Protein analysis
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
32 Terms
How are proteins obtained for separation?
Proteins are extracted from microbial cultures, plants, or animal tissues, then released into a crude extract containing many proteins.
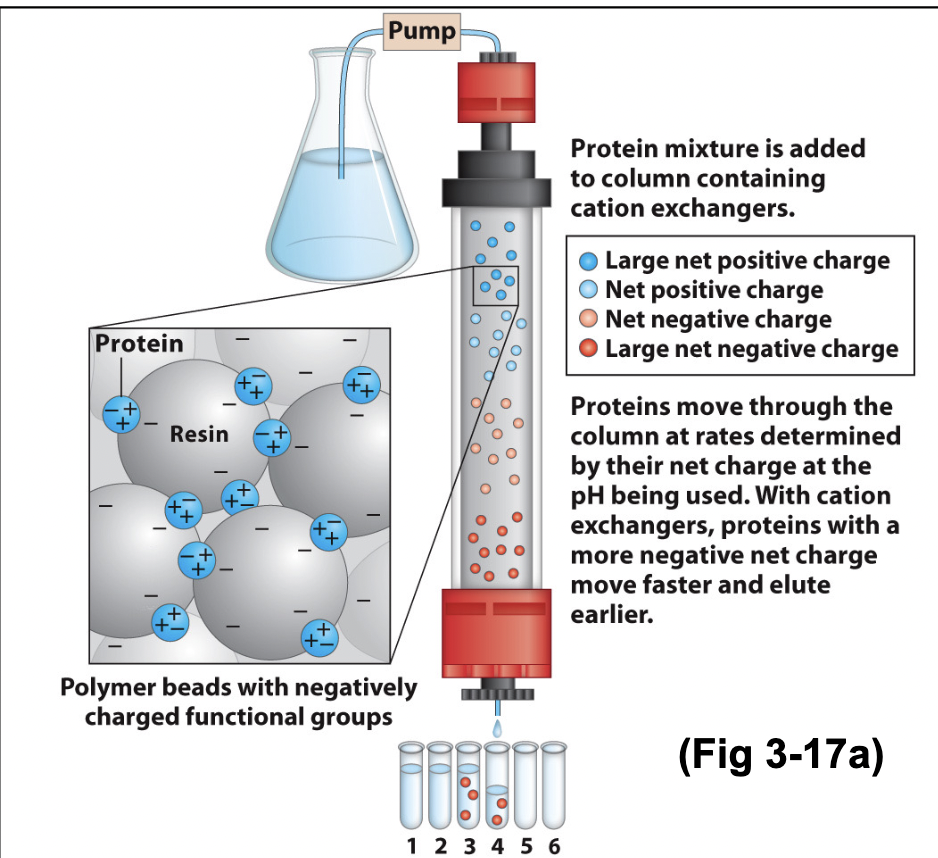
How does ion exchange chromatography separate proteins?
It separates proteins based on net charge, which depends on their amino acid composition and pH.
Cation exchange chromatography for proteins
Affinity chromatography
Protein purification technique where proteins bind to a specific ligand attached to beads, allowing seperation based on binding affinity.
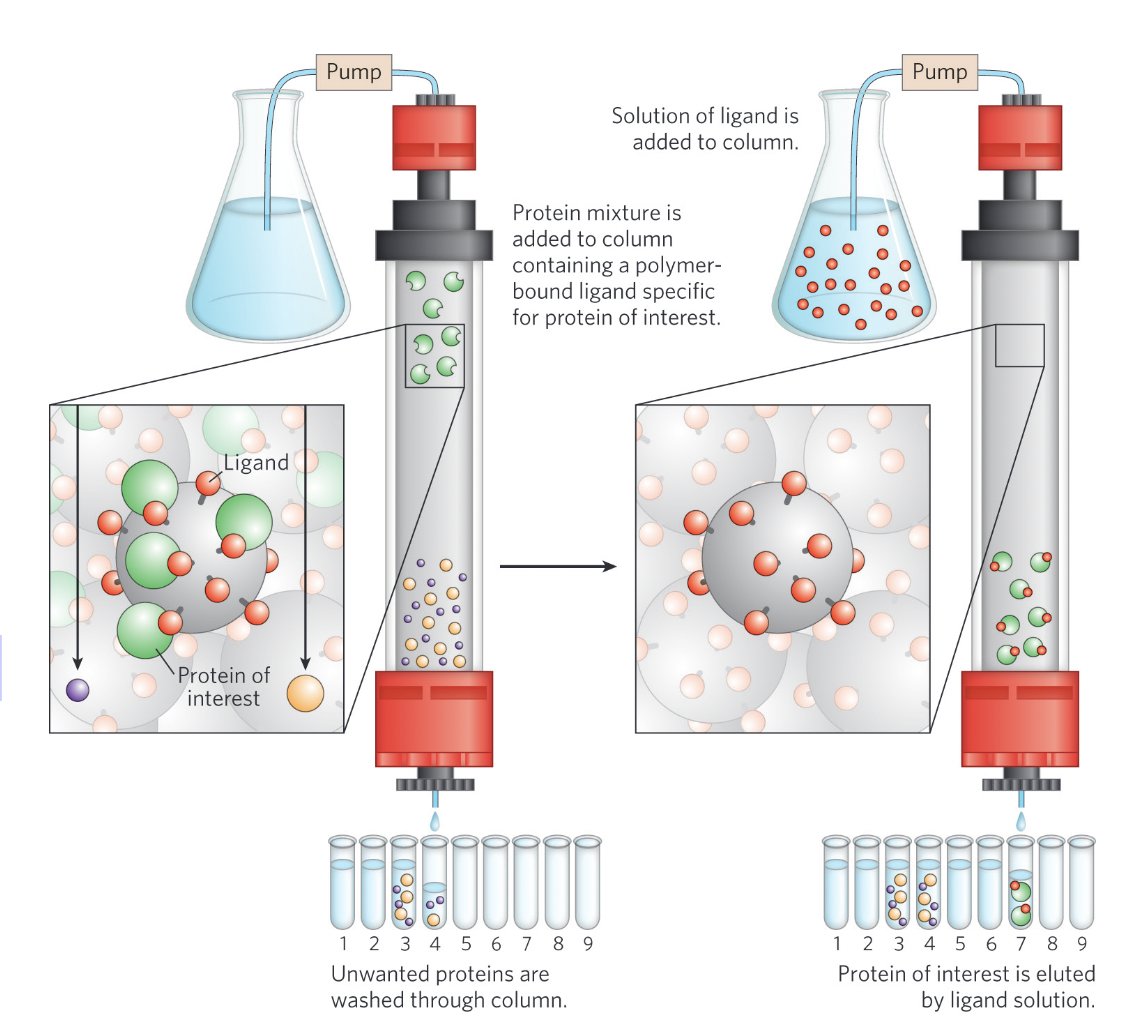
the second photo shows the elution of proteins due to addition of a ligand (the same ligand or very similar)
What is the key component of affinity chromatography?
A ligand covalently attached to beads in the column.
How do proteins bound to the ligand elute?
by the addition fo high concentration of salt (weakens binding) or ligand (competes with attached ligands)
What is a "Tag" in genetic engineering for protein purification?
A Tag is a peptide or protein that binds a ligand with high specificity that is fused to the gene encoding the target protein, allowing its purification by affinity chromatography.
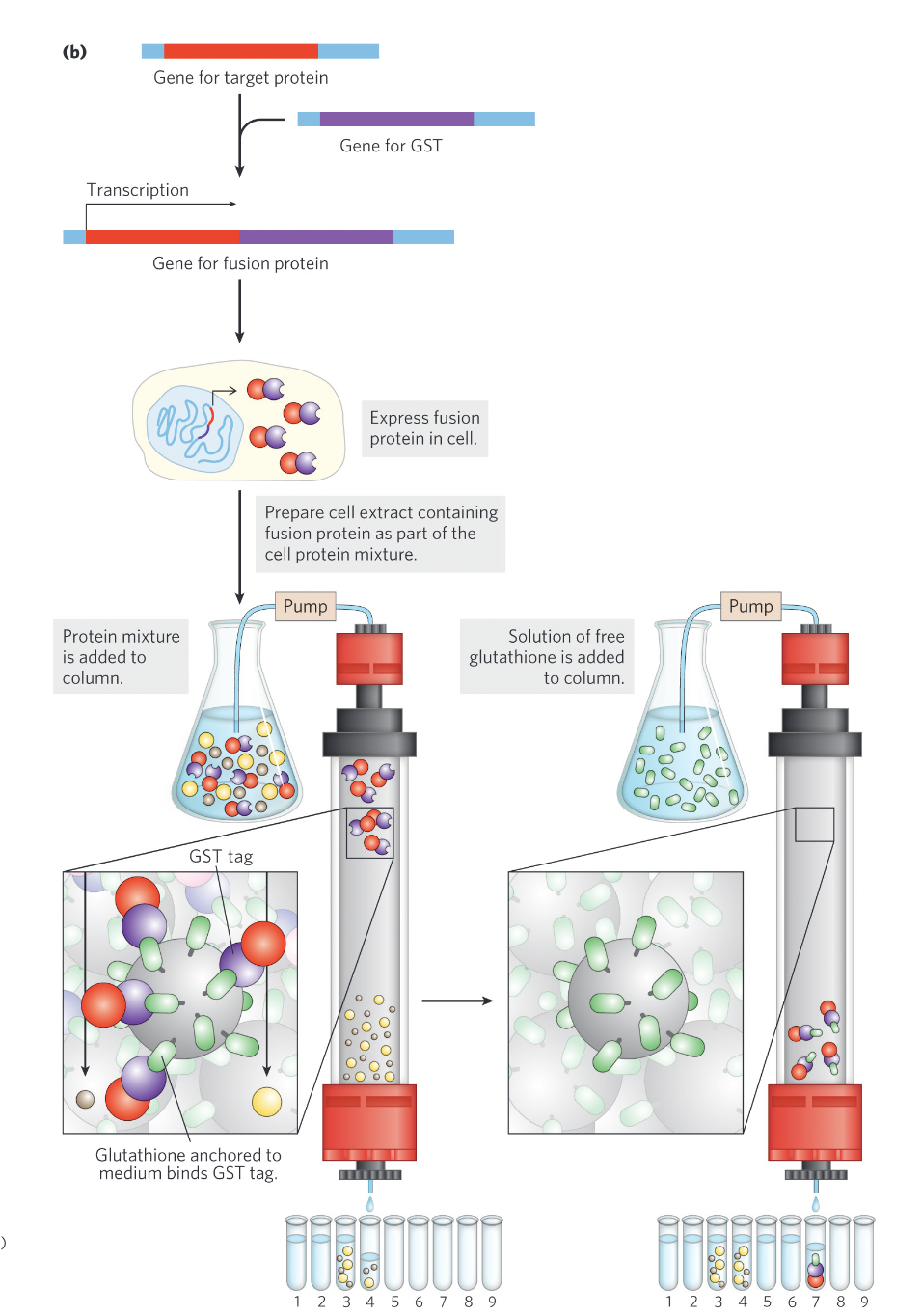
Protein tagging explained
Imagine you're studying a protein X and you want to purify it from a cell extract. To make purification easier, you genetically engineer protein X to have a GST tag attached to it.
GST Tagging: You add the gene for GST to the gene for protein X, so the final protein you produce is protein X-GST.
Purification: You pass the mixture through a column that has glutathione (the ligand for GST) attached to it.
The GST tag on protein X binds tightly to the glutathione in the column.
Other proteins in the mixture flow through, since they don't have the GST tag.
Elution: To release protein X-GST, you add excess glutathione.
The extra glutathione competes with the GST tag, causing protein X-GST to detach and flow out of the column.
Immobilized Metal Affinity Chromatography (IMAC)
protein purification technique that uses a metal ion (like Nickel (Ni²⁺) or Cobalt (Co²⁺)) to selectively bind proteins that have been tagged with a special sequence of amino acids (called a His tag).
How IMAC works
Adding a His tag: A small sequence of 6-8 histidine (His) amino acids is added to the target protein (this is called a His tag). The His tag allows the protein to bind to metal ions like Nickel (Ni²⁺).
Binding to the column:
The IMAC column contains resin that has Nickel ions (Ni²⁺) attached to it. When you run the mixture through this column, the His-tagged protein binds tightly to the Nickel ions in the column, while other proteins flow through.Elution:
To release the His-tagged protein from the column, you add imidazole (a chemical similar to the histidine tag). The imidazole competes with the His tag for the Nickel ions, causing the protein to detach and be collected.
What are the benefits and drawbacks of IMAC?
IMAC offers a high degree of purification in one step, though it may affect protein properties.
Separating proteins based on size method
Gel filtration/ exclusion chromatography
How does gel filtration work?
Polymeric gel beads have pores that allow proteins to enter if they fit. Larger proteins are excluded from the pores and elute first. We can then measure molar mass to identify protein.
Why do larger proteins elute first in gel filtration?
Larger proteins elute first because they cannot enter the pores, while smaller proteins enter the pores and elute later.
What is elution volume (Vₑ) in gel filtration?
The volume of buffer needed to move a protein from the top to the bottom of the column. t has a linear negative correlation with log molar mass.
How can gel filtration be used to determine the molar mass of a protein?
The elution volume (Vₑ) of an unknown protein is compared to proteins of known size to estimate its molar mass.
Measure its Vₑ, project it onto the log molar mass axis, and find the antilog. Alternatively, use the equation y = mx + b from known data.
What is electrophoresis used for in protein analysis?
It separates and characterizes proteins by moving charged molecules in an electric field.
Does electrophoresis purify proteins?
No, it does not contribute to purification because protein structure is often affected by the process.
instead it estimates
number of proteins in sample
purity of sample
isoelectric point
approx molecular weight
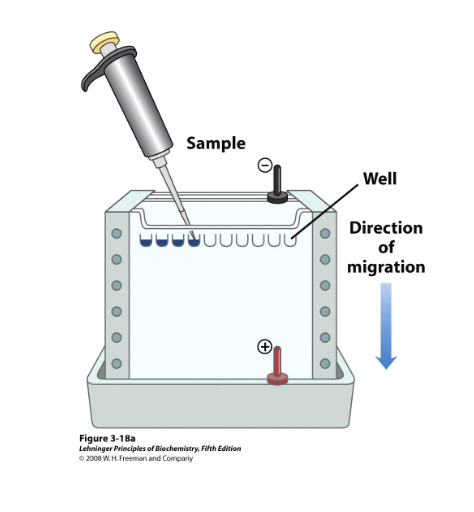
What factors affect protein movement in electrophoresis?
Size, shape, and charge determine how fast a protein moves in the gel.
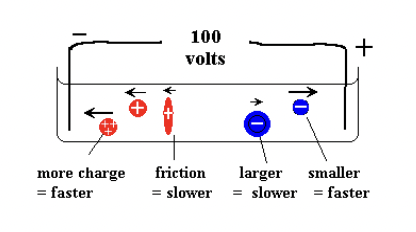
more charged and smaller molecules move faster.
What type of gel is used in electrophoresis?
Polyacrylamide gel, which has the right porosity to separate proteins ranging from 10 kDa to 1000 kDa.
How are proteins visualized after electrophoresis?
By staining with Coomassie blue, which binds to proteins.
What is SDS-PAGE and what does it do?
SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) is a method of gel electrophoresis that separates proteins based on size. SDS is used to denature proteins and give them a uniform negative charge, ensuring separation is strictly due to size.
How can the molecular weight of an unknown protein be estimated in SDS-PAGE?
By comparing the migration position of the unknown protein with known molecular weight standards.
What is Isoelectric Focusing?
Isoelectric Focusing is a technique that separates proteins based on their isoelectric point (pI): the pH at which a protein’s net charge is zero.
How does isoelectric focusing work?
Proteins move through a pH gradient in a gel, where at high pH they are deprotonated (negative charge), and at low pH they become protonated (positive charge), eventually stopping when their net charge is zero.
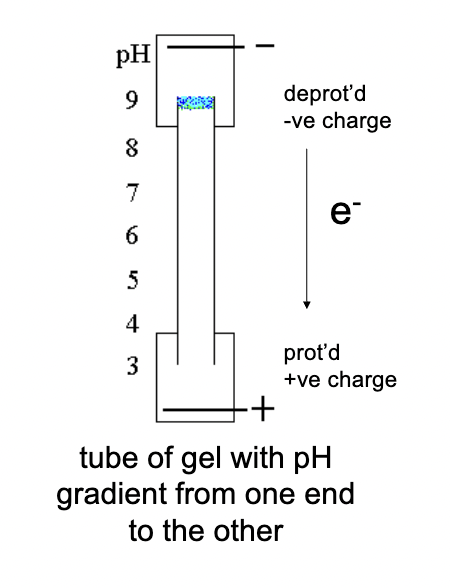
Why do proteins separate in isoelectric focusing?
ach protein has a different isoelectric point (pI), so they stop moving at different positions along the pH gradient, leading to separation.
What are Two-Dimensional Gels used for?
Two-Dimensional Gels combine isoelectric focusing and SDS-PAGE to separate complex mixtures of proteins based on both charge and size.
useful for analyzing large numbers of proteins from complex mixtures.
How does Mass Spectrometry identify proteins?
Mass Spectrometry vaporizes a protein with a laser beam, creating charged particles that are detected based on their mass. Particles travel towards detector.
larger particles move slower
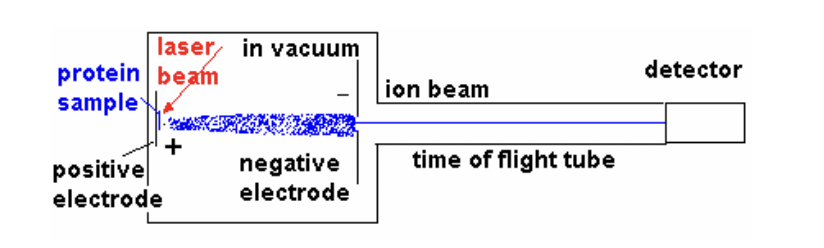
How is mass measured in Mass Spectrometry?
The time of flight to the detector provides an accurate mass measurement, with five significant figures of accuracy.
How is the protein identified in Mass Spectrometry?
The measured mass is compared to a database of proteins with known masses to identify the protein.
Purification of a specific protein from a crude mixture requires the use of multiple seperation methods
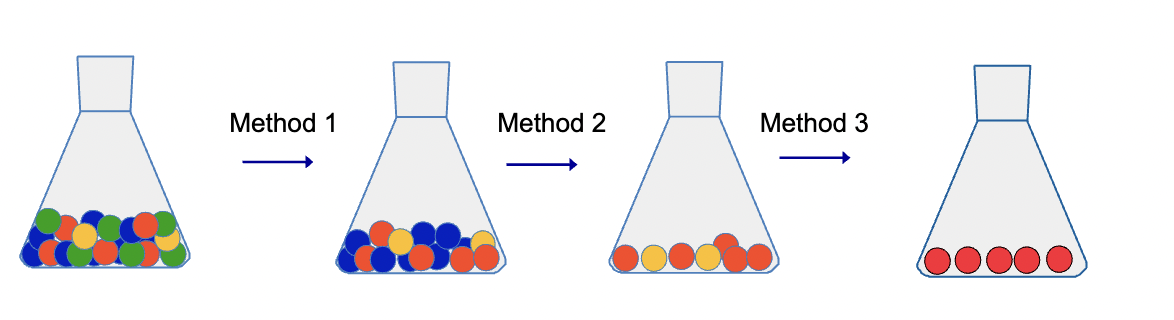
All protein identification/separation/purification methods
Ion Exchange Chromatography (IEX)
Affinity Chromatography
Immobilized Metal Affinity Chromatography (IMAC)
Gel Filtration / Molecular Exclusion Chromatography
SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis)
Isoelectric Focusing
Two-Dimensional Gel Electrophoresis
Mass Spectrometry