MBIO 2700 / Topic 8a: Nucleotides, Nucleic Acids, and DNA
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
32 Terms
What are the monomers that make up nucleic acids?
What are the four jobs of a nucleotide?
Nucleic acids.
The five jobs of a nucleotide are as follows:
Constituents of nucleic acids.
Energy currency in metabolic transactions.
Molecular links in response of cells to hormones and other extracellular stimuli.
Structural components of an array of enzyme cofactors and metabolic intermediates.
What are the two key types of nucleic acids? What’s the difference between them in both structure and function?
Deoxyribonucleic acid (DNA) → 2’ deoxy-D-Ribose.
Ribonucleic acid (RNA) → D-Ribose.
Both carry genetic information, but RNA can be a catalyst while DNA cannot.
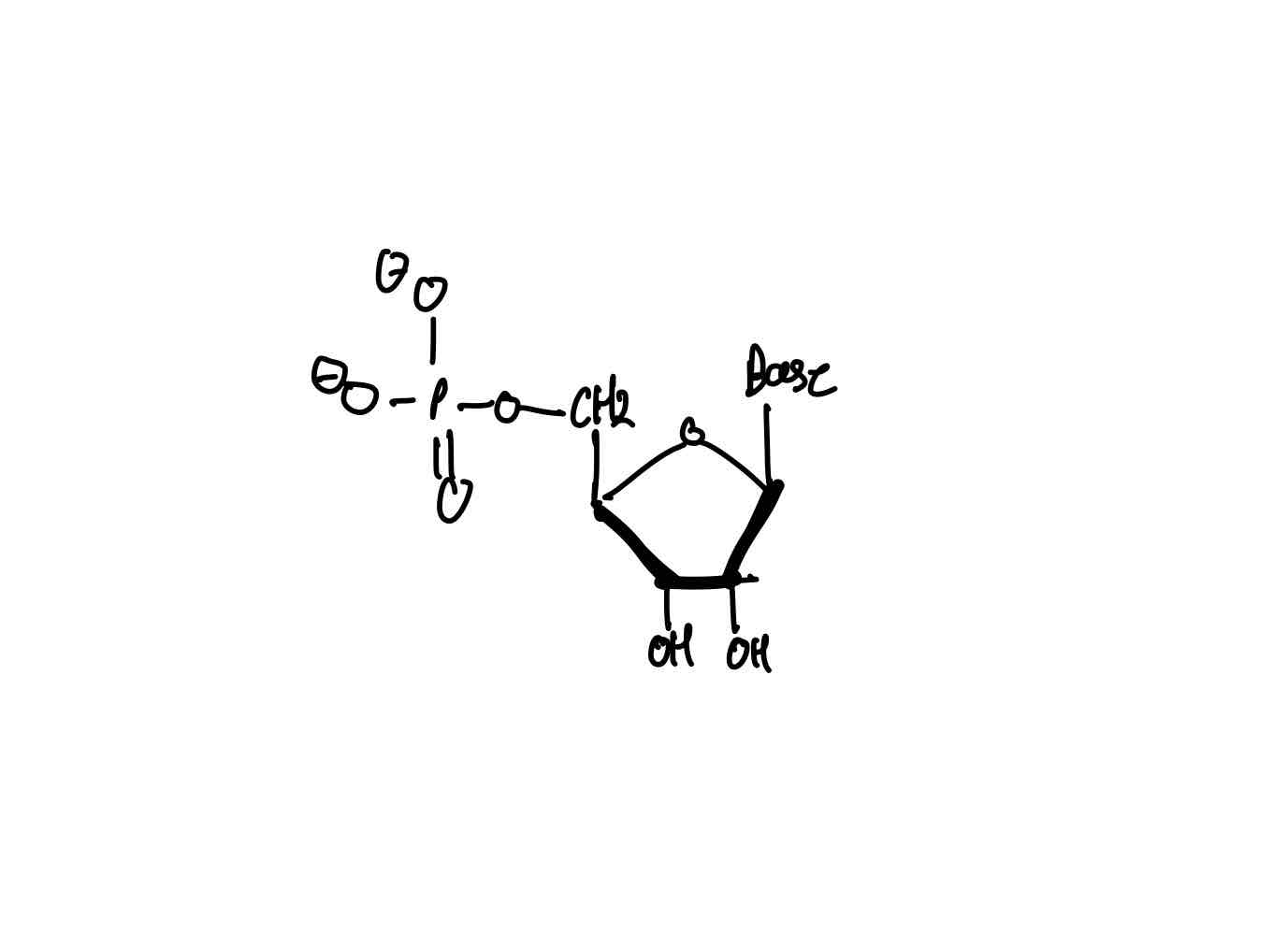
What are the three key components of a nucleotide?
What is a nucleoside?
> Nitrogenous (nitrogen-containing) base.
> Pentose.
> 1/2/3 phosphates.
> Nucleoside = nucleotide w/o PO43−.
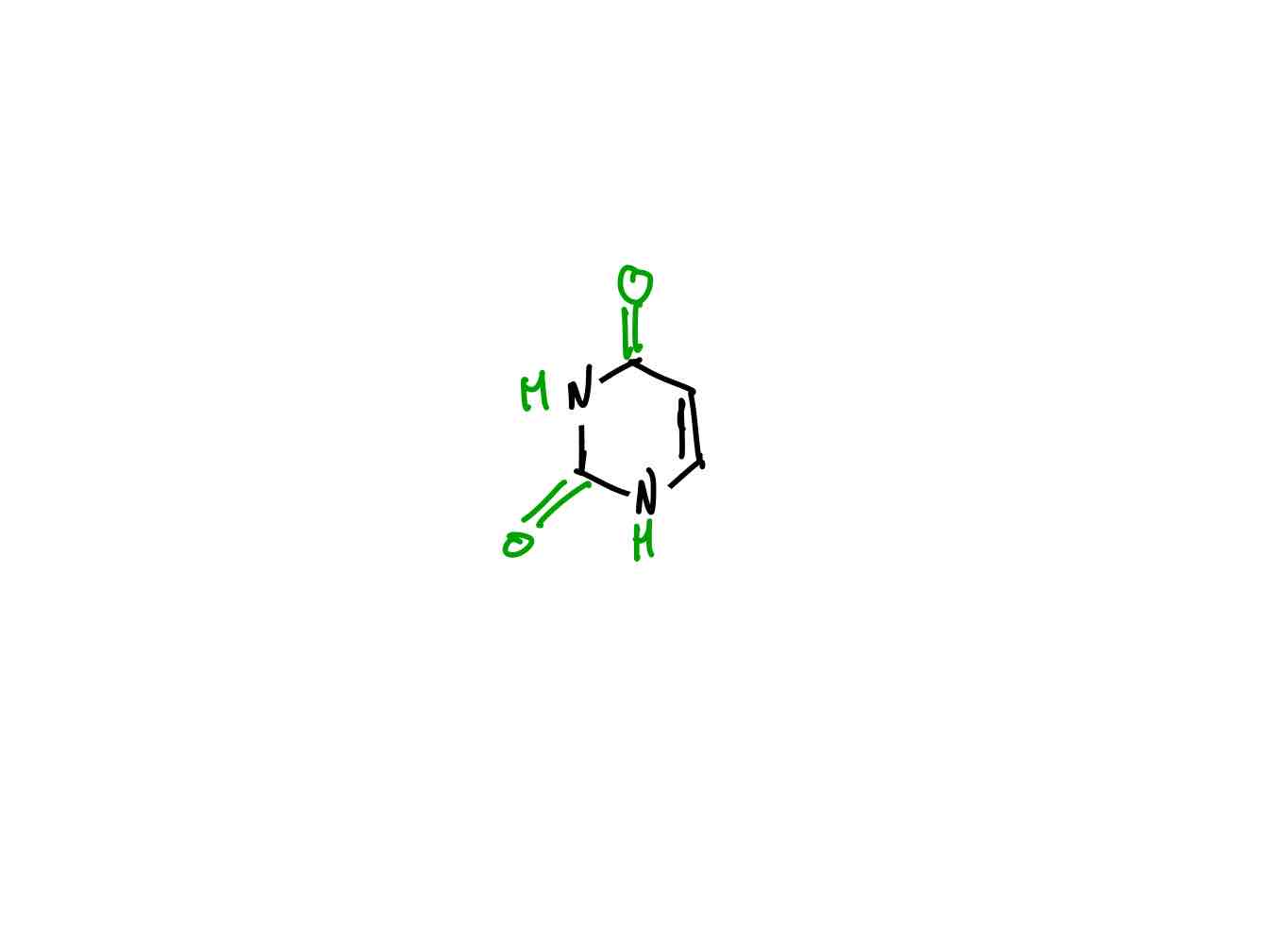
What are the five possible bases for a nucleotide?
What are the two groups for these five bases? Why are they grouped as such?
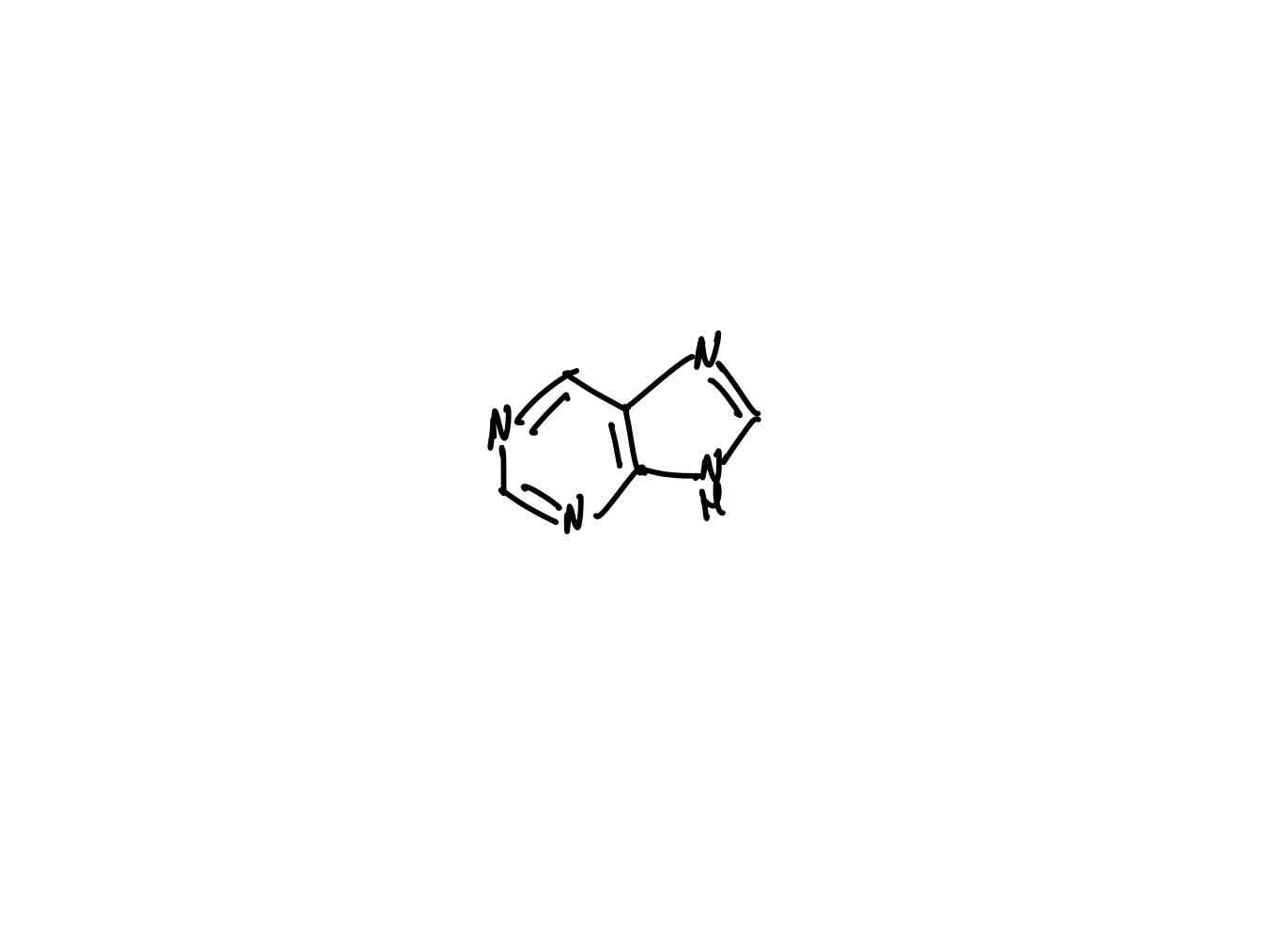
The purine bases (adenine and guanine) are called purine bases because their parent compounds are purines.
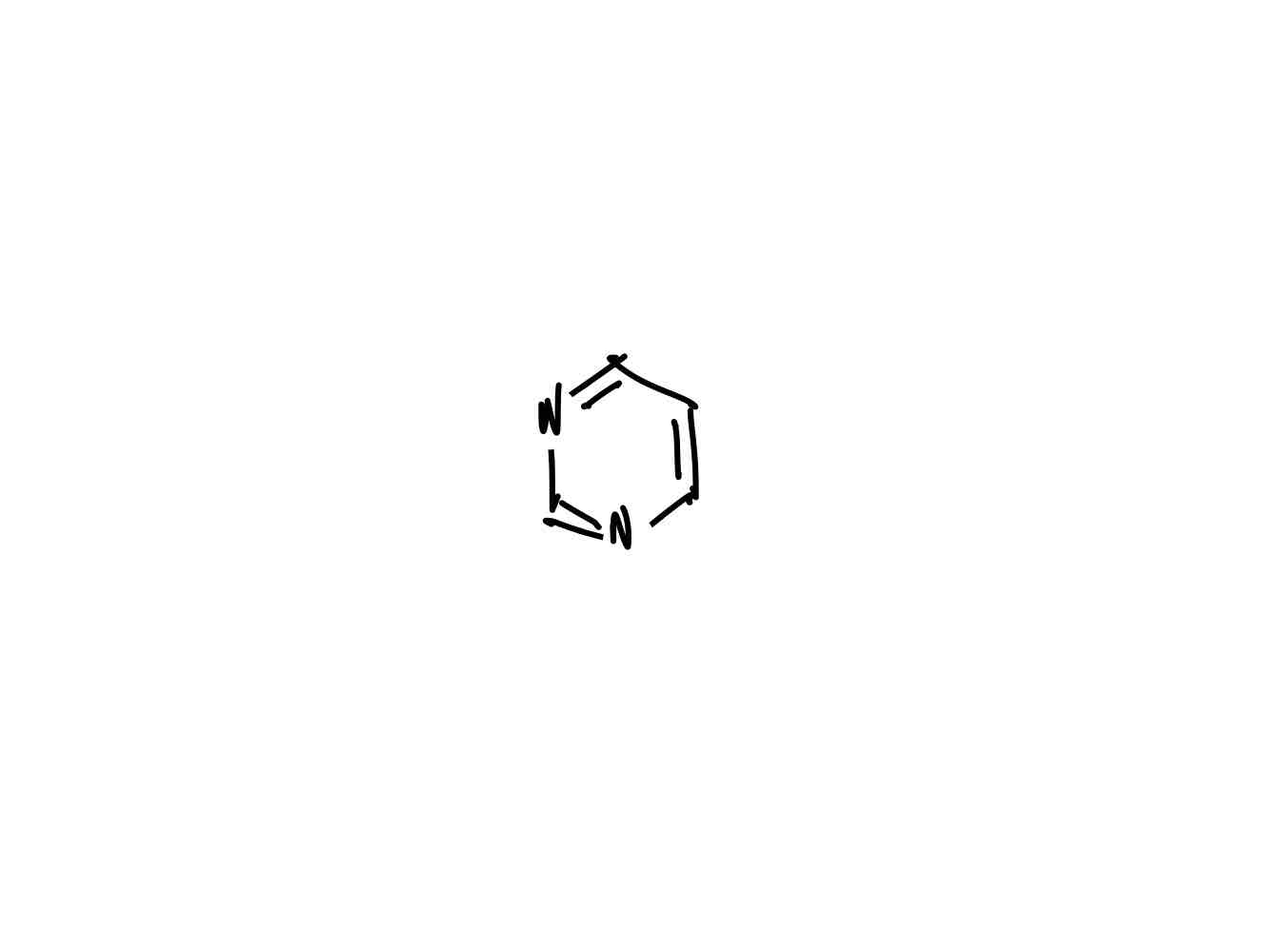
The pyrimidine bases (cytosine, thymine, and uracil) are called pyrimidine bases because their parent compounds are pyrimidines.
What is the name of the RNA nucleoside for a pentose with:
An adenine base.
A guanine base.
A cytosine base.
A uracil base.
For each of these nucleosides, once it has been monophosphorylated, what is the name of the RNA nucleotide? Provide each’s symbols.
The names are as follows:
Adenosine.
Guanosine.
Cytidine.
Uridine.
The names are as follows:
Adenosine 5’-monophosphate or adenylate (A, AMP)
Guanosine 5’-monophosphate or guanylate (G, GMP).
Cytidine 5’-monophosphate or cytidylate (C, CMP).
Uridine 5’-monophosphate or uridylate (U, UMP).
What is the name of the DNA nucleoside for a pentose with:
An adenine base.
A guanine base.
A cytosine base.
A thymine base.
For each of these nucleosides, once it has been monophosphorylated at the 5’ position, what is the name of the DNA nucleotide? Provide each’s symbols.
The names are as follows:
Deoxyadenosine.
Deoxyguanosine.
Deoxycytidine.
Deoxythymidine.
The names are as follows:
Deoxyadenosine 5’-monophosphate or deoxyadenylate (A, dA, dAMP)
Deoxyguanosine 5’-monophosphate or deoxyguanylate (G, dG, dGMP).
Deoxycytidine 5’-monophosphate or Deoxycytidylate (C, dC, dCMP).
Deoxythymidine 5’-monophosphate or deoxythymidylate (T, dT, dTMP).
On the pentose sugar, where does the purine and pyrimidine base attach to, in what conformation, and from which atom number on the base?
> Bases linked to C1 of sugar.
> Via B-linkage.
> w/ N9 of purine or N1 of pyrimidine.
How do you name a nucleotide with a phosphate that makes its own little ring with the pentose sugar? Take the RNA nucleoside adenosine for example with a phosphate group attached to 2’ and 3’.
Adenosine 2’,3’-cyclic monophosphate.
At this point in biochemistry, what should you know about any available OH group in regards to reactivity and enzymology?
OH → dehydration requiring enzymes + enzyme specificity.
What is a pyrophosphate (PPi)? If attached to a nucleoside, what does it give?
What is a triphosphate? If attached to a nucleoside, what does it give?
How much energy is stored in each phosphate bond P-O?
Two phosphate groups bonded together can give a nucleoside diphosphate.
Three phosphate groups bonded together can give a nucleoside triphosphate.
The phosphate bond has 30.5kJ/mol of energy.
Nucleic acids are made of nucleotides. However, what holds each nucleotide together and in what way?
How are nucleotide sequences usually written? 3’ to 5’ or 5’ to 3’?
What is our backbone and side groups on nucleic acids?
What part of our nucleic acid is hydrophilic and how is it hydrophilic?
What part of our nucleic acid is ionic and how is it ionic?
What part of our nucleic acid is hydrophobic and how is it hydrophobic?
• Nucleotides are held together by phosphodiester linkages from the 3’ of one nucleotide to the 5’ of another.
• Nucleotide sequences are always written 5’ to 3’.
• Backbone = alternating phosphates and nucleotides while our side groups would be our bases.
• Hydrophilic part = OH + PO43− of sugar residues, as they form H-bonds w/ water.
• pKa of PO43− = 0 → negatively charged at pH of 7.
• Hydrophobic region = bases, due to the aromatic ring's non-polar and hydrophobic character.
Bases pair up with each other in a base-pair stacking fashion via vdW and dipole:dipole forces.
Are the two coiling strands of DNA parallel or antiparallel? Do they form left- or right- handed double helices?
What are Chargaff’s rules?
What are the base pairs in DNA? How about in RNA?
Antiparallel + right-handed.
#dA = #dT and #dC = #dG.
In DNA, A:T and G:C. In RNA, A:U and G:C.
How far are two base-pair stacks apart in Å?
How long is one complete turn of a DNA helix in Å? How many base pairs is in one complete turn?
3.4A (0.34nm)
36A (3.6nm) in 10.5 base pairs
Through what molecular forces do bases pair and thus keeps the double helix held together?
H-bonding btwn bps.
What are the two key stabilizing features of the double helix?
(1) Metal cations shielding (-) charges of PO43−.
(2) Base-stacking interactions of successive GC pairs.
• Why are GC pairs stronger than AT pairs?
• What happens @ the major + minor grooves of DNA?
• GC = 3×H-bonds ; AT = 2×H-bonds.
• Proteins bind to specific nt sequences in these grooves → gene expression regulation.
> As opposed to proteins, how many how many flexible bonds in NAs can provide new conformations of DNA?
> What do these conformations significantly allow?
> 7 flexible bonds in one nt can rotate.
> Allows diff DNA conformations for 2º + 3º structure.
In order to have a double helix, you’d have to twist your primary structure (linear), and, in order to twist it, you’d have to rotate said bonds.
Is DNA static? Why or why not?
> DNA = random, dynamic, localized separation of bps and b-stacks and rapid reforming of bps and b-stacks.
> Not static.
> What is DNA melting? What change in parameters are adjusted to achieve DNA melting?
> Are covalent bonds broken in DNA melting?
> Two strands separated into single strands, done by extreme pH or high temperatures disrupting bps + bp stacking.
> No covalent bonds in DNA are broken.
> In partial denaturation, when temp or pH is returned to the optimal range, does the rewinding of unwound segments go through one or two steps? What is the step(s) called?
In partial denaturation, one step occurs.
> Remaining unpaired bases successively come into register as bps = annealing → duplex is zipped up.
> In complete denaturation, when temp or pH is returned to the optimal range, does the rewinding of unwound segments go through one or two steps? What is the step(s) called?
In complete denaturation, two steps occur.
> (1) Two strands finding each other by random collisions to form a short segment of a complementary duplex.
> (2) Annealing.
How does one measure DNA denaturation? Explain how it works in detail.
> mono-nts absorb UV light at 260nm due to the presence of aromatic conjugation.
> bp stacks do not absorb as much UV light bec of how the light interacts w/ e-’s in this fashion = hypochromic effect.
> DNA melting → denatures dsDNA into ssDNA → unpaired nts mimic fully free nts.
> UV absorbed @ 260nm → lots of denaturation.
> Little UV absorption @ 260nm → don’t have lots of denaturation.
• What is the melting point in DNA denaturation? (tm)?
• What is the relationship and reasoning behind the relationship between C:G pairs and denaturation?
• What can you estimate with denaturation temp?
• Temp @ 50% ssDNA and 50% dsDNA.
• ↑ %G:C ↑ tm = due to 3×H-bonds and stronger stacking interactions.
• The mp of any DNA molecule ≈ %G:C composition.
What are bubbles?
> Denaturation conditions controlled → AT-rich region will denature while most of DNA remains dsDNA.
> Denatured regions = bubbles.
Explain how DNA replication works step-by-step.
> Helicase unwinds.
> ssbs stabilize unwound parent strands.
> Primase synthesizes RNA primers on each strand.
> DNA synthesis occurs.
> On the leading strand, DNA pol III synthesizes the leading strand continuously in 5' to 3'.
> On the lagging strand, DNA pol III synthesizes from 3’ end of RNA primer to the 5’ end of the next RNA primer.
> DNA pol I removes primers on both leading + lagging strands and replaces with DNA nts.
> DNA ligase seals the gaps between Okazaki fragments.
What is the structure of purine?
What is the structure of pyrimidine?
What is the structure of adenine?
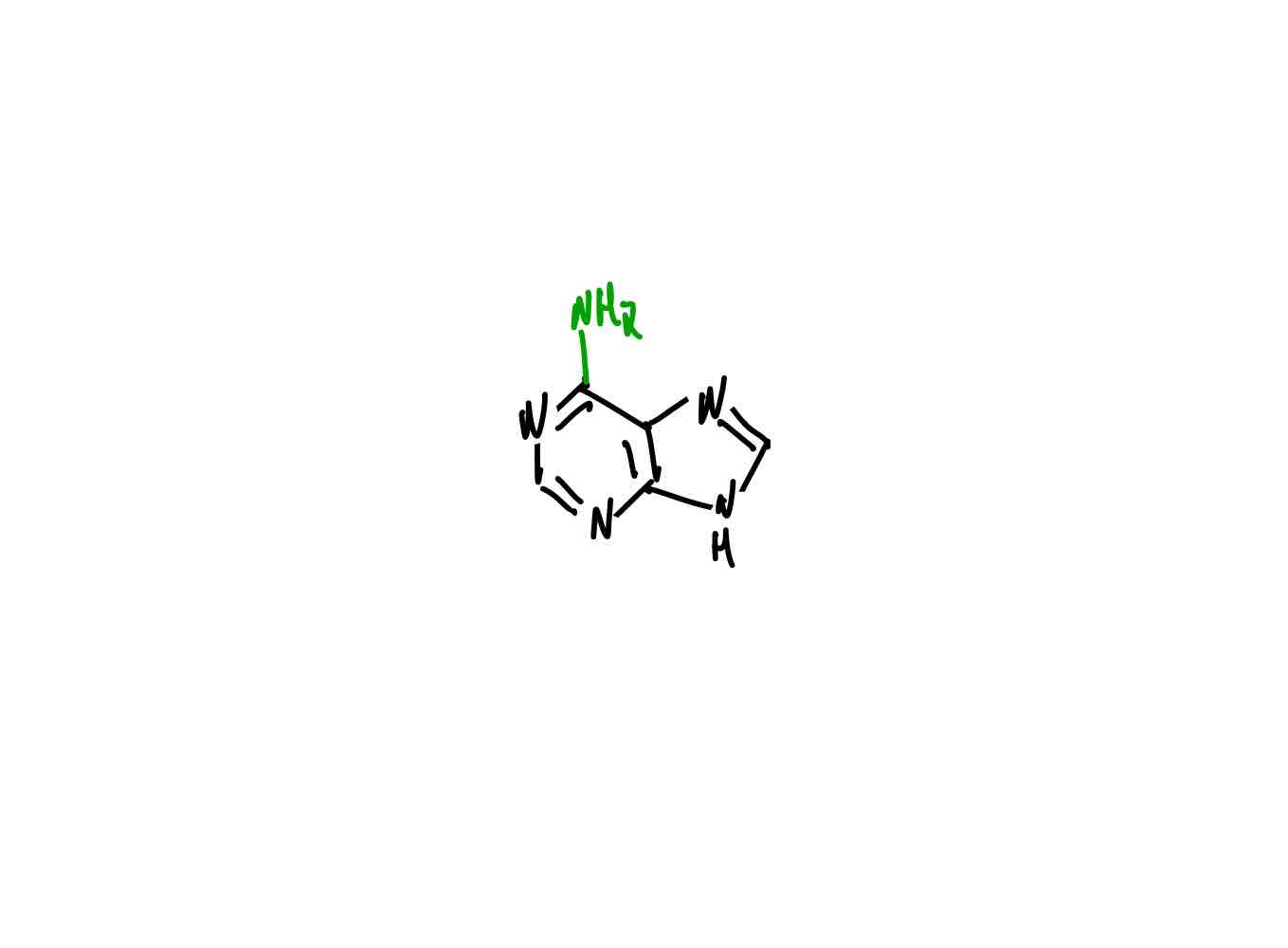
What is the structure of guanine?
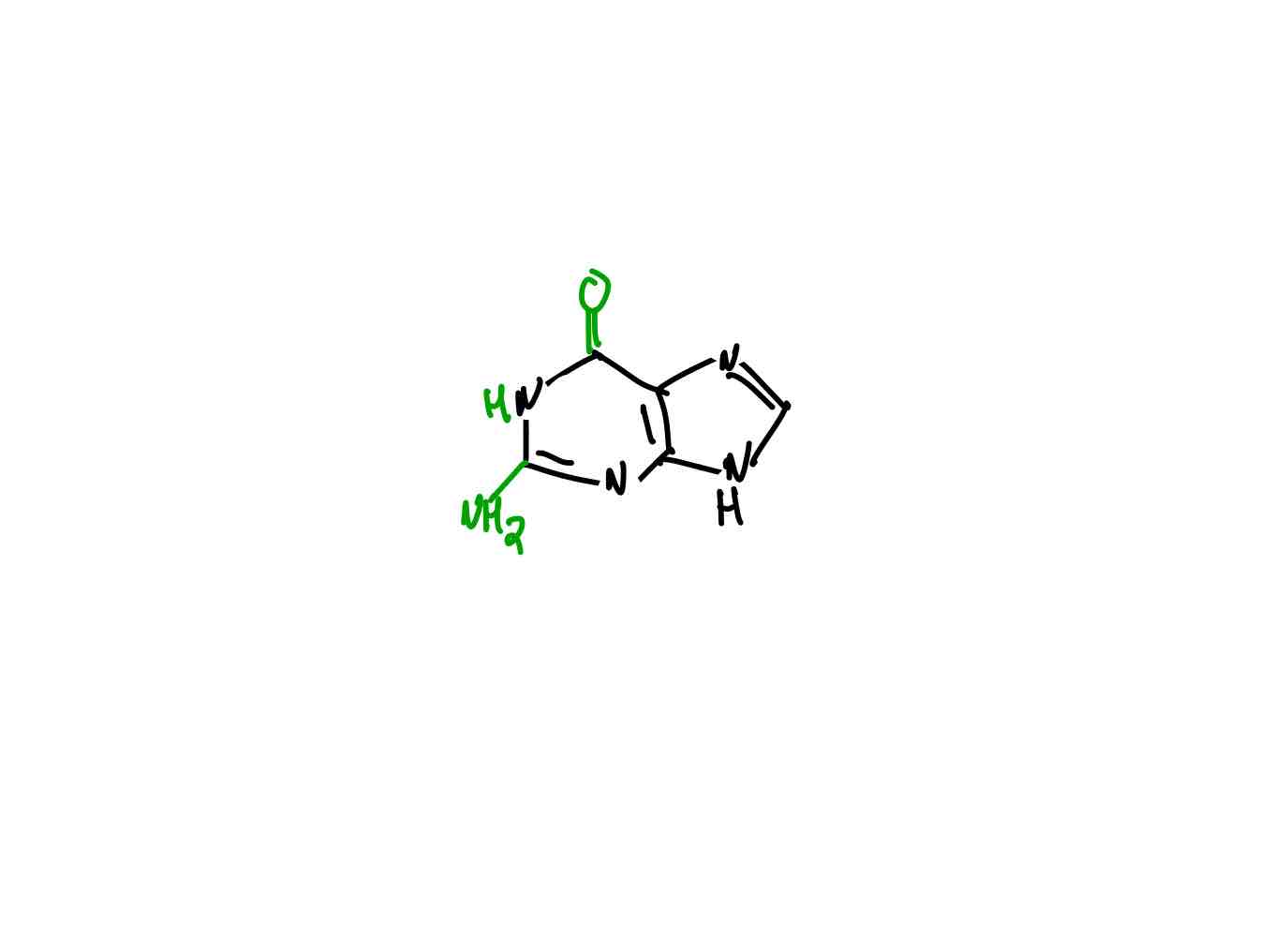
What is the structure of cytosine?
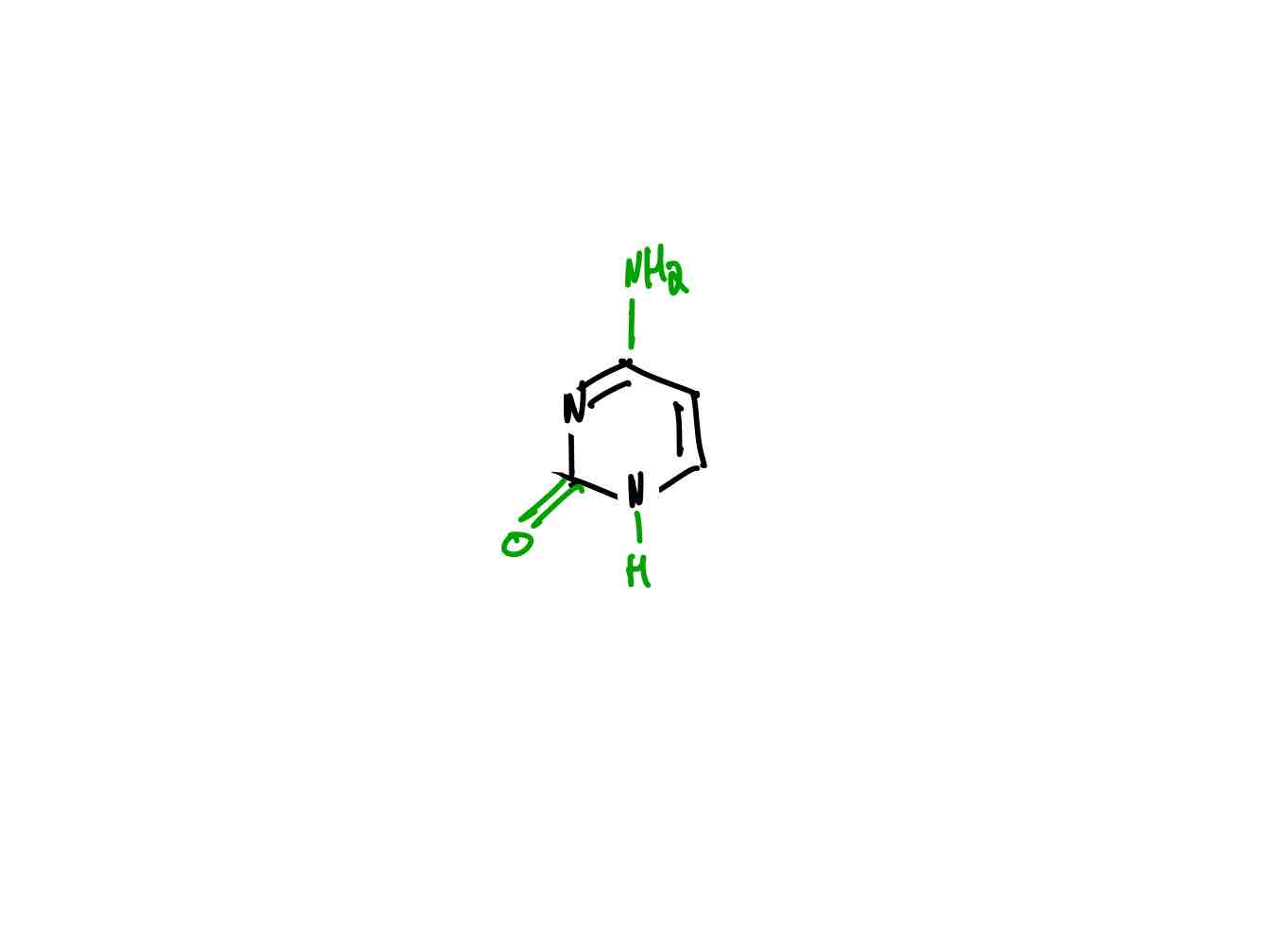
What is the structure of thymine?
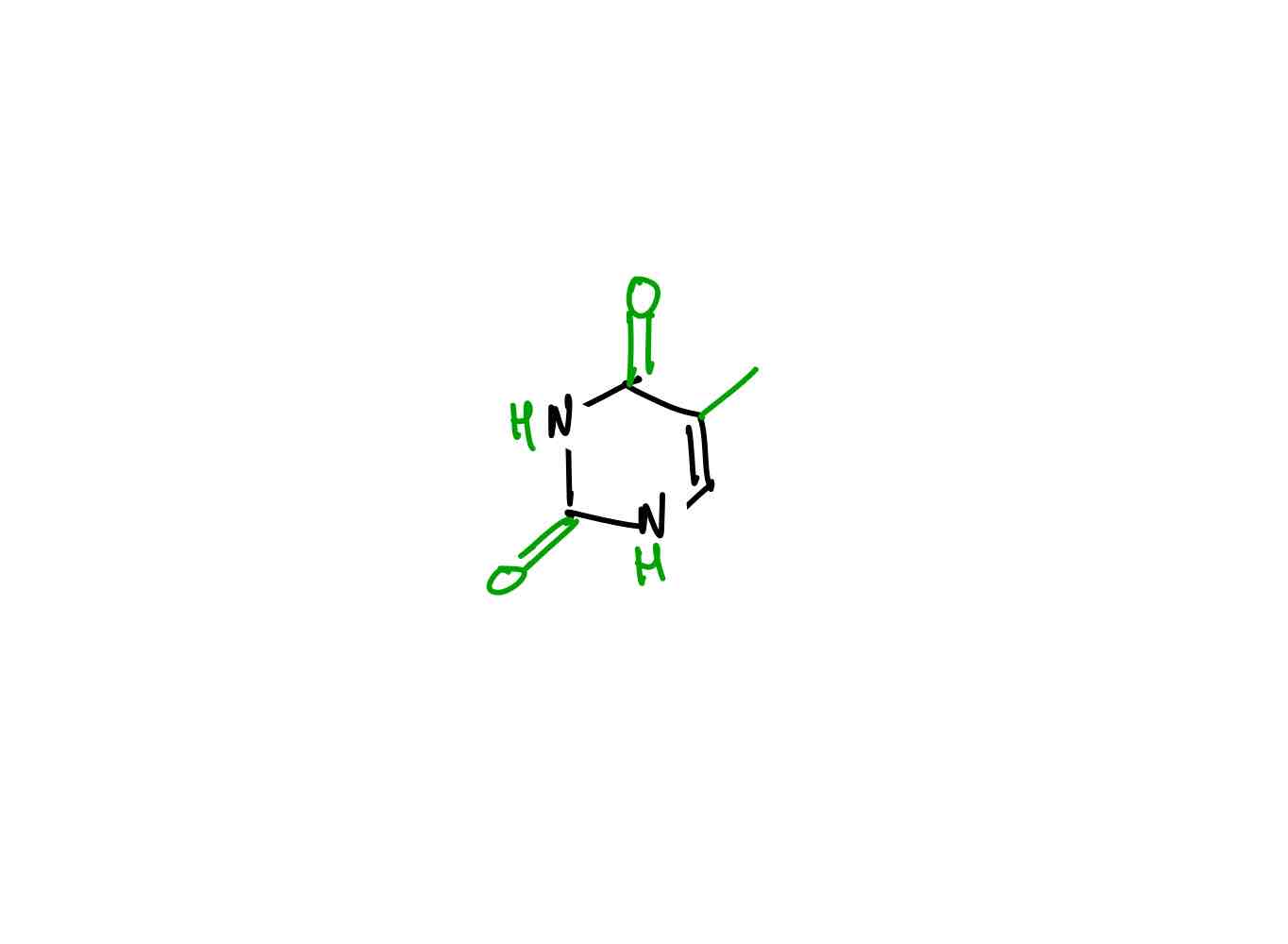
What is the structure of uracil?