DNA replication and Cell cycle
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
22 Terms
Who first discovered the cell
Robert Hooke 1665
Schwann 1839: “ the cell is the unit of structure, physiology and organisation in living things”
Who discovered that all cells come from existing cells?
1855- Rudolph Virchow
What did Fleming identify
1882- First descriptions of mitosis, identifying stages and interphase vs mitosis
What did the cell cycle look like until 1950’s

1950s radioactivity experiments
1950’s - used radioactive phosphorus
found that while the quantity of RNA and proteins increased steadily during the cell cycle, DNA synthesis only happened at one point during interphase
Divided interphase into G1(46 single chromosomes), S (each chromosome is 2 chromatids) and G2
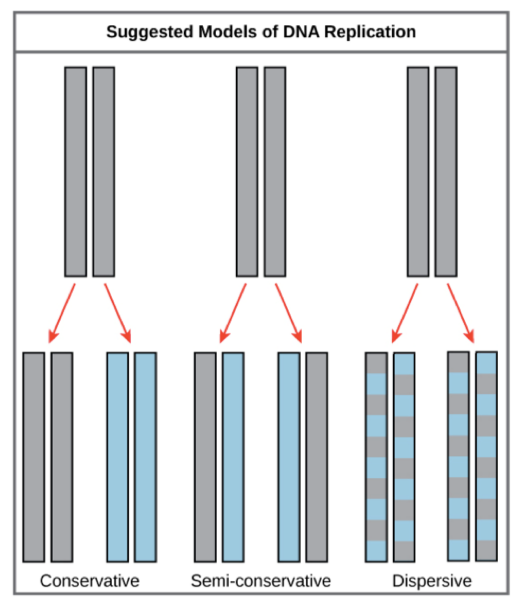
What 3 models were proposed for replication
Conservative
Semi-conservative
Dispersive
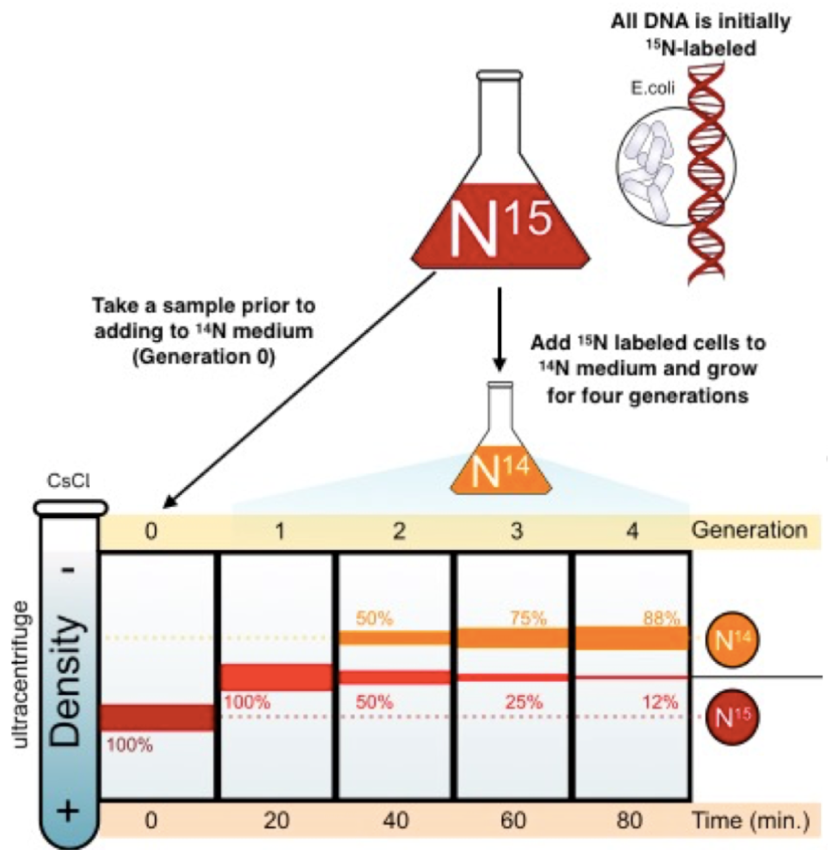
Meselson and Stahl procedure
used E.coli bacteria to model, grew it in a medium containing heavy isotope N15
E.coli took this up and used it to synthesise DNA
Repeated until all the E.coli contains N15
THEN
bacteria switched to a medium containing N14
DNA made after the switch would’ve only had N14 available for DNA synthesis
Took samples after each new generation
Meselson and Stahl results
used a density gradient centrifuge
Separates DNA into bands by spinning at high speeds
Saw that DNA replicated semi-conservatively
Each DNA strand serves as a template for a new one
1 gen = 100%
2nd gen = 50:50
3rd = 75 N14 & 25 N15
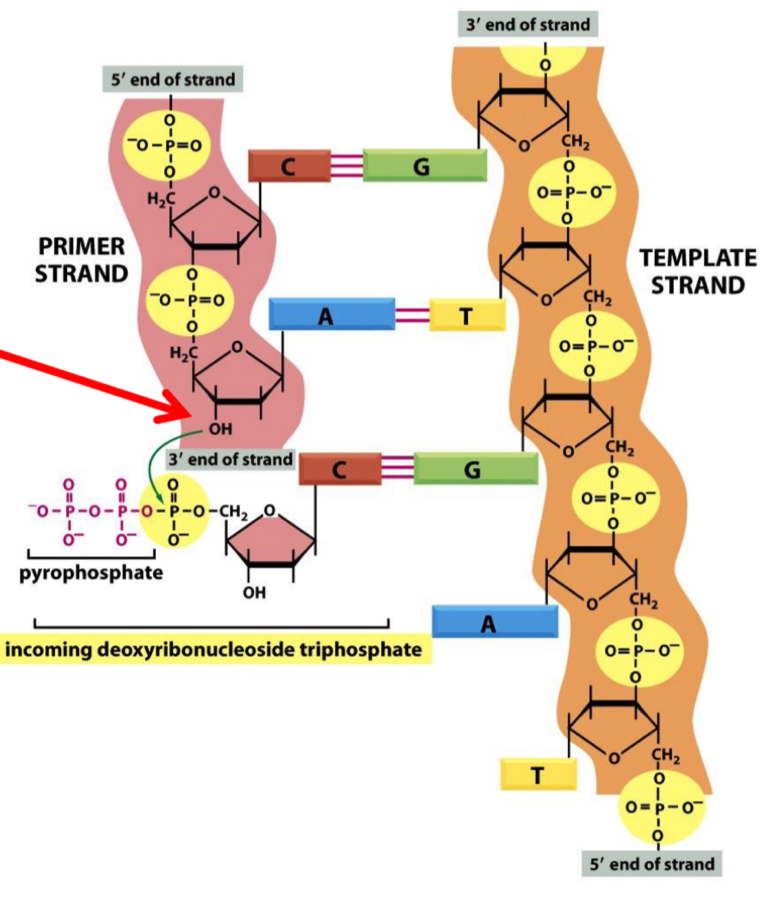
DNA synthesis requirements
DNA polymerase + Mg2+
DNTPs (building blocks and energy)
Template DNA
Primer 3’-OH
DNA synthesis = 5’→3’
What are dNTPs
Molecular building blocks (A, T, C, G) used in DNA synthesis
base
Deoxyribose sugar
3 phosphates
Breakage of dNTP phosphodiester bond provides energy for polymerisation
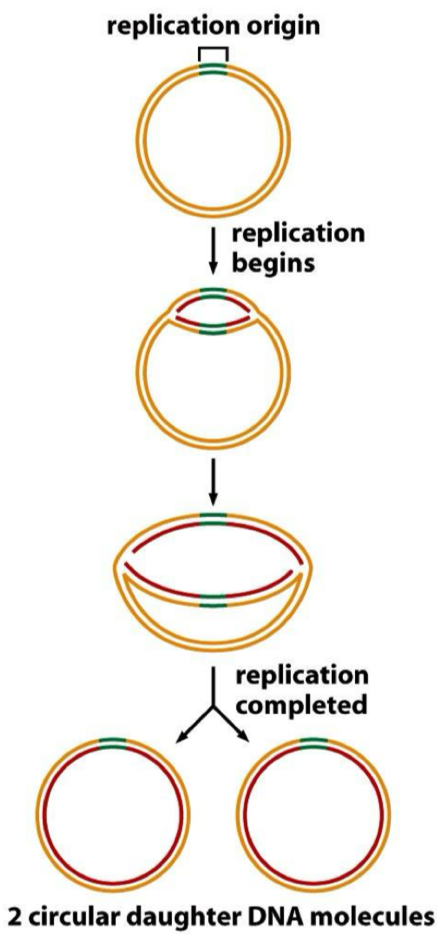
Initiation: prokaryotes
bacteria have circular DNA & plasmids
Each chromosome or plasmid has an ‘origin of replication’- a special sequence where it begins
Bidirectional replication
Theta structure
Makes 2 circular DNA molecules
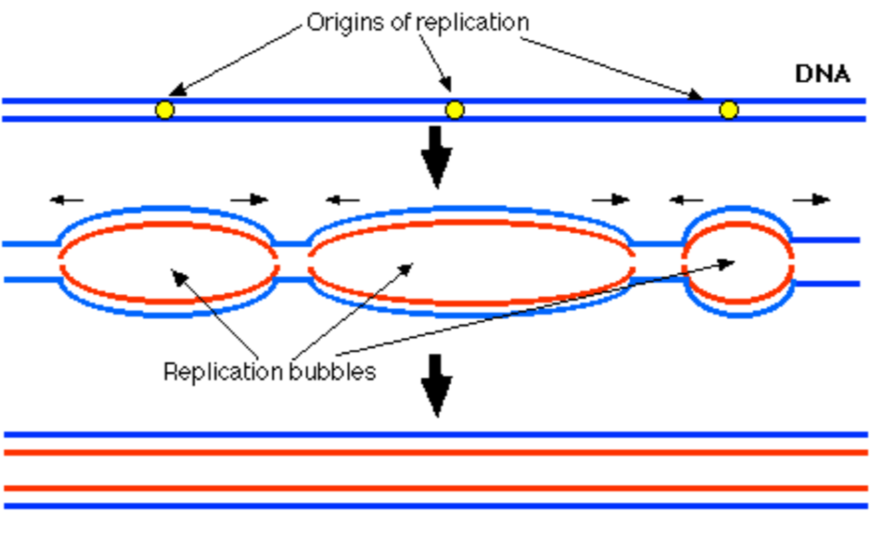
Initiation: eukaryotes
chromosomes are much longer
Each will have many origin
Bidirectional replication creates ‘bubbles’ of replications
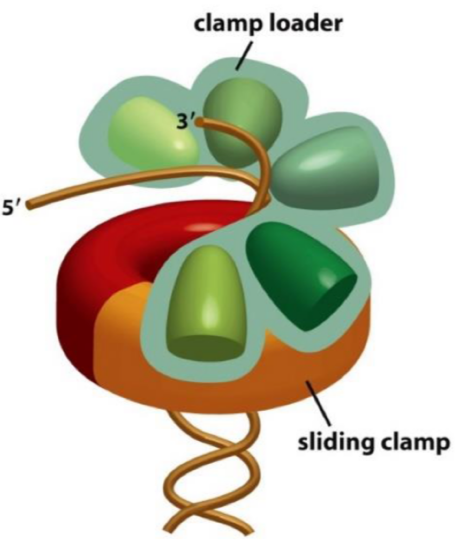
Elongation: clamp loader
challenge = loading and holding onto DNA
A ‘clamp loader’ loads the sliding clamp onto DNA
Elongation: twisting
helicase unwinds double stranded DNA using ATP
This causes supercooling (twists) which need to be unwound
Topoisomerases unwind the twists
They break and reform the phosphodiester bonds to untwist the DNA
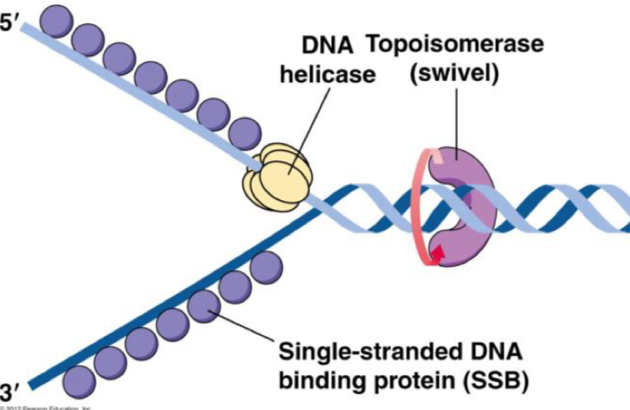
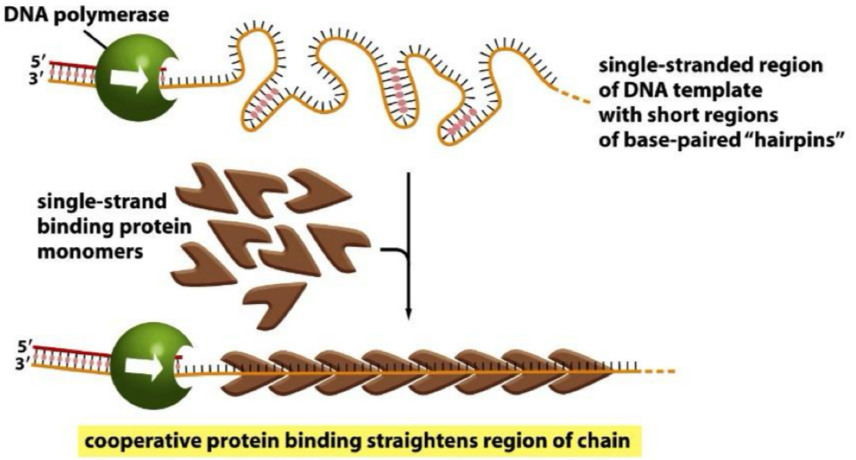
Elongation: single stranded binding protein
single stranded DNA forms H bonds with itself
Single-stranded binding protein prevents this
Monomers bind but bases are left exposed
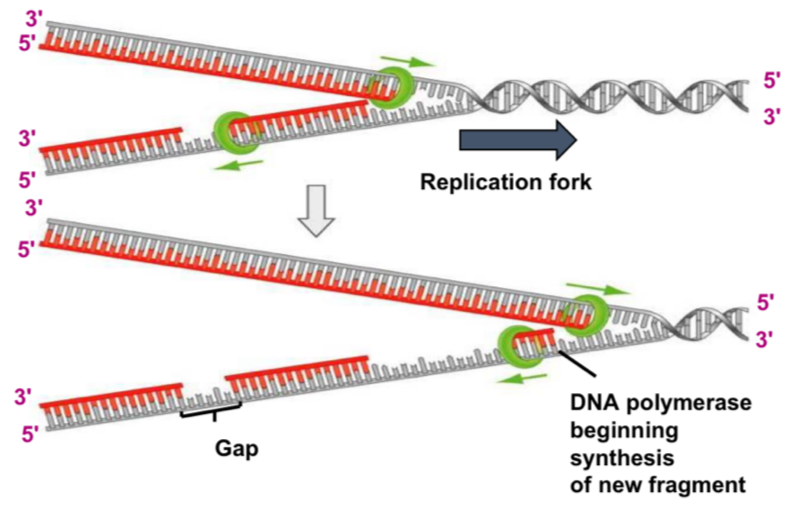
Leading vs lagging synthesis
replication can only happen from 5’ to 3’
Laggin strand synthesis = discontinuous
Primase adds lots of RNA primers(oligonucleotides) as it unwinds, and DNA polymerase synthesises sections= Okazaki fragments
DNA ligand will then join the fragments together
= semi-discontinuous
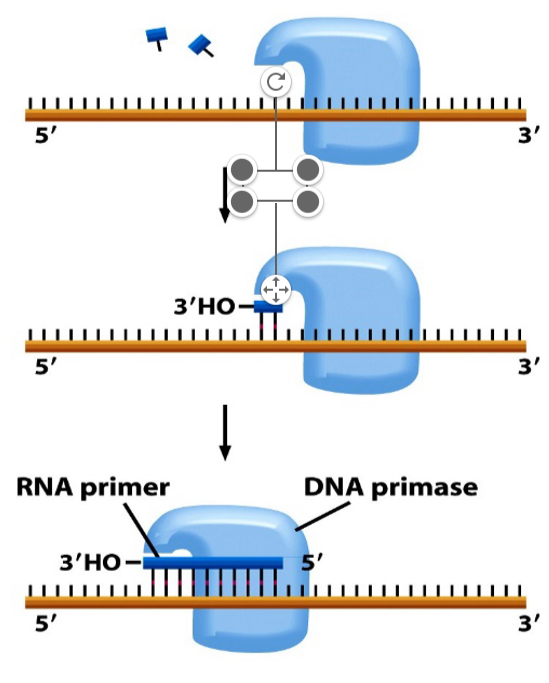
Elongation: primase
Challenge on lagging stand is that DNA polymerase requires an OH overhang.
Primase synthesises short RNA primers about 10 nucleotides long with the OH overhangs
Elongation: DNA hloenzyme
•DNA polymerase holoenzyme is an asymmetric dimer that catalyses both leading and lagging strand synthesis.
•It synthesises 1000 bp/second
•100 turns of the helix each second.
Double stranded breaks
Chromosomes damages with double-stranded breaks are repaired by:
End joining→ joins two ends making a small deletion
Homologous recombination→ copies a similar sequence to repair DNA
A double stranded break at the end of a chromosome would result in them being joined together = cell would likely die
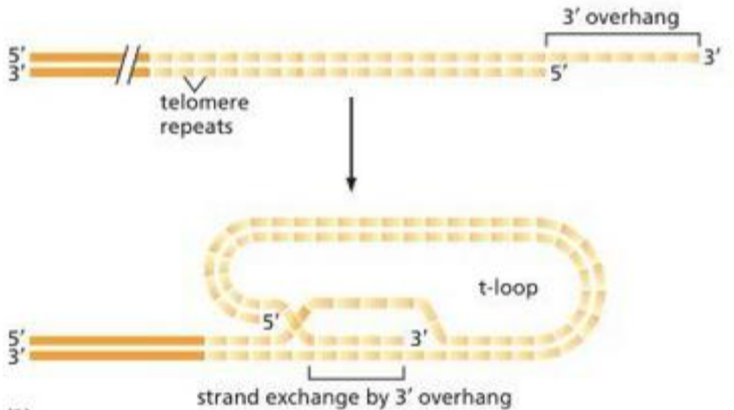
Telomere structure
3’ overhang of 100s of the same short DNA sequence = 5’TTAGGG-3’ in humans
T-loop (overhang tucks in/hides) protects the ends from the DNA repair pathways for double stranded breaks
Shelterin- a protective protein protects the ingle stranded DNA against other repair mechanisms
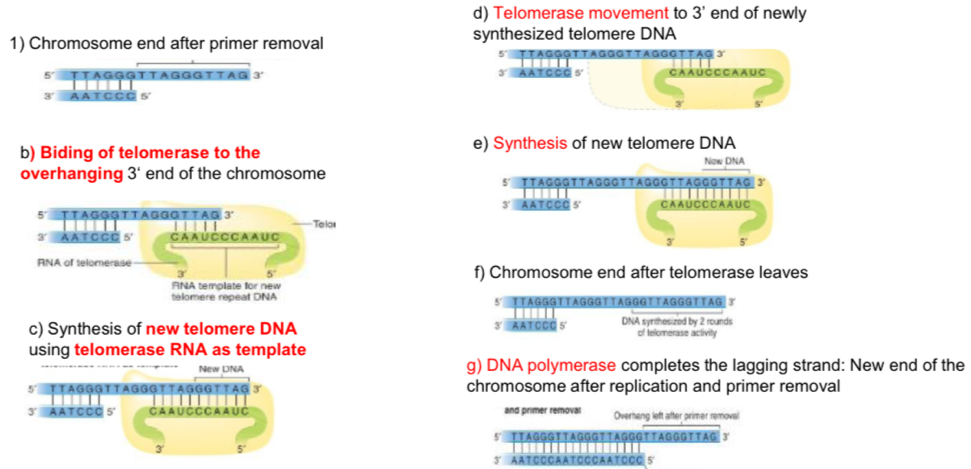
Telomere replication
Binding of telomerase to the overhang 3’
Synthesis of new telomere DNA using telomerase RNA as template
Telomerase movement to 3’ end of Newley synthesised telomere DNA
Synthesis of new telomere DNA
Telomerase laves and chromosome ends
DNA polymerase completes the lagging strand
Maintaining DNA integrity
DNA polymerase can also ‘proofread’
if an incorrect base is added, DNA only erase can remove it
Polymerisation continues
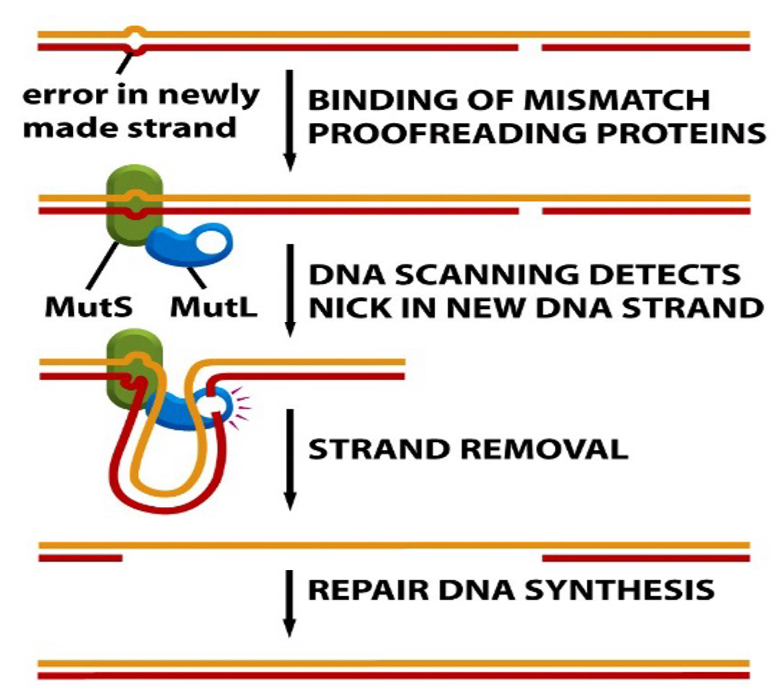
Mismatch repair
if an incorrect base is added, mismatch repair detects a bump in the helix
Proteins repair mismatches in DNA involving MutS and MutL proteins