IMED1003 - Nucleotide Metabolism 1 and 2 (L25 and 26)
1/35
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
36 Terms
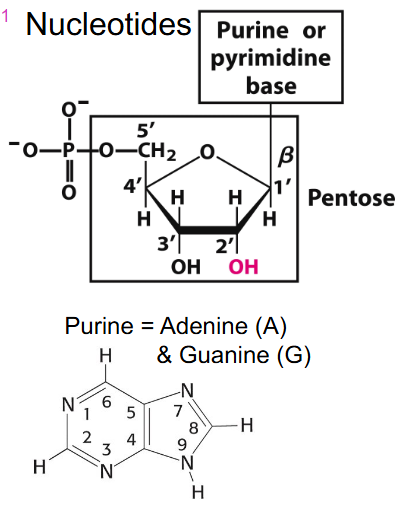
Nucleotides
- not only in DNA - making RNAs and proteins
- Metabolic - ATP is the cellular energy "currency"
- "activated" intermediates e.g UDP-glucose
- cofactors in biochemical reactions - e.g NAD+
- cell signalling - e.g cAMP
PURINE: Adenine (A) and Guanine (G)
PYRIMIDINE: Cytosine (C), Thymine (T), Uracil (U)
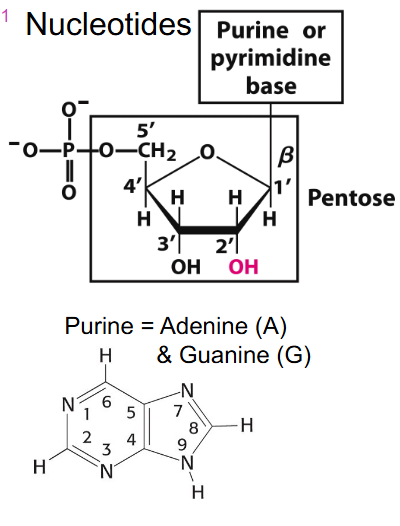
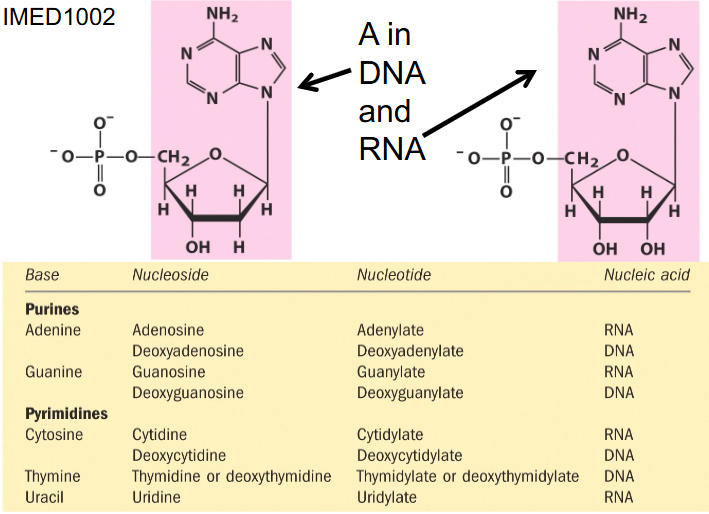
Purine and Pyrimidine binding
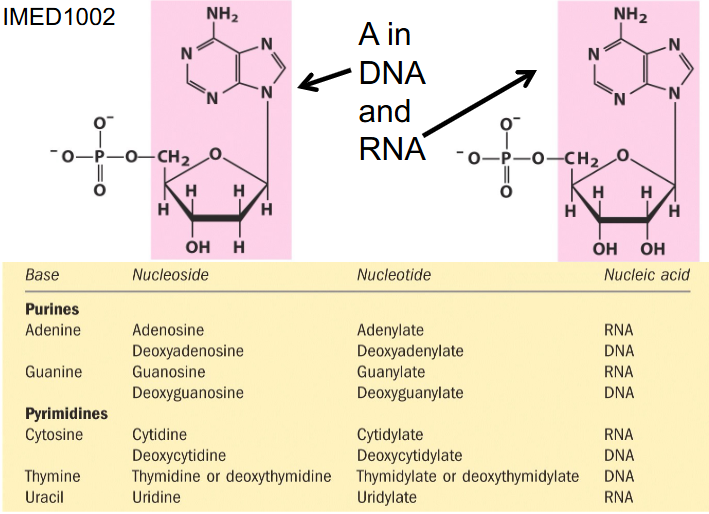
- if we have a base and sugar together its called nucleoside
- if we add a phosphate it is nucleotide
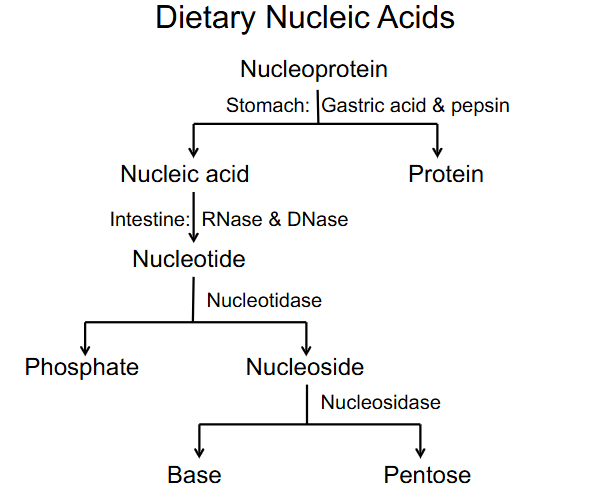
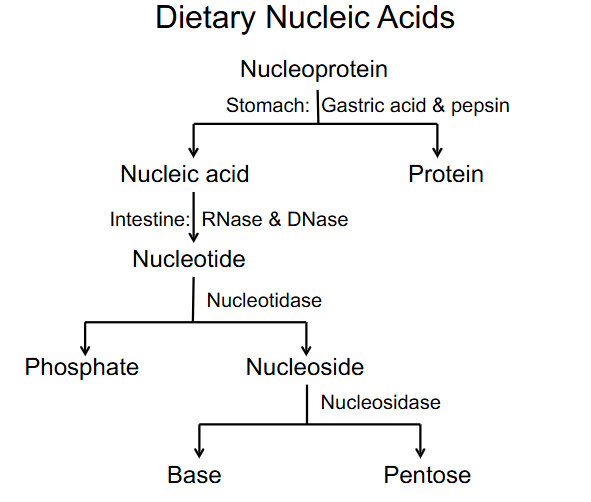
Dietary Nucleic acids
- nucleoprotein breaks off into nucleic acids and proteins (stomach: gastric acid and pepsin (protease))
- Nucleic acid turns to nucleotide (intestine: RNase and DNase) (these are nucleases)
- nucleotide breaks off into phosphate and nucleoside
- nucleoside turns into base and pentose (by nucleosidase)
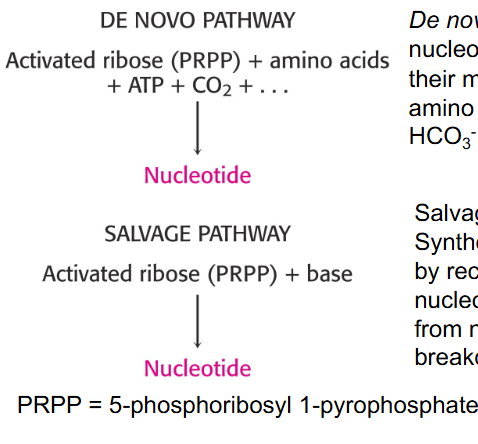
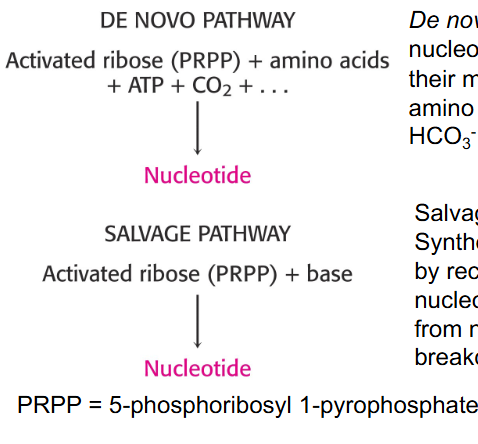
Nucleotide Synthesis de novo or via salvage pathways
- De novo: synthesis of nucleotides begins with their metabolic precursors: amino acids, ribose-5P, HCO3-, and one carbon units
- Salvage pathways: synthesis of nucleotides by recycling of bases, or nucleosides released from nucleic acid breakdown
Nucleotide Biosynthesis
- main sites: liver, small intestine, and thymus - cytosol
- Nucleotide pools small, so cells must continually synthesise, or may limit rates of transcription and DNA replication
- nucleotides can be synthesised de novo and this is very similar in all organisms studied
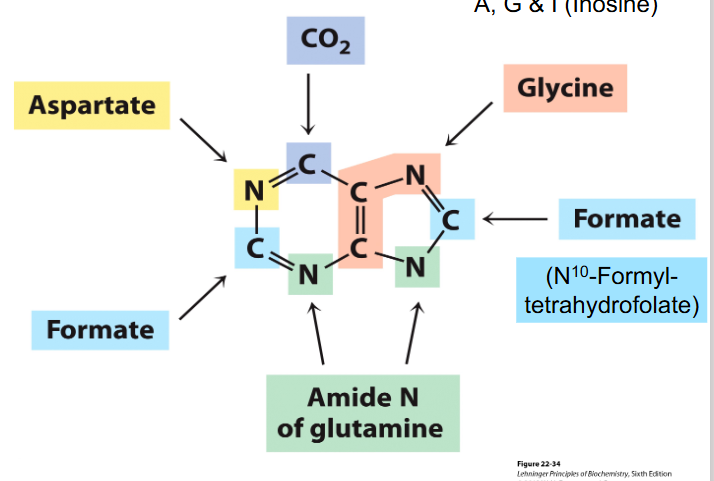
- Glutamine provides most amine groups
- Glycine is a precursor for purines
- Aspartate is precursor for pyrimidines
- purine bases synthesised while attached to ribose, pyrimidines not initally attached to ribose
- make ribo-nts, then reduce to deoxyribo-nts
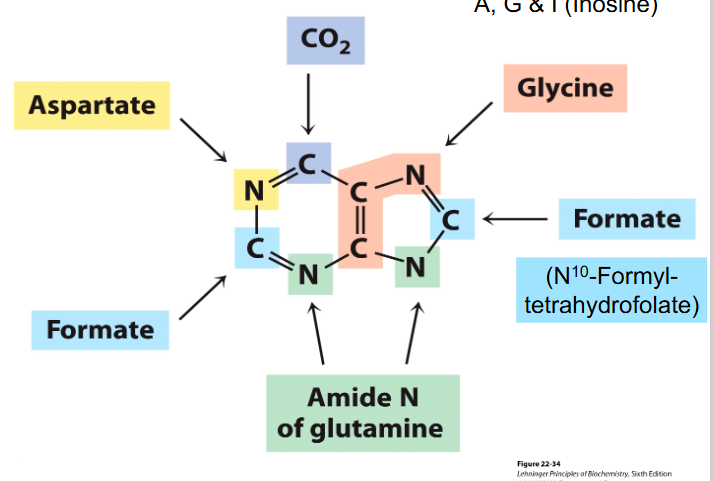
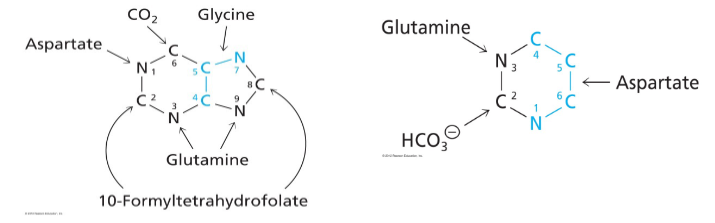
Purine Bases
- we need folate to produce purines, and thats a vitamin
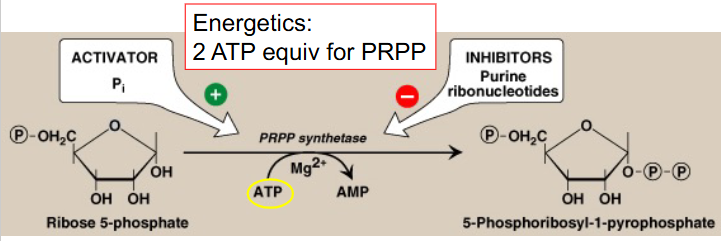
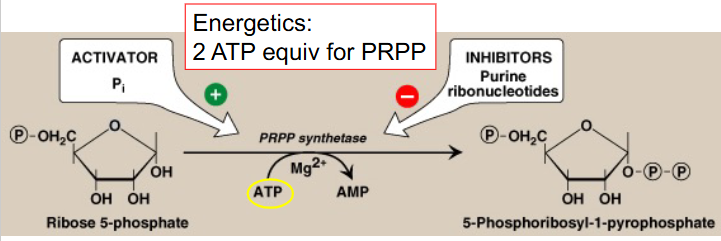
Conversion of Ribose-5P to PRPP
basic pathway for biosynthesis of purine ribonucleotides:
- starts with ribose-5-phosphate (R-5-P)
- Occurs primarily in liver cytosol
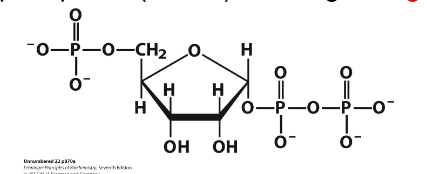
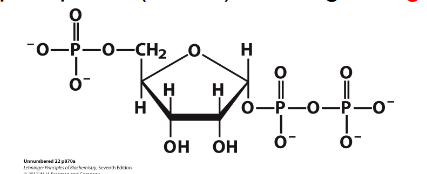
- PRPP = 5-phosphoribosyl-1-pyrophosphate
- PRPP is an "activated R-5-P" and provides the foundation on which the purine bases are constructed step by step
- Mg++ is a cofactor
.
- uses PRPP synthetase and has 2 ATP equivalents (converts to AMP)
- activated by Pi, inhibited by purine ribonucleotides
De Novo biosynthesis of purines begins with PRPP
- Begins with activated ribose-5P = 5-phosphoribosyl 1-pyrophosphate (PRPP) reacting with glutamine
- purine ring builds up in a series of reactions
- first intermediate with full purine ring = inosine monophosphate (IMP, inosinate)
- Then adenine and guanine are synthesised as AMP and GMP from IMP
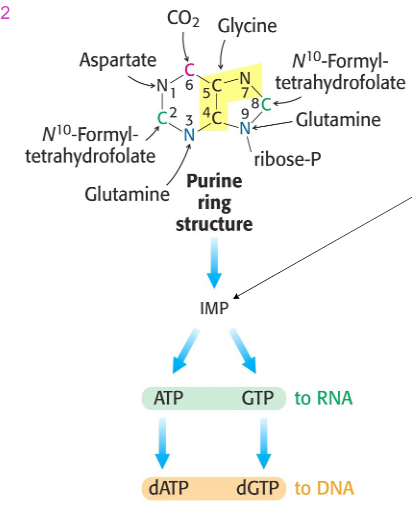
Formation of Purines: A and G
- IMP (inosinate) is synthesised first, then AMP and GMP ... phosphorylated to ATP and GTP, then converted to deoxy (dATP and dGTP)
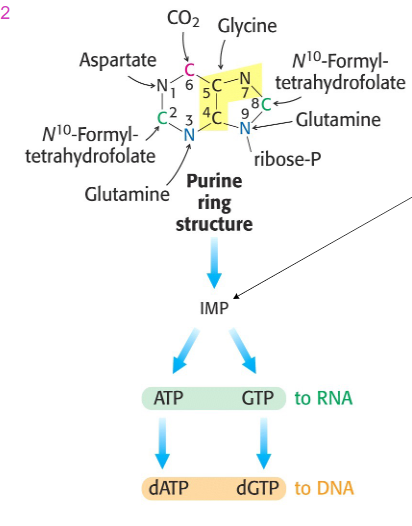
Purine Synthesis
- u dont need to know reaction details, have to know energetics of reaction
- 10 reactions + synthesising PRPP = 11 steps overall
- Energetics: 2 ATP eq. for PRPP, 5 more ATP
Synthesis of IMP (NO LEARNING OUTCOME)
- reaction details not examinable
- First intermediate with a full purine ring = IMP
- Energetics: 2ATP eq. for PRPP, 5 more ATP
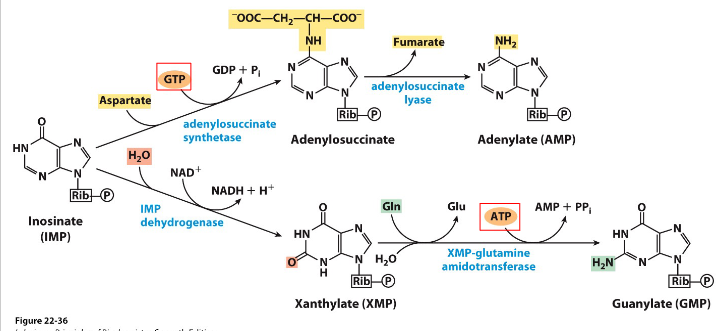
Synthesis of AMP and GMP from IMP
- ATP used for GMP synthesis, GTP used for AMP synthesis
.
- reaction details not examinable
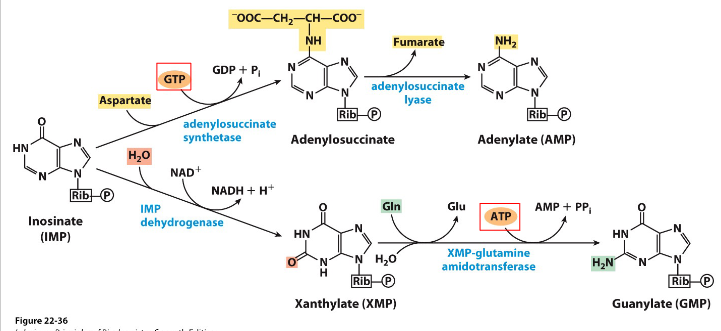
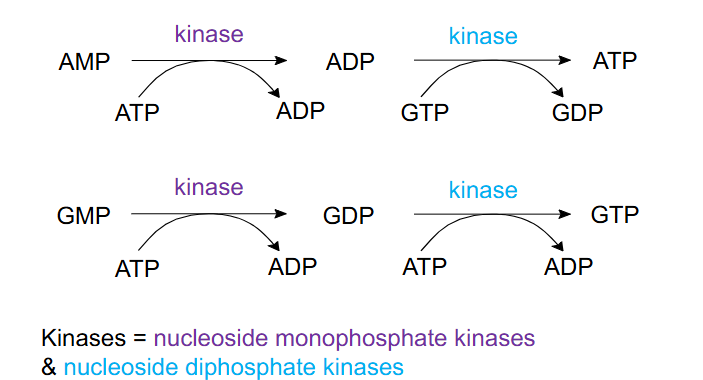
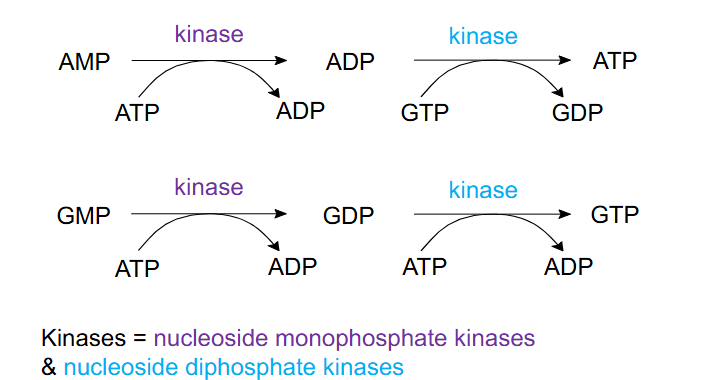
ADP, ATP, GDP and GTP via phosphorylation
- Kinases = nucleoside monophosphate kinases and nucleoside diphosphate kinases
- reaction details not examinable
- this meets learning outcome that states: "describe how nt DiP and nt TriP are made"
.
- AMP -(ATP -> ADP, action of kinase)-> ADP -> (GTP -> GDP, action of kinase)-> ATP
- GMP -(ATP -> ADP, action of kinase)-> ADP -> (ATP -> ADP, action of kinase)-> ATP
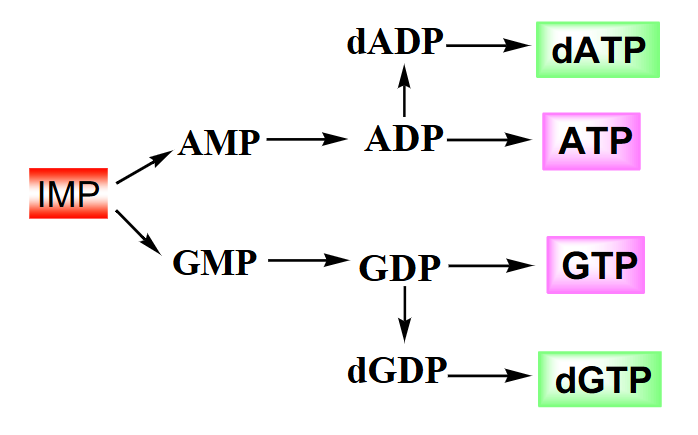
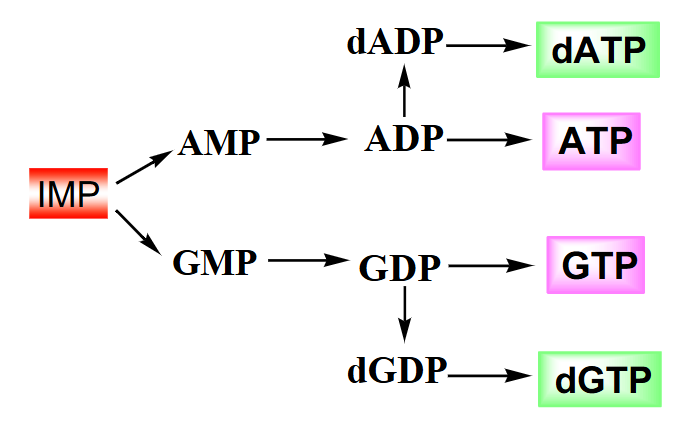
Summary of Purine Biosynthesis
- IMP converted to AMP with one ATP equivalent
- reduction to deoxyribose when the nucleoside diphosphate is converted (look at dGDP and dATP)
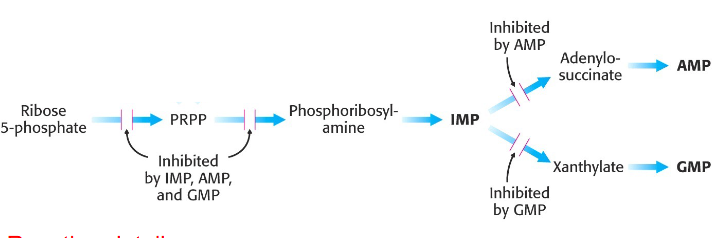
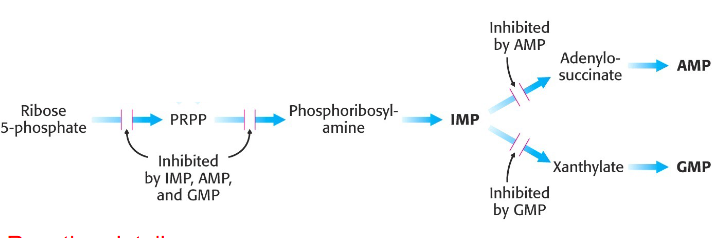
Regulation of de novo Purine synthesis
- must meet needs of the body, without wasting
- AMP and GMP control their respective synthesis from IMP by feedback inhibition
- reaction details not examinable
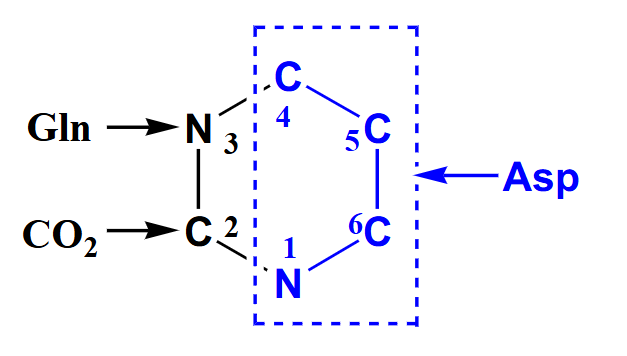
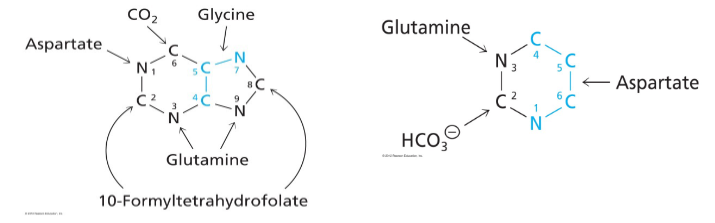
De Novo Pyrimidine Synthesis
- Precursors: aspartate, glutamine, HCO3-
- Shorter pathway than for purines, 6 steps
.
- rather than starting with the ribose and making the ring attached to the ribose in purine synthesis we are going to start with the base of the ring and attach ribose later with pyrimidine synthesis
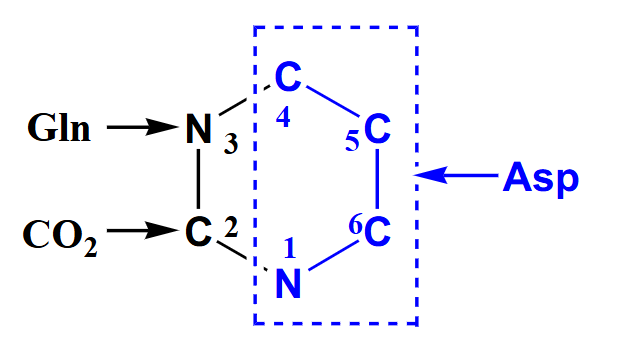
De Novo Pyrimidine Synthesis ALL STEPS
- reaction details not examinable
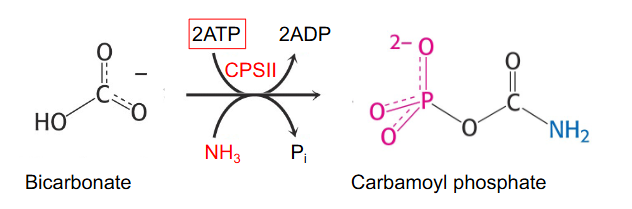
De Novo Synthesis of Pyrimidines
First step is synthesis of carbamoyl phosphate from HCO3- and NH3 (from glutamine) catalysed by carbamoyl phopshate synthetase 2 (CPS2)
- two different enzymes can make carbamoyl phosphate: CPS1 - in mitochondria for urea cycle and CSP2 - in cytosol for pyrimidine synthesis
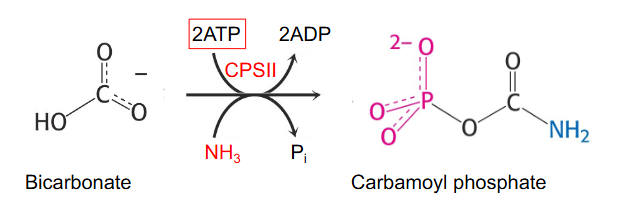
De Novo Synthesis of Pyrimidines up to first pyrimidine base (orotate)
- unlike purine synthesis, pyrimidine synthesis proceeds by first making the pyrimidine ring (in the form of orotate) and then attaching it to Ribose-5P
- aspartate and carbamoyl phosphate = substrates in first reaction
- Regulation: ATCase - inhibited by end product CTP and accelerated by ATP
.
- reaction details NOT examinable
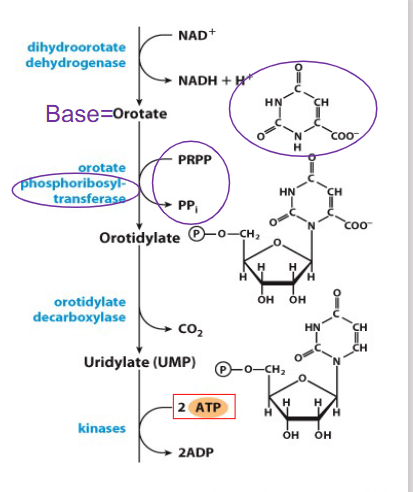
De Novo Synthesis of Pyrimidines continuing from orotate
- after addition of ribose-5-phosphate to the base uses PRPP to form nt (orotidylate)
- reaction catalysed by phosphoribosyltransferase
- then decarboxylated to form uridylate (UMP), then first pyrimidne nt
- reaction details not examinable
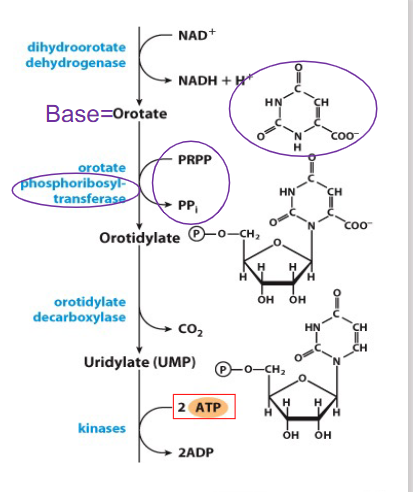
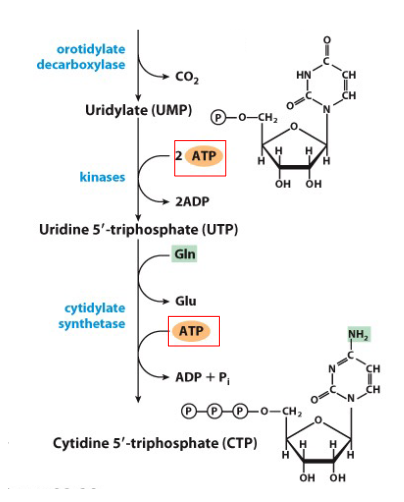
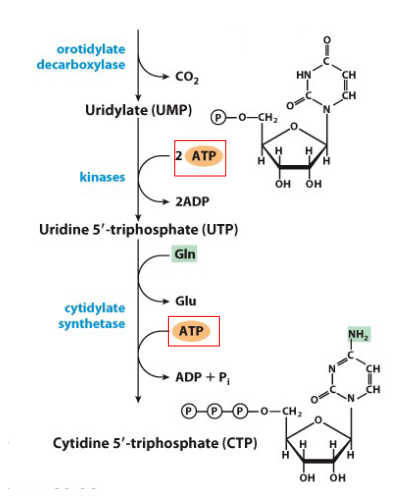
De Novo Synthesis of Pyrimidines continuing from Uridylate
- UMP phosphorylated to UTP
- after formation of UTP, amination can convert UTP to CTP
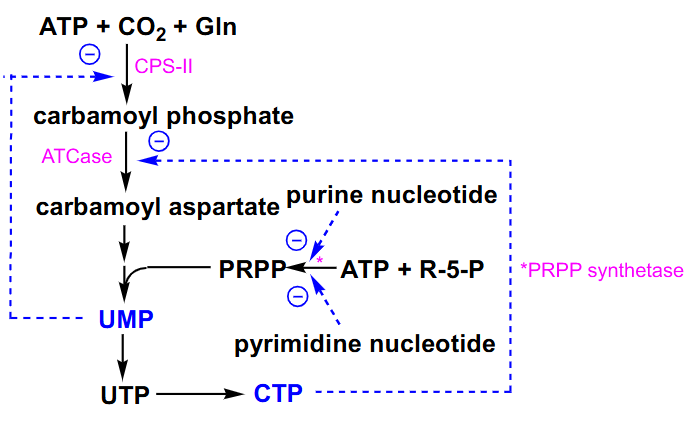
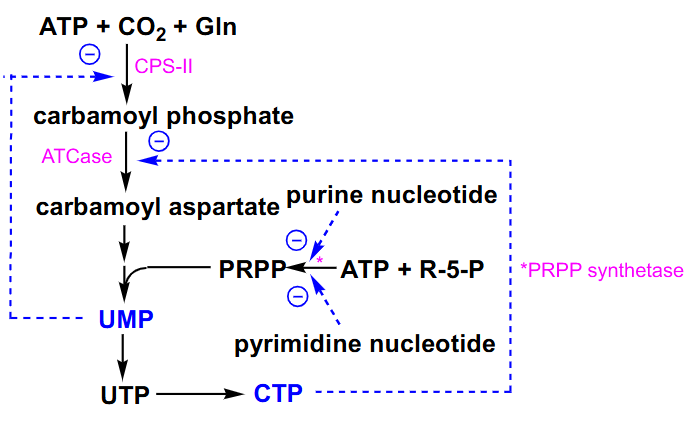
Regulation of De Novo Pyrimidine Synthesis
- we can inhibit carbamoyl phosphate synthetase 2 by the cytosolic end product of our uridine monophosphate
- aspartate transcarboxylase can be inhibited by end product CTP
- PRPP synthetase is inhibited by both types of nucleotides so its another control point
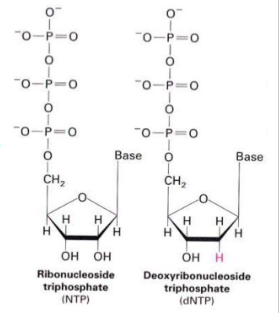
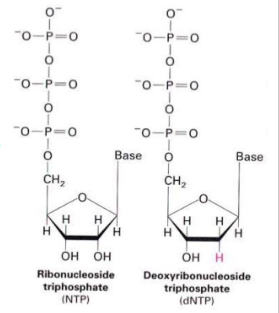
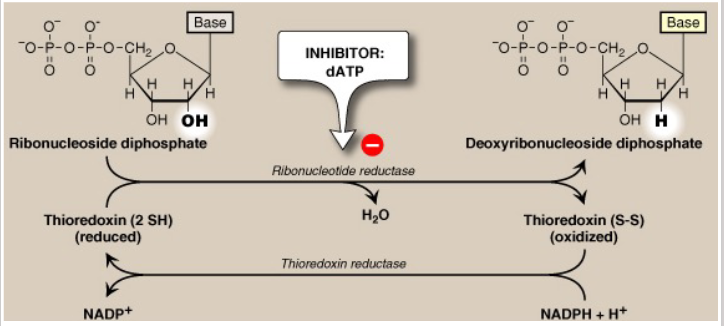
Ribonucleotides (NTP) are precursors of Deoxyribonucleotides (dNTP)
- 2'C-OH bond is directly reduced to 2'C-H bond catalysed by ribonucleotide reductase
- Mechanism: 2 H atoms donated by NADPH and carried by proteins thioredoxin or glutaredoxin
- NADPH serves as the electron donor
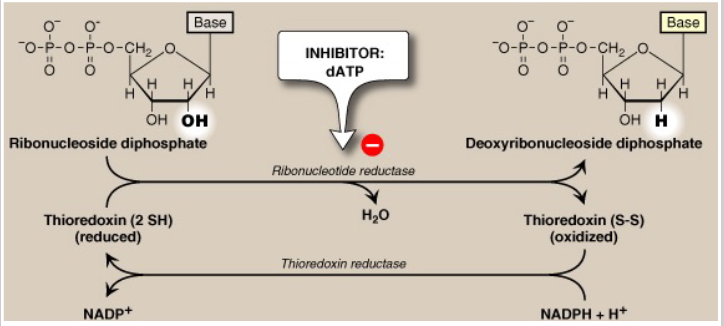
Deoxyribonucleosides from ribonucleosides
- ribonucleotide reductase: inhibited by dATP and activated by ATP
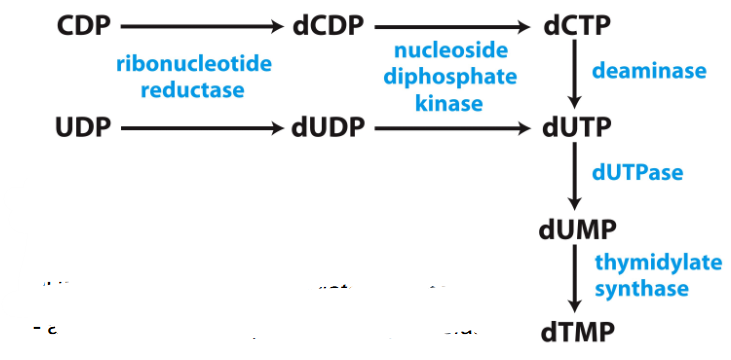
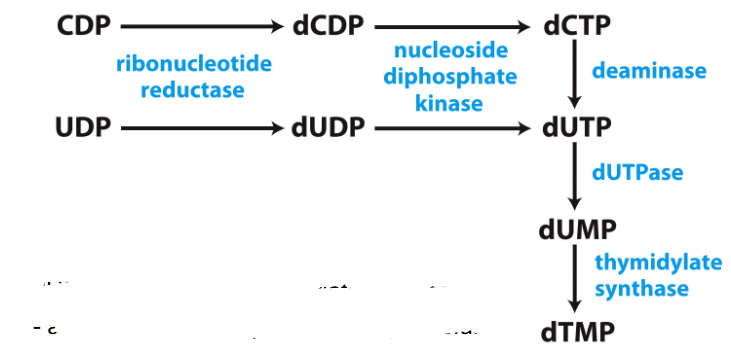
Biosynthesis of dTMP
- roundabout pathway...
- dUTP made.
- dUTP -> to dUMP by dUTPase
- dUMP -> dTMP by thymidylate synthase
- adds a methyl group from tetrahydrofolate
.
- thymidylate synthase is a target for some anticancer drugs
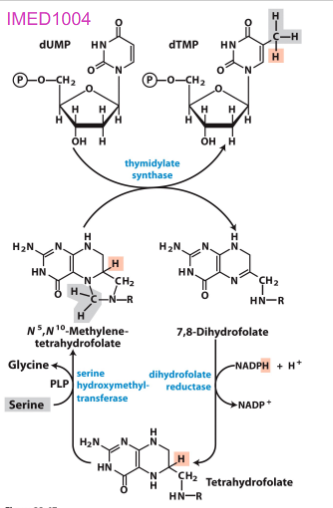
Biosynthesis of dTMP vitamin
- folic acid deficiency widespread, leads to reduced dTTP synthesis and results in uracil incorporation into DNA
- repair mechanisms remove uracil by creating strand breaks that affect structure of DNA: associated with cancer, heart disease, neurological impairmente
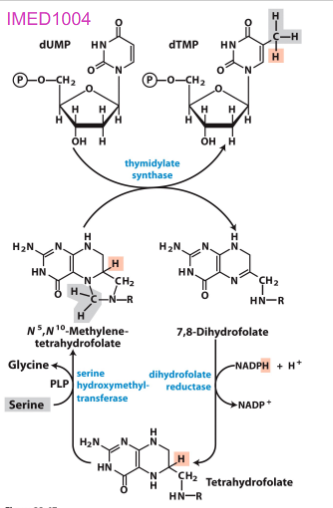
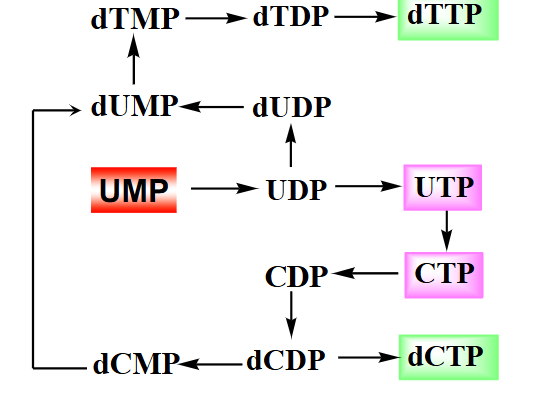
Summary of Pyrimidine Biosynthesis
DIAGRAM ON SLIDE 33
Purine vs Pyrimidine Biosynthesis
PURINE BIOSYNTHESIS:
- salvage is a major pathway
- base synthesised while attached to ribose
- IMP is common intermediate for AMP and GMP, but itself is not a typical nucleotide
.
PYRIMIDINE BIOSYNTHESIS:
- de novo is a major pathway
- base is synthesised, then attached to ribose
- UMP, a typical nucleic acid nt, is converted to other pyrimidines
DIAGRAM ON SLIDE 34

Nucleotide Interconversions
- Fast, reversible, driven by high [ATP]
- NMP -> NDP catalysed by specific nucleoside monophosphate kinases
- NDP -> NTP catalysed by less specific kinases
- AMP + ATP -> ADP + ADP important in energy balance
![<p>- Fast, reversible, driven by high [ATP]</p><p>- NMP -> NDP catalysed by specific nucleoside monophosphate kinases</p><p>- NDP -> NTP catalysed by less specific kinases</p><p>- AMP + ATP -> ADP + ADP important in energy balance</p>](https://knowt-user-attachments.s3.amazonaws.com/e9b884d5-c60f-4057-bfa0-d5525fd18eeb.png)
Nucleotide Catabolism
- less important than other catabolic processes
- not a major energy source
- lots of salvage
- serves to clear excess
.
- in humans, purines -> uric acid (excreted)
- in humans, pyrimidines -> acetyl CoA, succinyl CoA - some energy gain
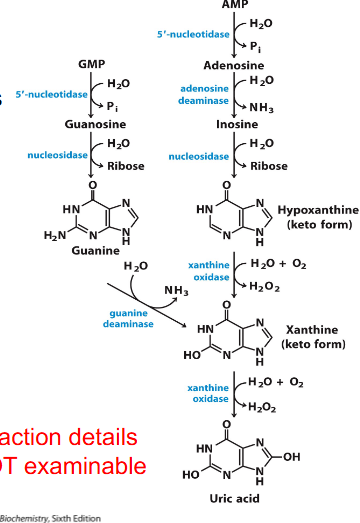
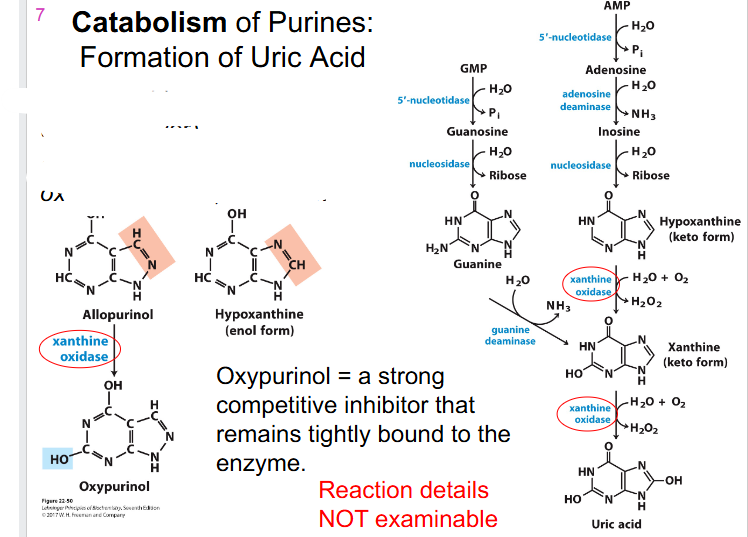
Catabolism of Purines: Formation of Uric Acid
- degredation of purines proceeds through dephosphorylation (via 5' nucleotidase)
- adenosine is deaminated to inosine and then hydrolysed to hypoxanthine and ribose
- Guanosine yields xanthine via these hydrolysis and deamination reactions
- Hypoxanthine and xanthine are then oxidised to uric acid by xanthine oxidase
.
- reaction details not examinable
Uric Acid
- uric acid = excreted end product of purine catabolism in humans, plus minus 0.6g/24h, arising in part from ingested purines and in part from turnover of purine nucleotides of nucleic acids
- normal serum [uric acid] = 3-7 mg/dL
- Gout is a disease of joints, usually in males, caused by an elevated [uric acid] in blood and tissues
- joints become inflamed, painful and arthritic, owing to abnormal deposition of crystals of sodium urate
- kidneys also affected, as excess uric acid is deposited in kidney tubules
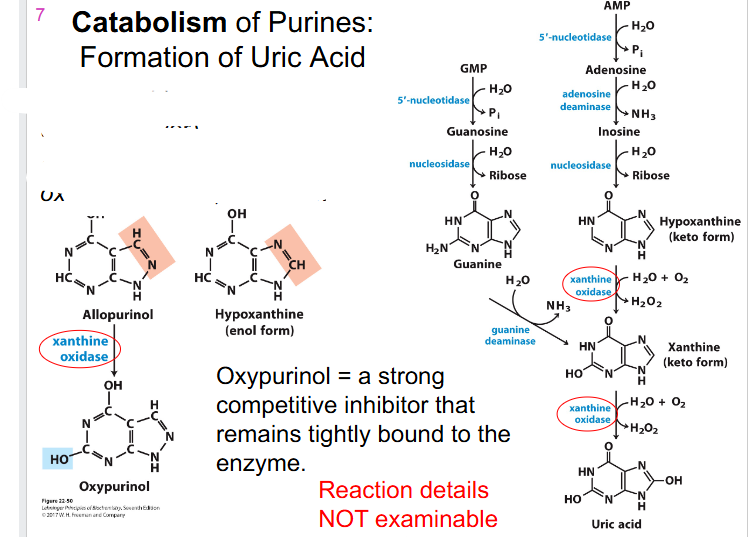
Treating Gout
- Gout: avoid purine-rich foods (seafood, liver)
- Also treated with xanthine oxidase inhibitor, allopurinol
.
- Oxypurinol = a strong competitive inhibitor that remains tightly bound to the enzyme
Catabolism of Pyrimidines
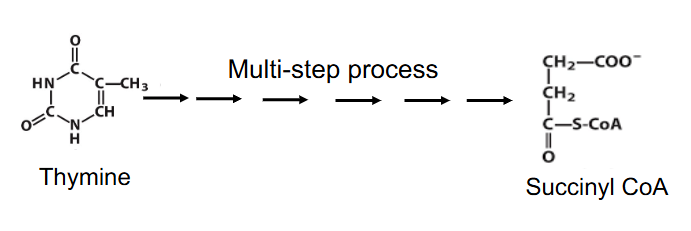
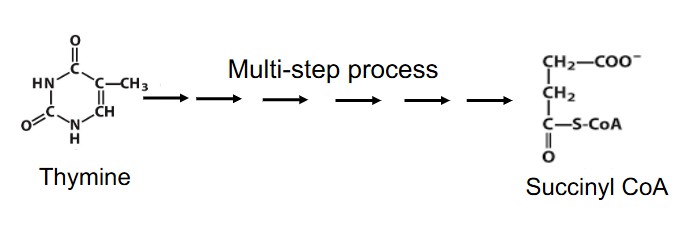
- Leads to NH4+, then urea
- can produce intermediates of TCA cycle e.g thymine is degraded to succinyl CoA
Bases Recycled by Salvage Pathways
- relatively few pyrimidine bases are salvaged in human cells
- Purine bases from diet or created by degredation of RNA or DNA can be directly converted to the corresponding nucleotides
- The significance of the salvage pathway: some tissues and organs (brain and bone marrow) are only capable of synthesising nucleotides by salvage pathway
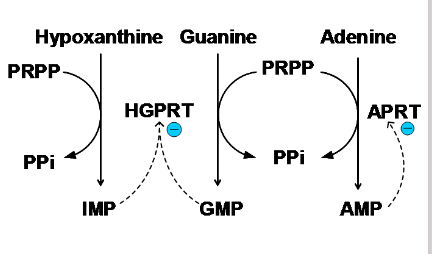
- Two phosphoribosyl transferases are involved: APRT (adenine phosphoribosyl transferase) for adenine and HGPRT (hypoxanthine guanine phosphoribosyl transferase) for guanine or hypoxanthine
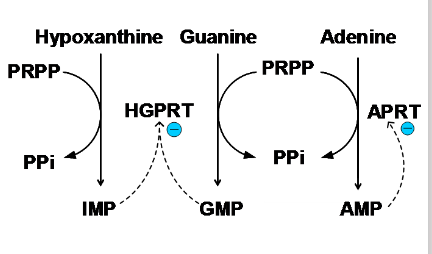
Purine Salvage Pathways
- Hypoxanthine-Guanine phosphoribosyl transferase (HGPRT)
- hypoxanthine + PRPP -> IMP + PPi
- Guanine + PRPP -> GMP + PPi
.
- adenine phosphoribosyl transferase (APRT)
- adenine + PRPP -> AMP + PPi
.
- are reversible reactions
- so, AMP, IMP, GMP do not need to be synthesised de novo