Step 1 First Aid - Biochemistry
1/554
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
555 Terms
Chromatin structure
Negatively charged DNA loops twice around histone octamer (2 each of the positively charged H2A, H2B, H3, and H4) to form nucleosome bead. H1 ties nucleosomes together in a string. (Think of "beads on a string"; H1 is the only histone that is not in the nucleosome core.) In mitosis, DNA condenses to form mitotic chromosomes.
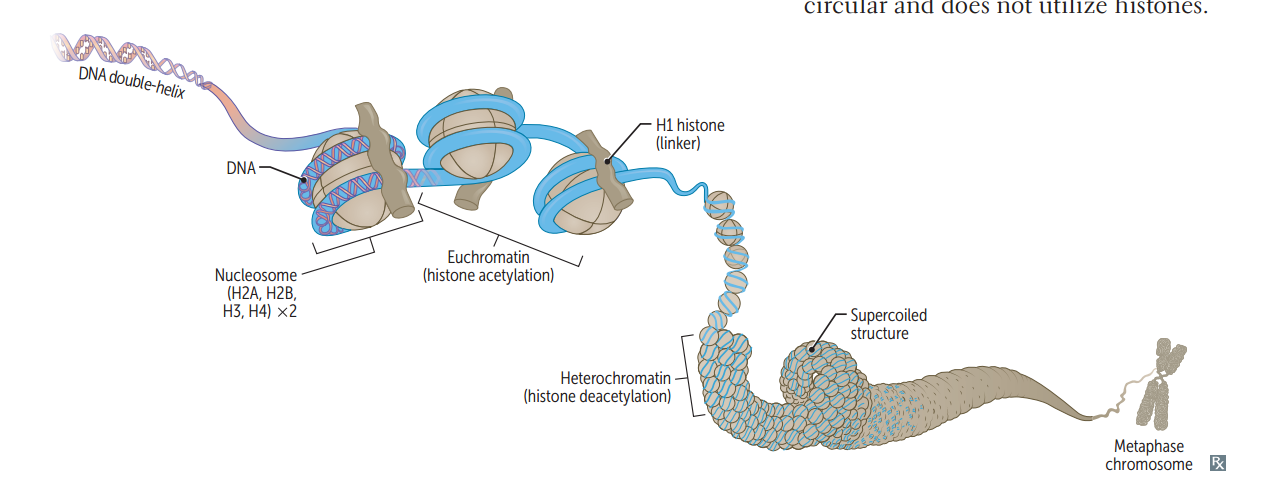
Heterochromatin
Condensed, transcriptionally inactive ("H eteroC hromatin = H ighly C ondensed.")
Euchromatin
Less condensed, transcriptionally active (Eu = true, "truly transcribed")
Purines
A, G 2 Rings ("PUR e A s G old = PUR ines")
Pyrimidines
C, T, U 1 ring ("CUT the PY (pie): PY rimidines")
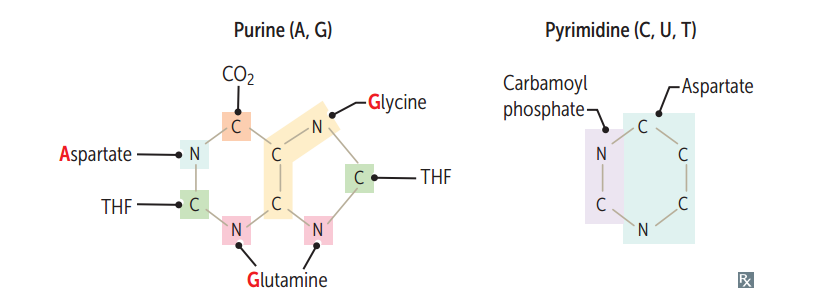
Functional groups of the nucleosides
Guanine has a ketone. Thy mine has a methy l. Deamination of cytosine makes uracil.
Base differences btw RNA and DNA
Uracil is found in RNA; Thymine in DNA
Base pair bonds
G-C bond (3 H-bonds) is stronger than A-T bond (2 H-bonds). Incr G-C content --< higher melting temperature.
AA's necessary for purine synthesis
G lycine A spartate G lutamine
Nucleoside
Base + ribose
Nucleotide
Base + ribose + phosphate; linked by 3'-5' phosphodiester bond.
Purines are made from...?
IMP precursor (see bottom/right)
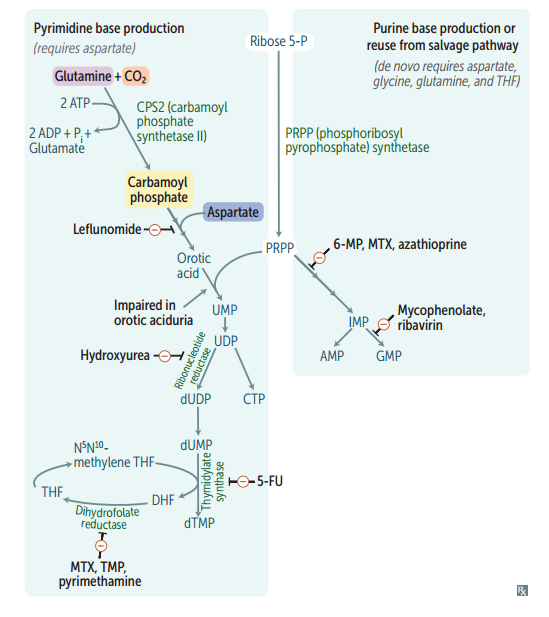
Pyrimidines are made from...?
Orotate precursor, with PRPP added later.
Deoxyribonucleotide synthesis
Ribonucleotides are synthesized first and are converted to deoxyribonucleotides by ribonucleotide reductase.
ABX and anti-neoplastic drugs that function by interfering w/ nucleotide synthesis (list)
Hydroxyurea 6-mercaptopurine 5-fluorouracil Methotrexate Trimethoprim

Hydroxyurea
Inhibits ribonucleotide reductase.
6-mercaptopurine (6-MP)
Blocks de novo purine synthesis.
5-Fluorouracil (5-FU)
Inhibits thymidilate synthase (decr dTMP).
Methotrexate
Inhibits dihydrofolate reductase (decr dTMP)
Trimethoprim
Inhibits bacterial dihydrofolate reductase (decr dTMP)
Transition vs. transversion
Transition: Substituting purine for purine or pyrimidine for pyrimidine ("TransI tion = I dentical type") Transversion: Substituting purine for pyrimidine or vice versa ("TransV ersion = conV ersion btw types")
Genetic code: unambiguous
Each codon specifies only 1 AA.
Genetic code: degenerate/redundant
< 1 codon may code for the same AA. (Methionine is encoded by 1 codon: AUG)
Genetic code: Commaless, nonoverlapping
Read from a fixed starting point as a continuous sequence of bases. *some viruses are an exception.
Genetic code: universal
Genetic code is conserved throughout evolution. *exceptions include mitochondria, archaebacteria, Mycoplasma , and some yeasts
Adenosine deaminase
deficiency.
ADA is required for degradation of adenosine
and deoxyadenosine. less ADA more dATP
Less lymphocytes. One of the major causes of autosomal recessive
SCID.
Lesch-Nyhan
syndrome
Defective purine salvage. Absent HGPRT
less GMP (from guanine) and less IMP (from
hypoxanthine) formation. Compensatory in
purine synthesis ( increase PRPP amidotransferase
activity) excess uric acid production. HGPRT:
Hyperuricemia
Gout
Pissed off (aggression, self-mutilation)
Red/orange crystals in urine
Tense muscles (dystonia)
Treatment: allopurinol, febuxostat.
Silent mutation
Same AA, often base change in 3rd position of codon (tRNA wobble)
Missense mutation
Changed AA (conservative -- new AA is similar in chemical structure) Glutamine > Valine: Sickle cell anemia.
Nonsense mutation
Change resulting in early stop codon ("Stop the nonsense !")
Frame shift mutation
Change resulting in misreading of all nucleotides downstream, usually resulting in a truncated, nonfunctional protein. can be addition & deletion.
Severity of damage in DNA mutations
Nonsense < missense < silent
Eukaryotic vs. prokaryotic DNA replication.
Eukaryotic DNA replciation is more complex, but uses many analogous enzymes. In both: DNA replication is semiconservative and involves both continuous and discontinuous (Okazaki fragment) synthesis. For eukaryotes, replication begins at a consensus sequence of base pairs.
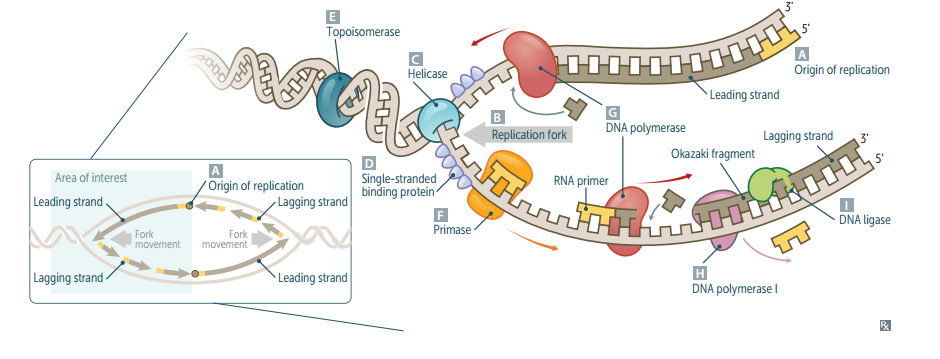
Origin of replication
Particular sequence in genome where DNA replication begins. May be single (prokaryotes) or multiple (eukaryotes). AT-rich sequences (eg, TATA box regions) are found in promoters (often upstream) and origins of replication (ori).
Replication fork
Y-shaped region along DNA template where leading and lagging strands are synthesized.
Helicase
Unwinds DNA template at replication fork.
Single-stranded binding protein
Prevents strands from reannealing.
DNA topoisomerases
Create a nick in the helix to relieve supercoils created during replication. *Fluoroquinolones inhibit DNA gyrase (a specific prokaryotic topoisomerase)
Primase
Makes an RNA primer on which DNA polymerase III can initiate replication.
DNA polymerase III
Prokaryotic only. Elongates leading strand by adding deoxynucleotides to the 3' end. Elongates lagging strand until it reaches primer of preceding fragment. 3'--<5' exonuclease activity "proofreads" each added nucleotide. (5'--<3' synthesis; 3'--<5' proofreading w/ exonuclease)
DNA polymerase I
Prokaryotic only. Degrades RNA primer and fills in the gap w/ DNA. (excises RNA primer w/ 5'--<3' exonuclease)
DNA ligase
Seals.
Single strand nucleotide excision repair
Specific endonucleases release the oligonucleotide-containing damaged bases; DNA polymerase and ligase fill and reseal the gap, respectively. (mutated in xeroderma pigmentosum)
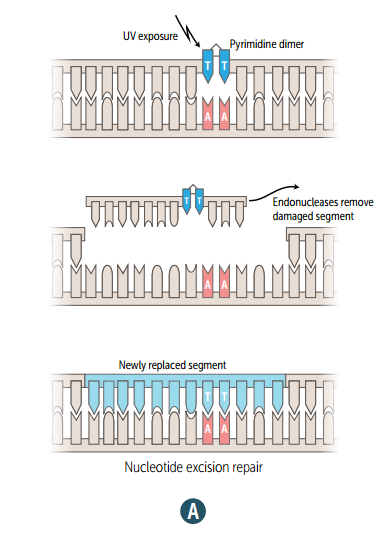
Xeroderma pigmentosum
Mutated single strand nucleotide excision repair gene, which prevents repair of thymidine dimers.; Dry skin w/ melanoma and other cancers ("children of the night").
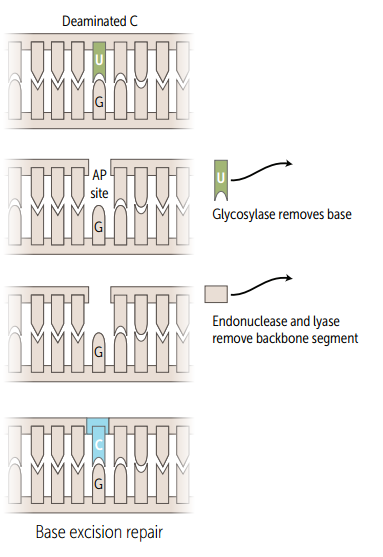
Single strand base excision repair
Specific glycosylases recognize and remove damaged bases, AP endonuclease cuts DNA at apyrimidinic site, empyty sugar is removed, and the gap is filled and resealed.
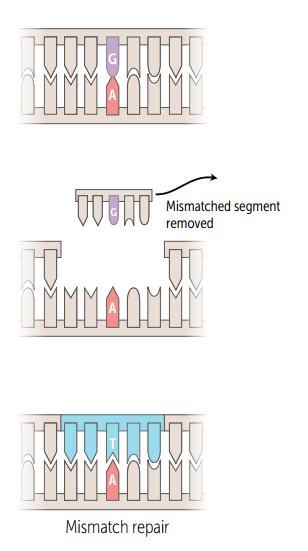
Single strand mismatch repair
Unmethylated, newly synthesized string is recognized, mismatched nucleotides are removed, and the gap is filled and resealed. Mutated in hereditary nonpolyposis colorectal cancer (HNPCC).
Double strand nonhomologous end joining
Brings together 2 ends of DNA fragments. No requirement for homology.
What direction is DNA/RNA made?
They are both synthesized in the 5'--<3' direction. Remember that the 5' of the incoming nucleotide bears the triphosphate (energy source for bond). The 3' hydroxyl of the nascent chain is the target.
What direction is mRNA read?
5'--<3'.
What direction is protein synthesized?
N--
3 Types of mRNA
rRNA(RNA poly 1) is the most abundant mRNA (RNA poly 2) is the longest tRNA (RNA poly 3)is the smalles ("R ampant, M assive, T iny")
mRNA start codon
AUG (or rarely GUG) ("AUG inAUG urates protein synthesis") In eukartyotes, codes for methionine, which may be removed before translation is completed. In prokaryotes, codes for formyl-methionine (f-Met).
mRNA stop codons
UGA, UAA, UAG UGA = U G o A way UAA = U A re A way UAG = U A re G one
Functional organization of the gene
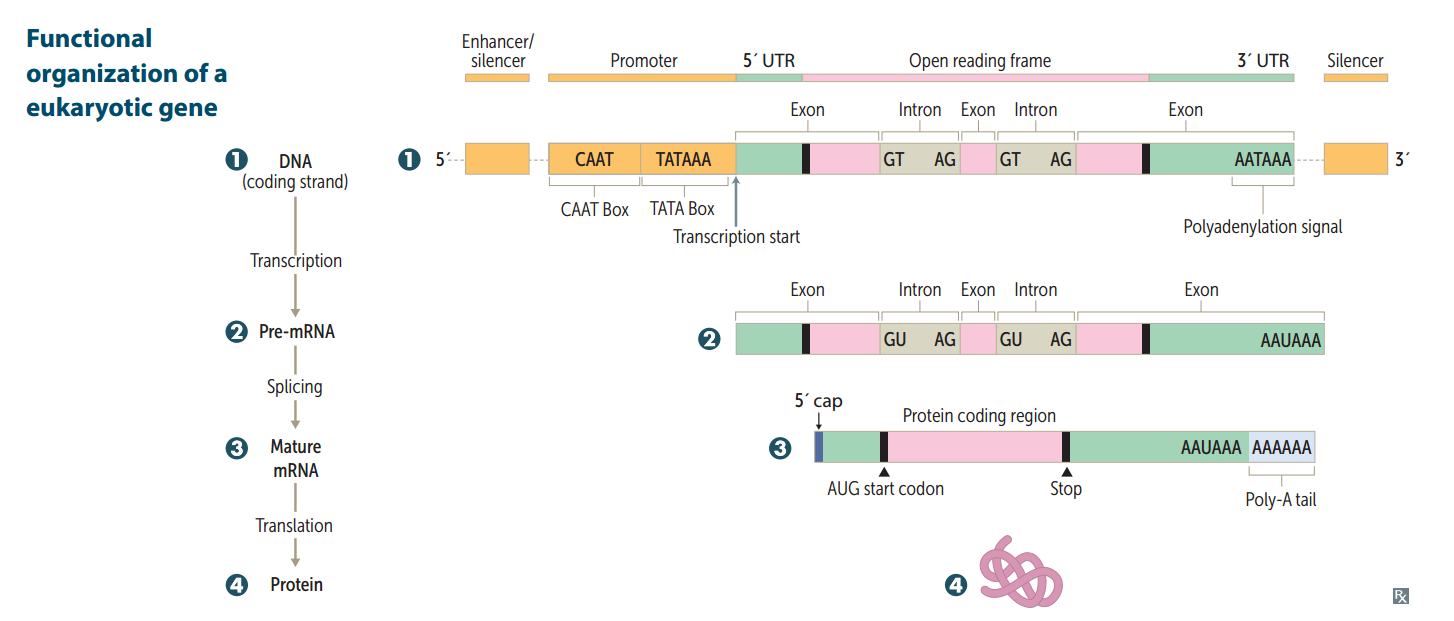
Promoter
Site where RNA polymerase and multiple other transcription factors bind to DNA upstream from gene locus (AT-rich upstream sequence w/ TATA and CAAT boxes). Mutation here commonly results in dramatic drop in amount of gene transcribed.
Enhancer
Stretch of DNA that alters gene expression by binding transcription factors.
Silencer
Site where negative regulators (repressors) bind.
Where are enhancers and silencers located?
May be close to, far from, or even within (in an intron) the gene whose expression it regulates.
Eukaryotic RNA polymerases
RNA pol I -- makes rRNA RNA pol II -- makes mRNA RNA pol III -- makes tRNA (I, II, and III are numbered as their products are used in protein synthesis) No proofreading fxns, but can initiate chains. RNA pol II opens DA at promoter site.
Prokaryotic RNA polymerase
One RNA polymerase (a multisubunt complex) makes all of the 3 kinds of RNA.
alpha-amantin
Found in death cap mushrooms. Inhibits RNA pol II.
RNA processing (in eukaryotes)
Occurs in nucleus. After transcription: 1.) Capping on 5' end (7-methylguanosine) 2.) Polyadenylation on 3' end (~200 A's) 3.) Splicing out of introns Only processed RNA is transported out of the nucleus.
hnRNA vs. mRNA
The initial transcript is called heterogeneous nuclear RNA (hnRNA) The capped and tailed transcript is called mRNA.
Polyadenylation signal
AAUAAA
Poly-A polymerase does not require...
...does not require a template.
pre-mRNA splicing (occurs in eukaryotes)
1.) Primary transcript combines w/ snRNPs and other proteins to form spliceosome 2.) Lariat-shaped intermediate is generated 3.) Lariat is released to remove intron precisely and join 2 exons.
Introns vs. exons
Exons contain the actual genetic information coding for protein. Introns are intervening noncoding segments of DNA. ("IN trons stay IN the nucleus, whereas EX ons EX it and are EX pressed")
Alternative splicing
Different exons are combined to make unique proteins in different tissues (e.g., beta-thalassemia mutations)
tRNA structure
75-90 nucleotides, secondary structure, cloverleaf form, anticodon end is opposite 3' aminoacyl end. All tRNAs, both eukaryotic and prokaryotic, have CCA at 3' end along w/ a high percentage of chemically modified bases. The AA is covalently bound to the 3' end of tRNA.
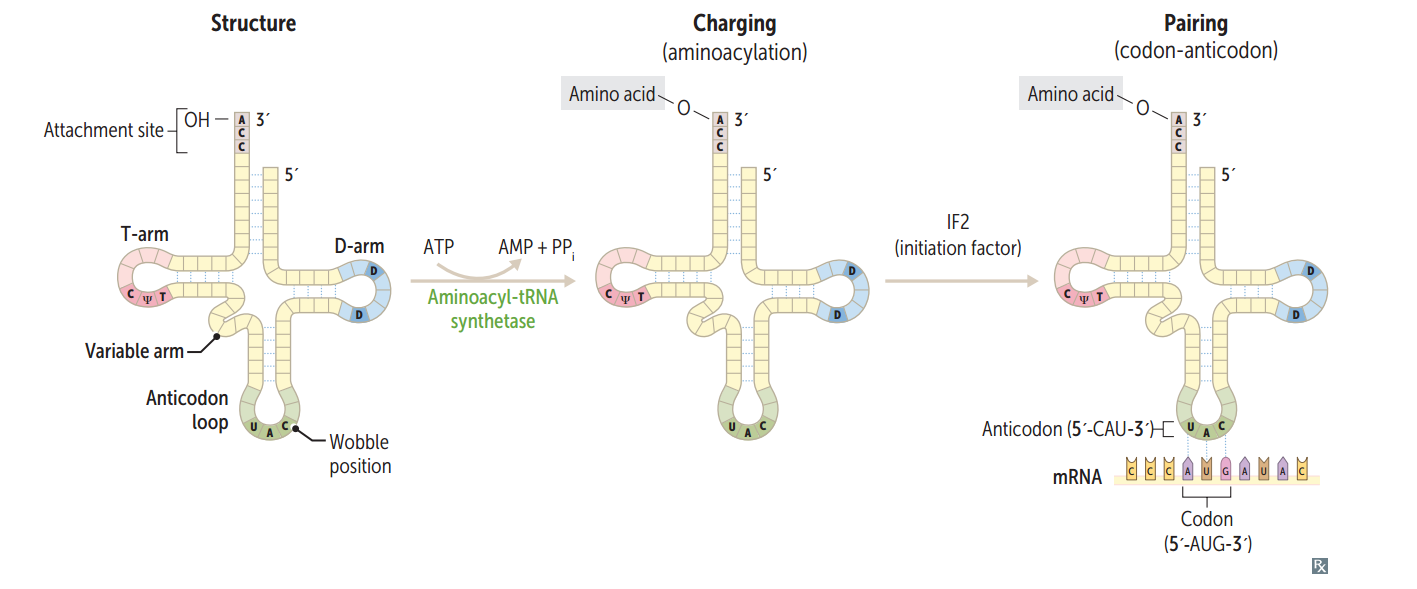
Charging of tRNA
Aminoacyl-tRNA synthetase (1 per AA, "matchmaker," uses ATP) scrutinizes AA before and after it binds to tRNA. If incorrect, bond is hydrolyzed. The aa-tRNA bond has energy for formation of peptide bond. A mischarged tRNA reads usual codon but inserts wrong AA.
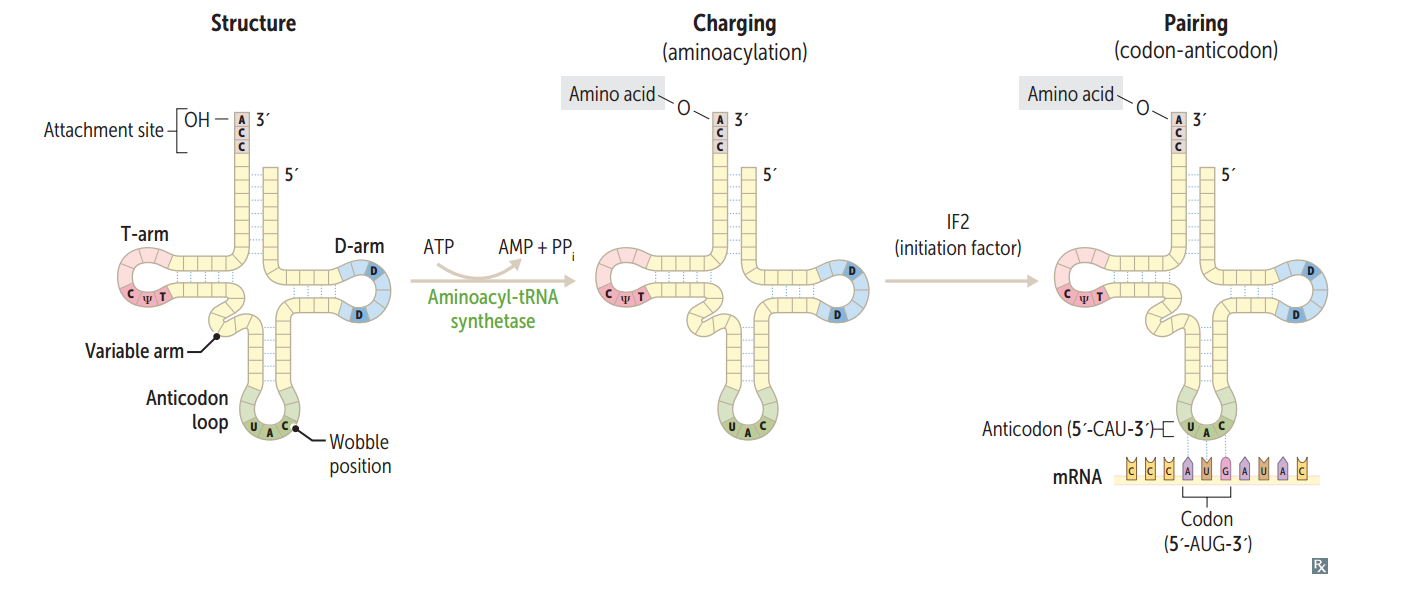
What is responsible for the accuracy of AA selection?
Aminoacyl-tRNA synthetase and binding of charged tRNA to the codon are responsible for accuracy of AA selection.
Mechanism of tetracyclines
Bind 30S subunit, preventing the attachment of aminoacyl-tRNA.
tRNA wobble
Accurate base pairing is required only in the first 2 nucleotide positions of an mRNA codon, so codons differing in the 3rd "wobble" position may code for the same tRNA/aa (due to degeneracy of genetic code).
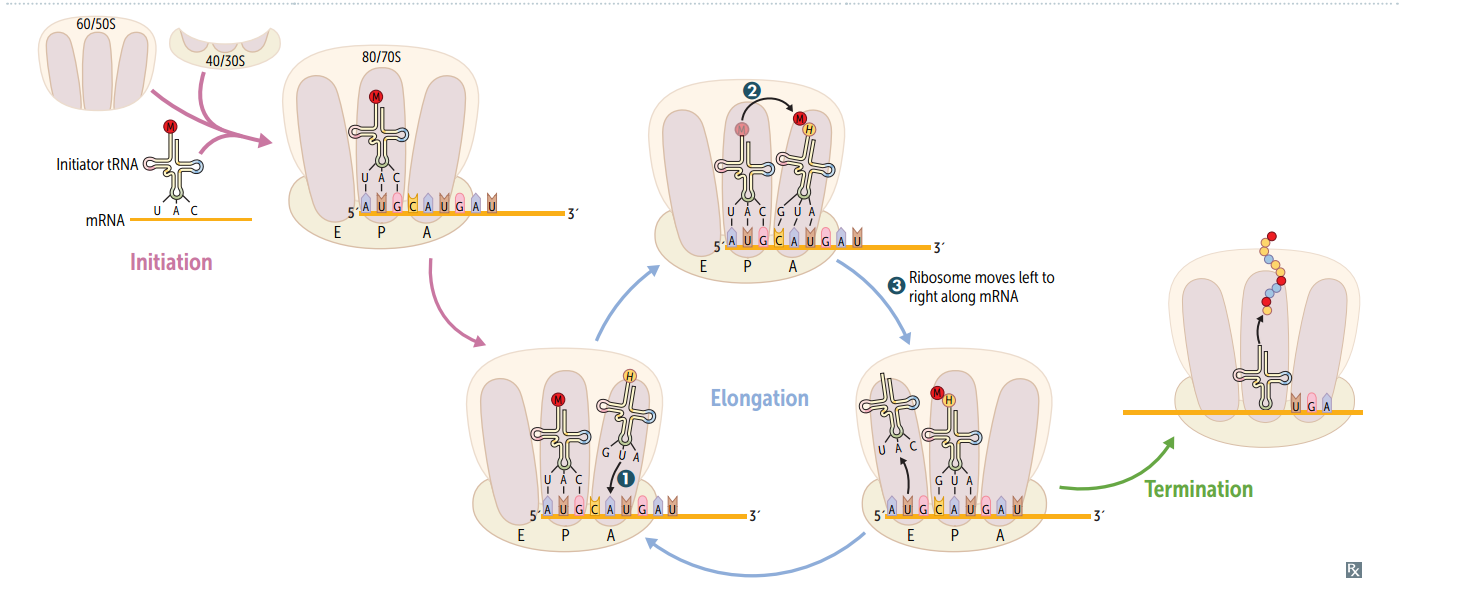
Protein synthesis: initiation
Activated by GTP hydrolysis, initiation factors (eIFs) help assemble the 40S ribosomal subunit w/ the initiator tRNA released when the mRNA and the ribosomal subunit assemble w/ the complex. E ukaryotes: 40S + 60S = 80S (E ven) PrO karyotes: 30S + 50S = 70S (O dd)
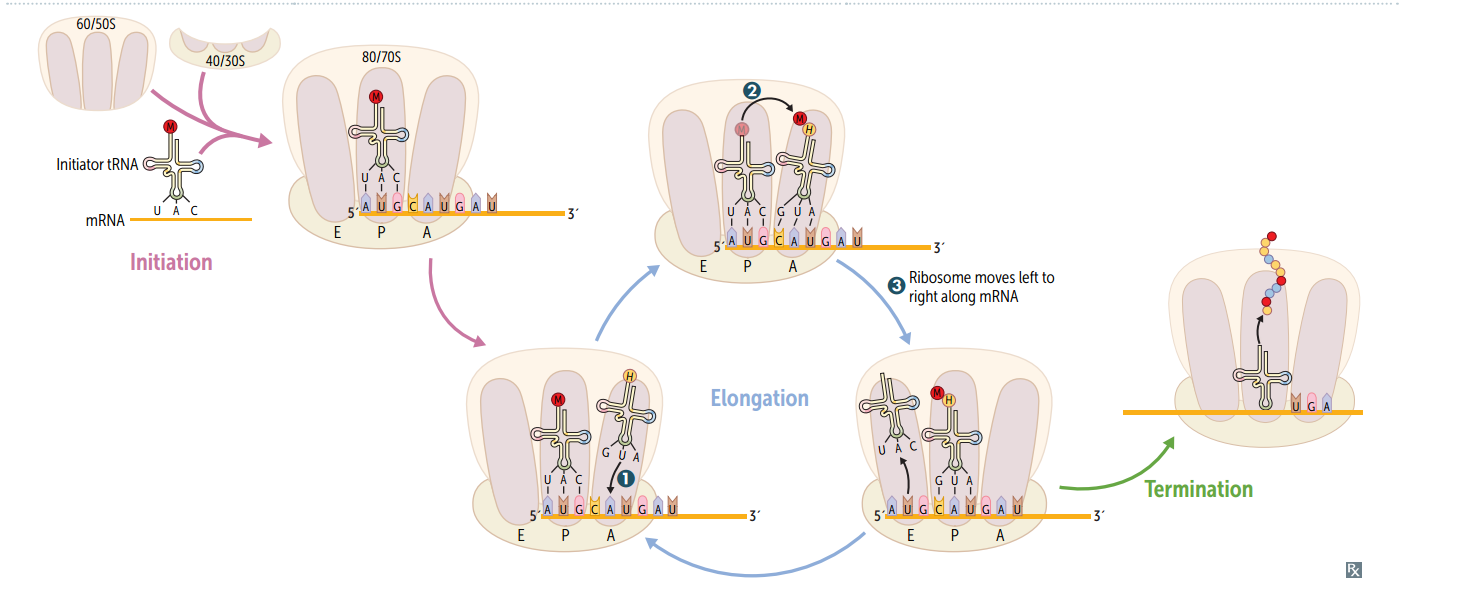
Protein synthesis: step 1 in elongation
Aminoacyl-tRNA binds to Aa site (except for initiator methionine)
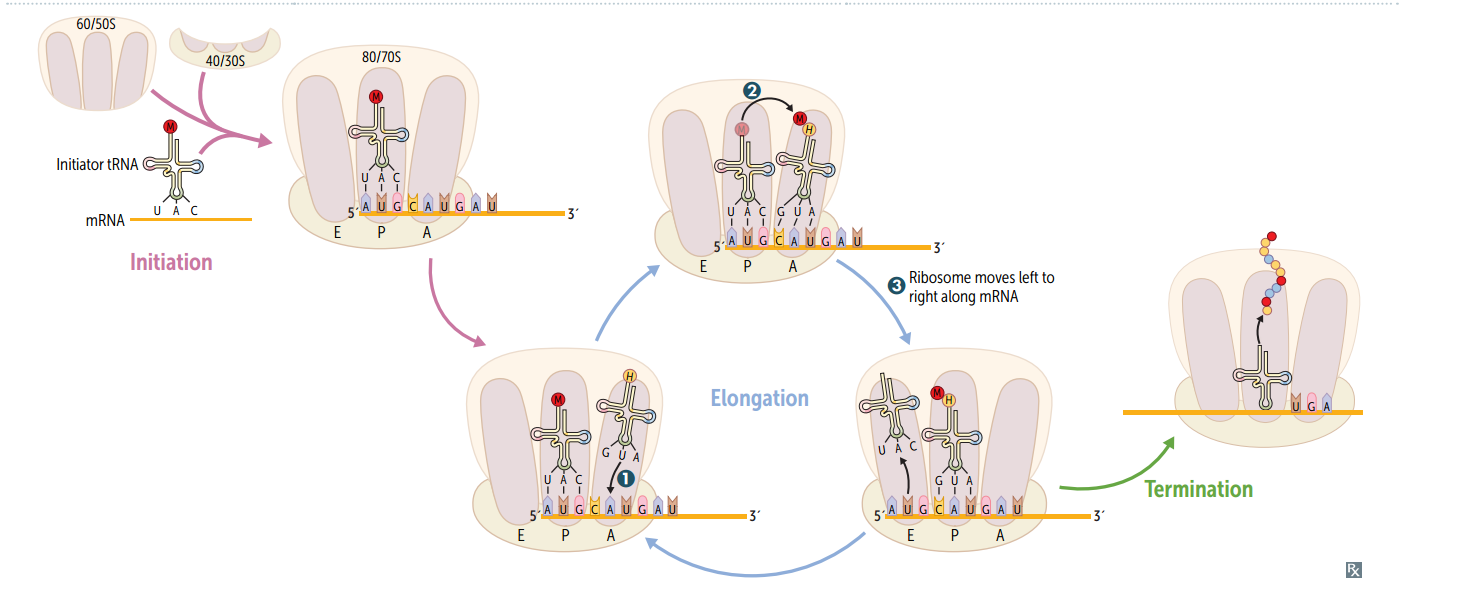
Protein synthesis: step 2 in elongation
Peptidyltransferase catalyzes peptide bond formation, transfers growing polypeptide to amino acid in A site.
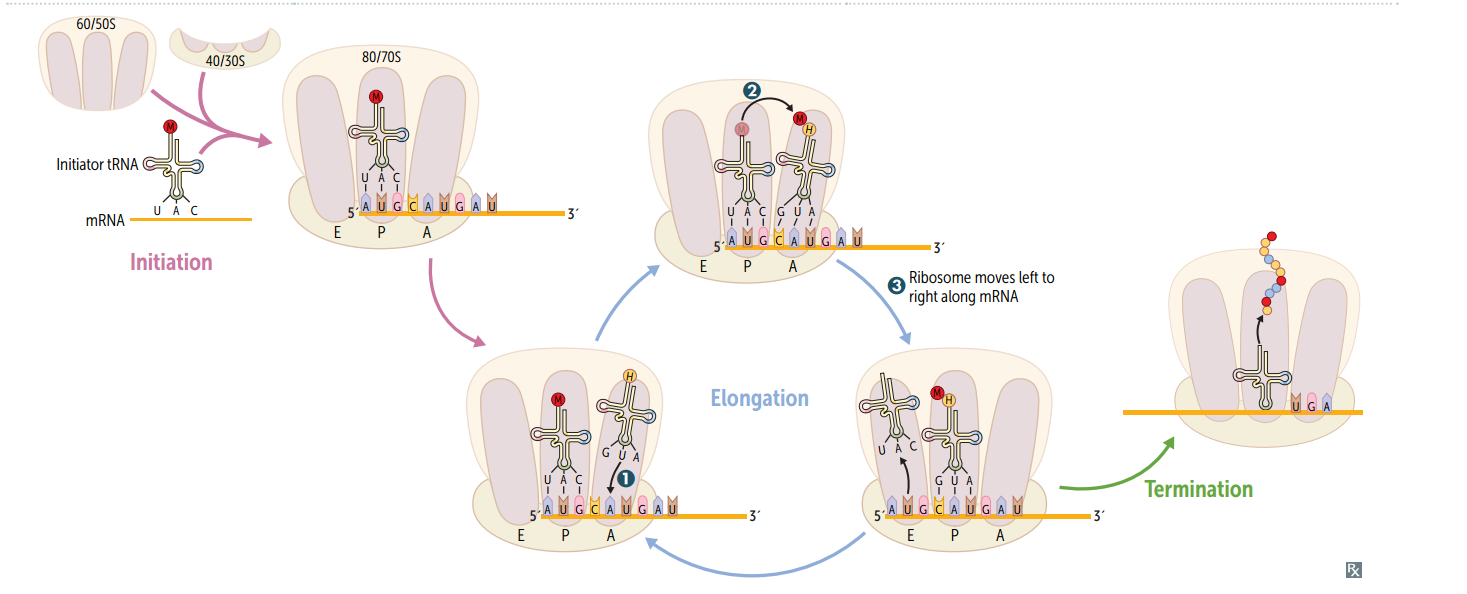
Protein synthesis: step 3 in elongation
Ribosome advances 3 nucleotides toward the 3' end of RNA, moving peptidyl RNA to P site (translocation)
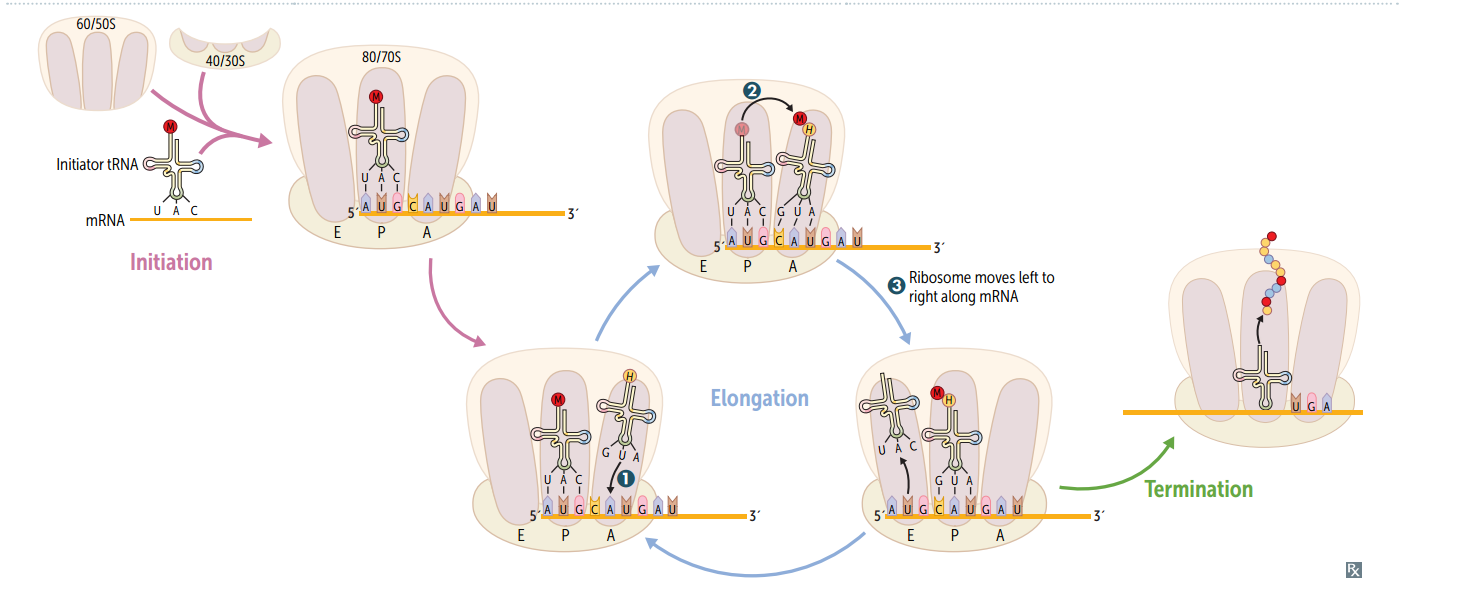
mnemonic for the 3 sites in the ribosome
"going APE " A site = incoming A minoacyl tRNA P site = accommodates growing P eptide E site = holds E mpty tRNA as it E xits
Protein synthesis: termination
Completed protein is released from ribosome thru simple hydrolysis and dissociates.
Aminoglycosides as protein synthesis inhibitors
Inhibit formation of the initiation complex and cause misreading of mRNA.
Chloramphenicol as a protein synthesis inhibitor
Inhibits 50S peptidyltransferase.
Macrolides as protein synthesis inhibitors
Bind 50S, blocking translocation.
Clindamycin as a protein synthesis inhibitor
Binds 50S, blocking translocation.
Energy requirements of translation
tRNA aminoacylation: ATP --< AMP (2 phosphoanhydride bonds) Loading tRNA onto ribosome: GTP --< GDP Translocation: GTP --< GDP Total energy expenditure = 4 high-energy phosphoanhydride bonds
Posttranslational modifications: trimming
Removal of N- or C-terminal propeptides from zymogens to generate mature proteins.
Posttranslational modifications: covalent alterations
Phosphorylation, glycosylation, and hydroxylation.
Posttranslational modifications: Proteasomal degradation
Attachment of ubiquitin to defective proteins to tag them for breakdown.
Enzyme regulation methods
Enzyme concentration alteration (synthesis and/or destruction) Covalent modification (e.g., phosphorylation Proteolytic modification (zymogen) Allosteric regulation (e.g., feedback inhibition) pH Temperature Transcriptional regulation (e.g., steroid hormones)
Cell cycle phases are regulated by what three things?
cyclins, CDKs, and tumor suppressors.
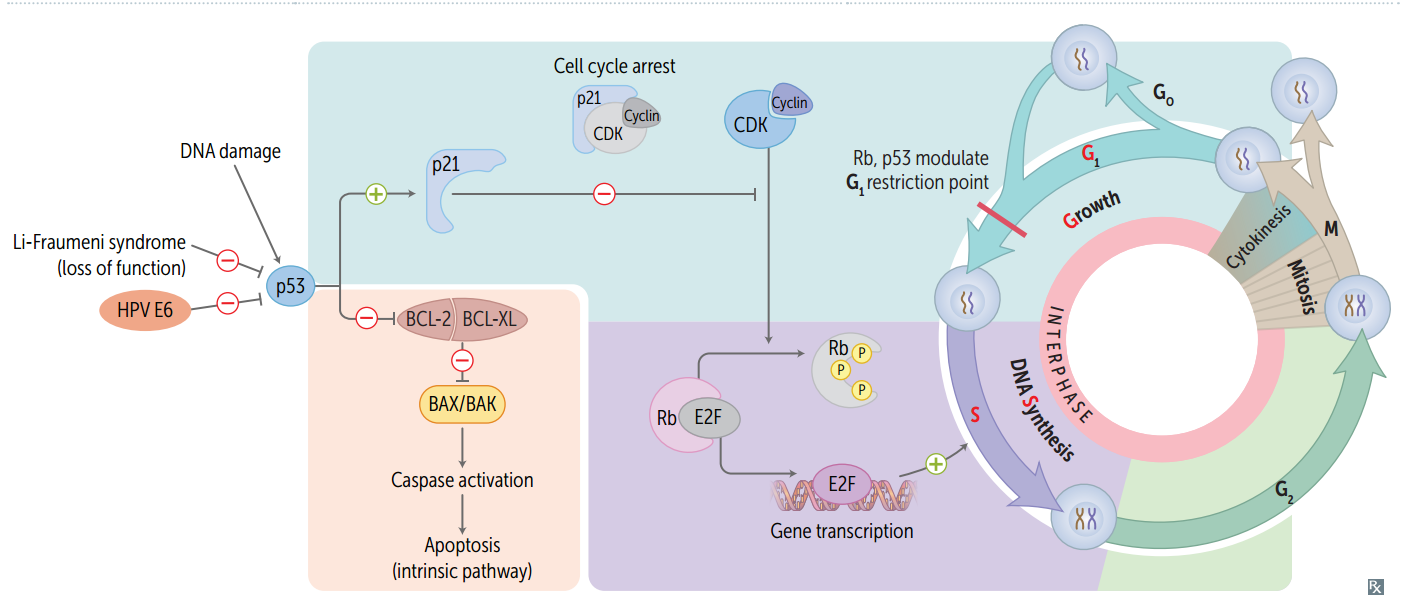
Order (and important lengths) of cell cycle phases
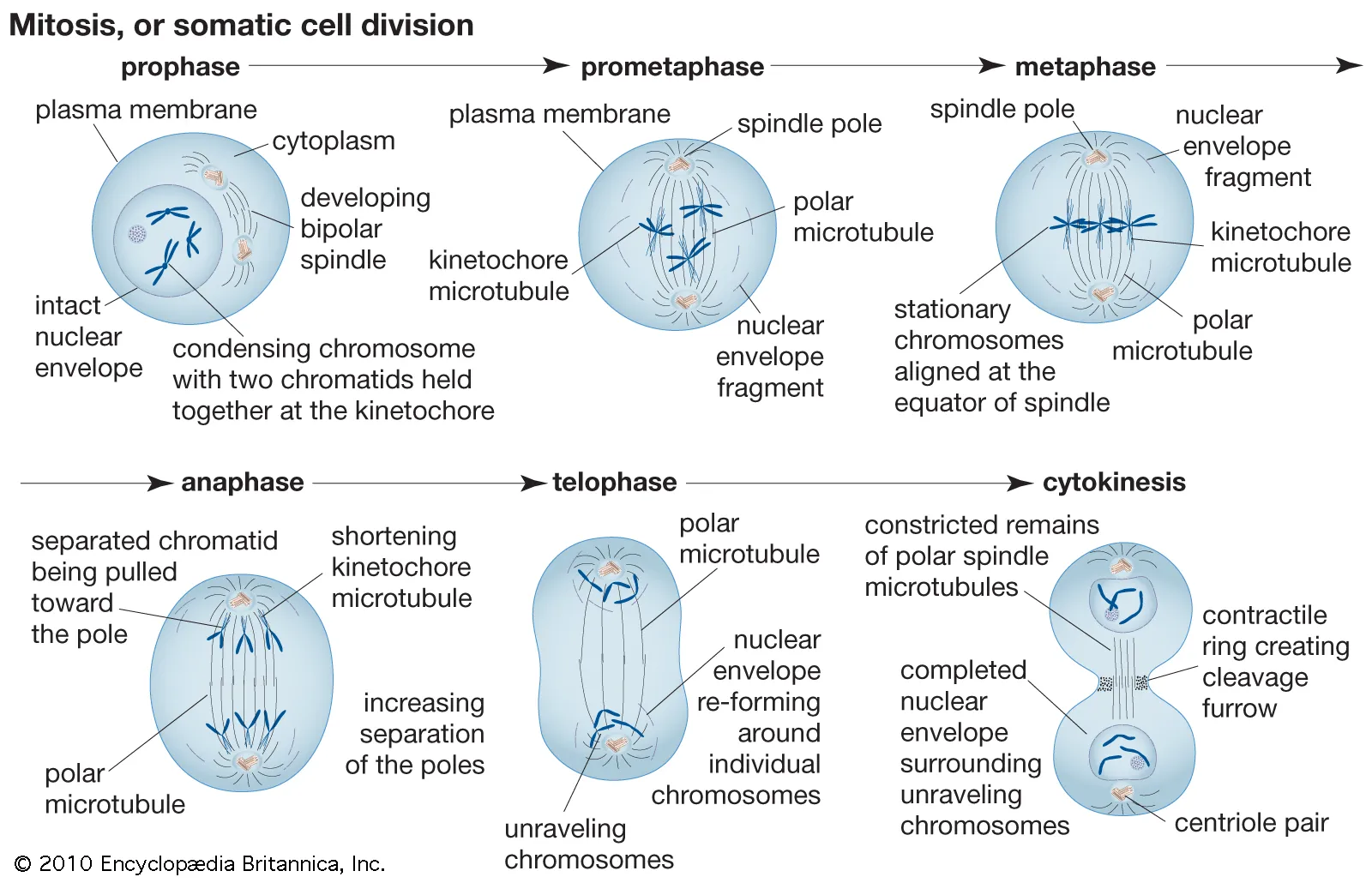
Mitotis (shortest phase): prophase - metaphase - anaphase - telophase. G1 and G0 are of variable duration. G = G ap or G rowth S = S ynthesis
CDKs
Cyclin-dependent kinases; constitutive and inactive.
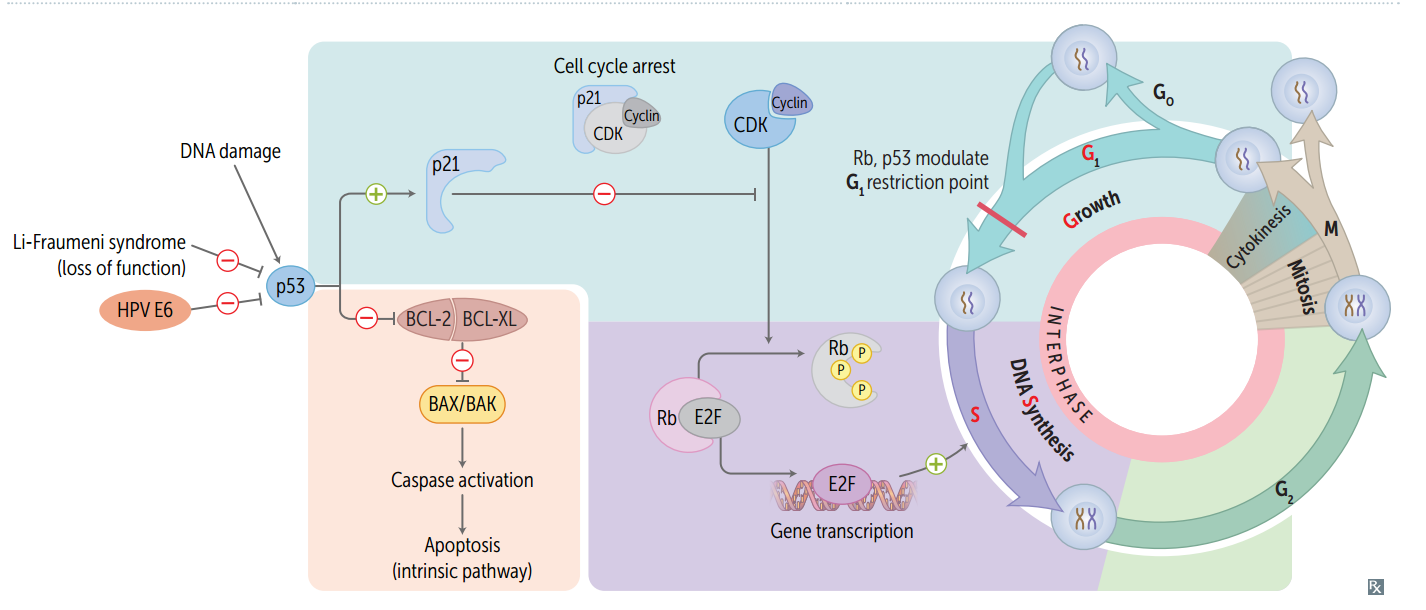
Cyclins
Regulatory proteins that control cell cycle events; phase specific; activate CDKs
Cyclin-CDK complexes
Must both be activated and inactivated for cell cycle to progress.
Tumor suppressors (and the cell cycle)
Rb and p53 normally inhibit G1-to-S progression; mutations in these genes result in unrestrained cell growth.
Permanent cells
Remain in G0, regenerate from stem cells. (e.g., neurons, skeletal and cardiac muscle, RBCs)
Stable (quiescent) cells
Enter G1 from G0 when stimulated (e.g., Hepatocytes, lymphocytes)
Labile cells
Never go to G0, divide rapidly w/ a short G1 (e.g., Bone marrow, gut epithelium, skin, hair follicles)
Rough Endoplasmic Reticulum (RER)
Site of synthesis of secretory (exported) proteins and of N-linked oligosaccharide addition to many proteins.
Nissl bodies
RER in neurons -- synthesize enzymes (e.g., ChAT) and peptide neurotransmitters.
Free ribosomes
unattached to any membrane; site of synthesis of cytosolic and organellar proteins.