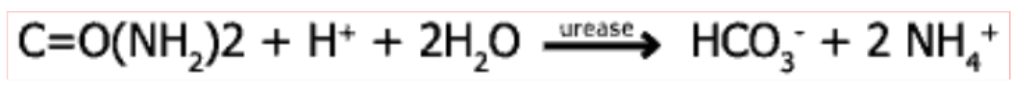
Treatment and prevention of H. pylori infection
Acid lowering drugs
Antibiotics
Group A Streptococcus (Streptococcus pyogenes) diseases, location
Location
skin and throat
Range of diseases
sore throat
localised common infections
cellulitis
impetigo (skin infection)
less common invasive infections
bacteraemia
toxic shock systems
necrotising fascilitis
post streptococcal sequelae - diseases after repeated infection with GAS
kidney failure and acute rheumatic fever (heart failure)
immune sequelae
GAS makes M protein which has anti phagocytic activity
similar to heart myosin - autoimmunity against heart myosin causing rheumatic heart disease
How do GAS infections align/misalign with Koch’s old postulates?
1 - Bacteria present in every case of disease and absent in healthy animals (NO FIT)
GAS present in normal flora
2 - Bacteria must be isolated from host with disease and grown in pure culture (FITS)
GAS can be cultured
3 - Specific disease must be reproduced when pure culture of bacteria is inoculated into a healthy susceptible host (NO FIT)
bacteria absent from post streptococcal sequelae so this doesn’t hold
different strains produce different things
4 - Bacteria must be recoverable from experimentally infected host and found to be same as original (NO FIT)
different strains of GAS
Koch’s molecular postulates
Identifying the gene or gene product responsible for virulence rather than the pathogen
Postulates
shows gene present in strains of bacteria that cause disease and not present in avirulent strains
disrupting the gene reduces virulence and reintroduction restores virulence
introduction of cloned gene into avirulent strain congers virulence
gene is expressed (not methylated)
specific immune response to gene protects against virulence
Is HtrA involved in GAS virulence?
thought HtrA involved in protecting GAS proteins during thermal stress
did test and found virulence disappeared when DNA added but didn’t return when it was returned to normal.
polar effect - DNA downstream affected (frame shift)
double crossover recombination
keeps reading frame the same
deleted mutant had no effect on virulence
Therefore HtrA doesn’t affect virulence
GAS virulence factors
M protein
helps resist phagocytosis
similar to heart myosin - immune sequelae
Fibronectin binding proteins (FBP)
allows GAS to bind to fibronectin in ECM of tissues and colonise that tissue
Phase variation
swap out FBPs
different combinations can contribute to different tissue binding - tissue tropisms
redundancy in FBPs allows infection of more than one tissue
What is a virus?
Structure evolved to transfer nucleic acid from one cell to another
obligate, intracellular parasites
cannot replicate outside host cell
possess one kind of nucleic acid (DNA or RNA)
limited genetic material
no ribosomes
viral nucleic acid and protein synthesis occur separately and come together
lipids and carbohydrates acquired from host cell
Origins of viruses
Pre LUCA (RNA world)
May have invented DNA
RNA viruses infected RNA cells
sensing of RNA evolved to combat viruses
viruses evolved DNA to counter
DNA from viruses integrated into cell genomes, more stable thus eventually takes over as genomic repository
Types of viral proteins
proteins are encoded in genome or acquired from host through budding
Structural
make up viral particle (virion)
involved in cell attachment and penetration
viral assembly and release
protect nucleic acid which is sensitive to restriction enzymes
Non-structural
not part of virion
involved in viral replication
e.g., polymerases, helicases
involved in virus assembly
Virus composition
Nucleocapsid
protein shell that surrounds viral genome
capsomers (structural protein subunits) in icosahedral or helical symmetry
icosahedral
capsomers form equilateral triangular faces (n-fold symmetry)
helical
capsomers wrapped around central genome core in helical pattern, tend to be rod shaped
Naked vs enveloped viruses
naked
only nucleic acid and protein
enveloped
acquire lipid membrane as they bud through membranes (ER or cell membrane often)
susceptible to inactivation from e.g., detergents
membranes have been transformed by virus proteins which were synthesised inside the cell, thus have these viral proteins
peplomers - spike proteins
helical nucleocapsids have flexible structure, can coil up to be
spherical/icosehedral
pleomorphic
filamentous
other shapes
rod (bacilform)
bullet
Complex viruses
neither helical or icosehedral
e.g., poxviruses
complex assembly processes
mulberry (multiple balls) or ball of yarn shapes
T - even bacteriophage
icosahedral head w genetic material
contractile tail/sheath
base plate
tail fibres
bind to receptor
pulls down and base plate goes into cell
tail contracts and genetic material injected
mimivirus - giant DNA virus
pandoravirus
Two main symmetries viruses use and examples, examples of enveloped and naked viruses
helical
orthomyxoviruses
influenza
paramyxoviruses
measles, mumps, hendra
filoviruses
marburg, ebola
icosahedral
togaviruses
ross river
rubella
flaviviruses
dengue
yellow fever
herpesviruses
chicken pox
herpes simplex
enveloped
influenza
COVID
HIV
naked
plant and bacteria viruses
polio
FDMV
Structure of virus determined by
size and coding capacity of genome
must fit inside nucleocapsid
most economic nucleocapsid symmetry
number of structural proteins available
functional requirements
protection of genome from environment (water vs air)
mode of attachment and cell entry
mode of replication
mode of virion assembly and release
burst or budding
Structural analysis of viruses
Electron microscopy
virus ID, cellular location
low res
X-ray crystallography
high resolution structure
virus/receptor interactions
identification of precise targets for antivirals
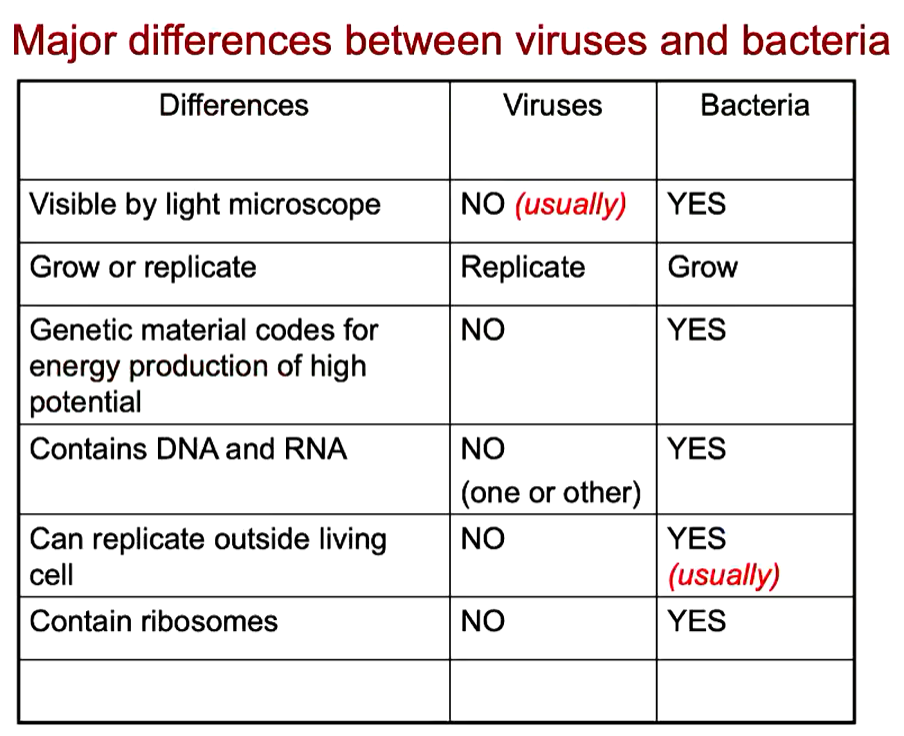
Virus vs Bacteria
Stages of viral infection
attachment
penetration
uncoating
transcription/translation (protein synthesis)
genome replication
assembly
release
Viral attachment
brownian (random) motion until collision w cell receptors which allows attachment
Virus attachment sites
enveloped viruses
spikes/peplomers
naked viruses
anywhere on virus surface
Cell receptors (attachment site on cells)
cell surface molecules
e.g., CCR5 binds to HIV peplomers, CD4 coreceptor pulls HIV towards membrane
Specific interaction
physical complementarity req for attachment
most viruses interact with specific receptors (only certain cell types)
host and tissue specificity
Virus entry/penetration methods
Endocytosis
virus engulfed into cytoplasmic vacuole
for enveloped viruses
virus endocytosed after binding to receptors
envelope fuses with endosome (vacuole) membrane
release of nucleic acid into cytoplasm
Membrane fusion (enveloped viruses)
envelope fuses with cell membrane
only nucleocapsid enters cell (envelope fuses)
Direct entry (some naked viruses)
capsid undergoes molecular rearrangement
whole virus or only genome enters
Exosomes (naked viruses)
membrane bound particles cells excrete as waste
naked viruses travel in these and enter new cells
Viral uncoating
Often spontaneous release of nucleic acid
carried out by host cell or viral enzymes (protease, lipases, etc)
Viral synthesis of proteins and nucleic acid
DNA viruses replicate in cell nucleus (except pox viruses which are too large to fit and carry their own replicative material)
RNA viruses generally replicate in cytoplasm, encode their own RNA dependent RNA polymerase
need ribosomes
need host cell enzymes e.g., to cleave polyproteins into individual proteins
How do viruses deal with competition?
Take over ribosomes to synthesis ONLY viral proteins
downregulate receptors so other viruses can’t enter
Baltimore classes of viruses
I = dsDNA
II = ssDNA
III = dsRNA
IV = ssRNA +ve sense
V = ssRNA -ve sense
VI = +ssRNA replicates w DNA intermediate
VII = dsDNA replicates w RNA intermediate
DNA viruses synthesis of viral proteins
dsDNA (I, VII)
direct transcription of mRNA
ssDNA (II)
synthesis of other strand
dsDNA intermediate
transcription onto mRNA
RNA viruses synthesis of viral proteins
dsRNA (III) and -ssRNA (V)
need own RNA dependent RNA polymerase to transcribe -RNA strand to create +RNA strand (mRNA)
+ssRNA (IV)
+RNA used directly as mRNA
Class VI retroviruses (+ssRNA with dsDNA intermediate)
reverse transcription to make dsDNA intermediate
dsDNA intermediate transcribed to make mRNA
Replication of +ssRNA virus (IV)
genome used directly as mRNA to make viral proteins
RNA dependent RNA polymerase (RNA replicase) made
-ve strand synthesised
more copies of +ve strand synthesised
Asymmetric strand synthesis
more +ve than -ve (only need some -ve as template)
Replication of -ssRNA virus (V)
RNA replicase enzymes brought into cell as part of virion
-ve strand RNA released into cytoplasm, replicase makes +RNA for translation of viral proteins
using +RNA, -ve strand genomic RNA produced
Virus assembly and release
Assembly
relatively spontaneous for simple viruses
stages for complex viruses
Release
enveloped
gradually bud from cell
naked
accumulate in cell until cell lyses
known as one step growth curve
may also be secreted gradually through exosomes
Examples of Class IV and V viruses
IV (+ssRNA)
flaviviruses (dengue)
togaviruses (rubella)
COVID
V (-ssRNA)
RSV
orthomyxoviruses (influenza)
paramyxoviruses (measles, mumps)
filoviruses (ebola)
Targets for antiviral agents
viral specific enzymes or nucleic acid
viral protease (necessary for viruses to cut up viral polyproteins)
RNA dependent RNA polymerase (RNA replicases)
viral DNA synthesis - acyclovir
affects host polymerase function
incorporation into DNA results in termination of viral replication
not toxic to host cells as activated by viral enzymes only produced in infected cells
Viral disease syndromes and what causes them
Cell damage due to viral replication
cell rupture during virus release
necrosis
cell death to control infection (apoptosis)
infected cells lose function
infectious cell transformed by virus, virus activates oncogenes by inserting into cell genome
causes tumours
Damage due to host response to infection
immunopathology - antibodies and immune cells destroy infected cells
tissue damage
fever invoked to stimulate immune response, inhibit virus replication
inflammation caused by immune cells infiltrating infection site
excessive cytokine production
can cause tissue damage and inflammation
Host factors affecting viral pathogenesis
Age
some viral infections more severe at different ages
immune system maturity/waning
hormonal influences
Genetics
e.g., cell receptor CCR5 doesn’t protrude through membrane, reducing susceptibility to HIV as it needs this coreceptor to CD4
Metabolic state (body condition)
generalised malnutrition or vitamin A deficiency increase susceptibility and severity
pregnancy (change in hormonal balance) alters susceptibility to some viruses
Altered immune responses
impaired immune system due to:
genetics
consequence of infection (HIV)
Iatrogenically/therapeutically acquired (after transplant)
enhanced immune system
auto-immunity
Routes of viral entry
Skin
mechanical trauma (microtears)
injection
infected mosquito bite
bite of infected animal
Genitourinary tract
tears or abrasions allow viral entry
sexually transmitted (HIV, Herpes Simplex)
host defence
cervical mucus
pH of vaginal secretions
chemical composition of urine
Respiratory tract
droplet infection in aerosols
generally enveloped viruses
GI tract
invasion of tissues underlying mucosal layer
virus survivability depends on:
acid stability
resistance to bile salts
resistance to inactivation by proteolytic enzymes
generally naked viruses
Conjunctiva - eyes
localisation vs systemic spread
localised infections
virus multiply in epithelial cells at site of entry
produce spreading infection then shed directly to exterior
infection back out apical side of cells since cells are polarised
none through basal layer
systemic spread
polarised infection of epithelial cells and spread
targeting of viral budding to apical or basal surfaced of polarised cells may define subsequent spread
Modes of viral transmission
Person-person transmission:
Respiratory/salivary
influenza, measles, rhinoviruses
Fecal-oral
enteroviruses, rotaviruses
Contact (sexual)
Herpes simplex 2, genital warts, HIV
No person-person transmission:
Vector (biting arthropod)
sandfly fever, dengue
Vertebrate reservoir
rabies, cowpox
vector-vertebrate
arbovirus
How to measure transmissibility/contagiousness of viruses?
Basic reproduction number (R0) = Attack rate (#ppl infected) * contacts
Acute vs chronic vs recurring infections
Acute
rapid onset of symptoms
usually complete recovery or death
influenza, cold
Chronic/persistent
long, slow infections (HIV, HCV)
may have insidious onset (no symptoms)
symptoms may be present most of time, sometimes for life
Recurrent/latent
infection occurs
virus replication dies down, minimal until triggered by stimuli
reoccurrence of symptoms from virus which has been latently present (herpes simplex)
Non-persistent vs persistent microbes
Non-persistent
retained briefly, transmitted quickly
don’t multiply within vector’s body
in small community will infect everyone acutely then die out
in large community infects susceptibles, spreads causing repeated outbreaks as fresh susceptibles appear
continuous circulation
Persistent
longer acquisition, can persist longer
replicates within vector’s body
microbe infects susceptibles
microbe remains latent
microbe reactivates, infects next generation of susceptibles - in large or small community
Plant virus structure
icosahedral or helical symmetry
most are naked
most have small +ssRNA genomes for better systemic spread btwn cells
some have circular DNA genomes
Multipartite plant viruses
Segmented genomes, each segment encased in individual nucleocapsid as opposed to segments within a single particle
need to infect plant with all segments for productive infection
way to regulate gene expression
control ratios of particles, can control the amounts of proteins each particle makes (e.g., make less of particle A which has gene for polymerase so less polymerase)
plant virus transmission
plants have rigid cell wall - no endocytosis or fusion
enter through vectors or infected seeds
How do plant viruses spread systemically and from cell to cell?
systemic infection via phloem
cell-cell
plasmodesmata connect cells, too small for viruses
movement protein
lining with protein tubule to wedge open (CPMV)
making viral proteins to wedge open plasmodesmata and allow virion to pass through (PVX)
Coevolution of virus, fungi and plant
Virus, fungi and plant live in hot spring
virus causes fungi to produce heat shock protein which protects plant