Proteins in Cells
1/171
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
172 Terms
Proteins
Proteins are linear heteropolymers of ⍺-amino acids.
What size defines a protein and peptide?
More than 10 kilodaltons (unit of weight (kDa)) means that it is a protein and less than this means that it is a peptide.
What are the 4 main functions of proteins?
Catalysis, transport, structure, and motion
Catalysis Protein Examples
Catalysis is the process of increasing the rate of a chemical reaction by adding a catalyst.
Ex. enzymes such as enolase and DNA polymerase
Transport Protein Examples
hemoglobin (transports O2 in the blood)
lactose permease (transports lactose across the cell membrane)
Structure Protein Examples
collagen (connective tissue)
keratin (hair, nails, feathers, horns)
Motion Protein Examples
myosin (muscle tissue)
actin (muscle tissue, cell motility)
Native Fold
The specific 3D conformation of a protein which has a cost in conformational entropy of folding the protein.
Protein Stages
Primary structure (amino acid residues) → secondary structure (alpha helix) → tertiary structure (polypeptide chain) —> quaternary structure (assembled subunits)
Primary Structure Overview
The linear sequence of amino acids in a polypeptide chain. It determines all higher levels of structure; even a single amino acid change can drastically affect function.
What order is a peptide chain read in?
Starts with the N-terminus (or amino-terminus) and ends with the C-terminus (or carboxyl-terminus).
Basic Properties of Amino Acids
Capacity to polymerize
Useful acid-base properties
Varied physical properties
Varied chemical functionality
Which groups of amino acids are hydrophobic and hydrophilic?
Hydrophobic includes nonpolar, aliphatic and aromatic.
Hydrophilic includes polar/uncharged, positively charged, and negatively charged.
L or D Amino Acids
Amino acids are either L- or D- amino acids which depend on their configuration. These are stereoisomers which means that they are mirror images of each other.
Proteins only contain L-amino acids and glycine
Peptide Bond Formation
The Carbonyl of one amino acid reacts with the Amino group of another amino acid to form an amide bond (peptide bond) which is one of the most stable bonds that exists in nature. It also releases 1 water molecule.
What does the backbone of amino acids contain?
An amino group (-NH2), a carboxyl group (-COOH), and an alpha-carbon (Cα) atom.
What is the pI of a molecule?
What amino acids have nonpolar, apliphatic R groups?
Glycine, alanine, proline, valine, leucine, isoleucine, and methionine.
Glycine and proline break any amino acid and methionine starts every protein
What amino acids have aromatic R groups?
Phenylalanine, tyrosine, and tryptophan.
These groups absorb UV light at 270-280 nm and stack well with DNA and RNA bases
Tryptophan is the largest amino acid
What amino acids have polar, uncharged R groups?
Serine, threonine, cysteine, asparagine, and glutamine.
These R groups can form hydrogen bonds
Cysteine can form disulfide bonds which are covalent bonds which help to stabilize protein structure.
What amino acids have positively charged R groups?
Lysine, arginine, and histidine.
What amino acids have negatively charged R groups?
Aspartate and glutamate.
What makes histidine special?
Histidine can have a hydrogen atom added to the N at the bottom which allows the double bond to switch between the two right sides of the ring. This causes a delocalisation of charge. This property allows proteins that contain histidine to degrade other proteins.
Uncommon Amino Acids
There are other uncommon amino acids that are not incorporated by ribosomes (aside from selenocysteine and pyrrolysine)
They arise by post-translational modifications of proteins
Reversible modifications, especially phosphorylation, are important in regulation and signaling
Resonance in the Peptide Bond
The carbonyl oxygen has a partial negative charge and the amide nitrogen a partial positive charge, setting up a small electric dipole.
What does the resonance of the peptide bond cause?
The bond:
to be less reactive compared to esters, for example
to be quite rigid and nearly planar
to exhibit a large dipole moment in the favored trans configuration
Rotation Around the Alpha-Carbon
A polypeptide is made up of a series of planes linked at alpha-carbons. Rotation around the peptide bond is not permitted but rotation around bonds connected to the alpha carbon is permitted.
Phi and Psi Angles
𝟇 (phi): angle around the a-carbon—amide nitrogen bond
𝟁 (psi): angle around the a-carbon—carbonyl carbon bond
Alpha helix and beta sheet phi and psi angles
Alpha helix has both negative angles and beta sheet has a positive phi and negative psi angles.
Favourable and Unfavourable Phi and Psi Angles
Some 𝟇 and 𝟁 combinations are very unfavorable because of steric crowding of backbone atoms with other atoms in the backbone or side chains
Some 𝟇 and 𝟁 combinations are more favorable because of chance to form favorable H-bonding interactions along the backbone
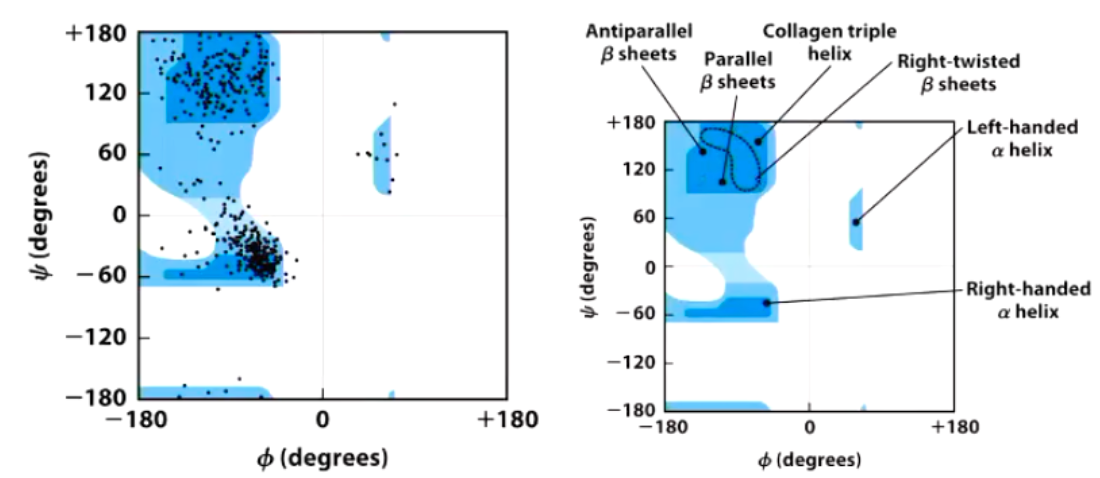
Ramachandran Plot
Shows the distribution of 𝟇 and 𝟁 dihedral angles that are found in a protein and therefore the common secondary structure elements.
Secondary Structure Overview
Secondary structure refers to a local spatial arrangement of the polypeptide backbone. They form due to hydrogen bonds between backbone atoms (not side chains).
Two regular arrangements of secondary structures are…
The ⍺ helix - stabilized by hydrogen bonds between nearby residues
The β sheet - stabilized by hydrogen bonds between adjacent segments that may not be nearby
What is the random coil?
Irregular arrangement of the polypeptide chain
Alpha Helix Overview
Helical backbone is held together by hydrogen bonds between the backbone amides of an n and n+4 amino acids
Right-handed helix with 3.6 residues (5.4 Å) per turn
Peptide bonds are aligned roughly parallel with the helical axis
Side chains point out and are roughly perpendicular with the helical axis
Alpha Helix Dipole
There is a positive end at the top due to the first 3 amino groups that are not involved in hydrogen bonding. This is the same at the bottom but it’s negative instead and overall this forms a dipole.
The positively charged amino acids are normally found at the start of amino acid chains and vice versa to enhance this
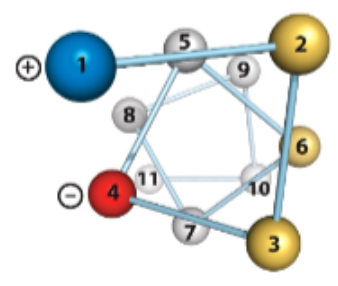
Top View of the Alpha Helix
The inner diameter of the helix (no side chains) is about 4–5 Å (too small for anything to fit “inside”)
The outer diameter of the helix (with side chains) is 10–12 Å
Happens to fit well into the major groove of dsDNA
Residues 1 and 8 align nicely on top of each other
Amino Acids and the Alpha Helix
Small hydrophobic residues such as Ala and Leu are strong helix formers
Pro acts as a helix breaker because the rotation around the N-Ca bond is impossible
Gly acts as a helix breaker because the tiny R- group supports other conformations
Attractive or repulsive interactions between side chains 3–4 amino acids apart will affect formation
Beta Sheets
Parallel and Antiparallel Beta Sheets
In parallel b - sheets the H-bonded strands run in the same direction resulting in bent H-bonds (weaker) (> 5 residues)
In antiparallel b - sheets the H-bonded strands run in opposite directions r esulting in linear H-bonds (stronger)
Beta Turns
The 180° turn is accomplished over four amino acids
The turn is stabilized by a hydrogen bond from a carbonyl oxygen to amide proton three residues down the sequence
Proline in position 2 or glycine in position 3 are common in b turns
Type I is rigid and type II is more flexible
Type I and II Beta Turns
Type I beta turns are the most common and are stabilized by a hydrogen bond between residues i and i+3.
Type II beta turns are characterized by a glycine residue in the third position (i+2)
Circular Dichroism (CD) Analysis
Measures the differential absorption of left and right circularly polarized light to determine protein folding, unfolding, and interactions with other molecules as well as the presence of different secondary structures.
Tertiary Structure
Refers to the overall spatial arrangement of atoms in a single polypeptide chain or protein. Stabilized by numerous weak interactions between amino acid side chains. Determines the protein’s functionality, especially the active or binding sites. Classified as fibrous or globular (water or lipid soluble).
Favourable Interactions in Proteins
Hydrophobic effect, hydrogen bonds, london dispersion, electrostatic interactions
Hydrophobic Effect
Release of water molecules from the structured solvation layer around the molecule as protein folds increases the net entropy.
Hydrogen Bonds in Proteins
Interaction of N-H and C=O of the peptide bond leads to local regular structures such as a-helices and b-sheets
London Dispersion
Medium-range weak attraction between all atoms contributes significantly to the stability in the interior of the protein.
Electrostatic Interactions
Long-range strong interactions between permanently charged groups
Salt-bridges, esp. buried in the hydrophobic environment strongly stabilize the protein
Quaternary Structure
Quaternary structure is formed by the assembly of 2 or more polypeptide chains into a larger functional cluster.
Found in some proteins: eg Haemoglobin (4 units)
Interactions include hydrogen bonding and disulfide bonds. Locks the complex into a specific geometry.
Quaternary Structure: Structural and Functional Advantages
Stability due to reduction of surface to volume ratio
Genetic economy and efficiency
Bringing catalytic sites together
Cooperativity
Thermodynamics of Protein Folding
Proteins fold into secondary and tertiary structures with the lowest possible free energy.
A protein’s internal residues (side chains) direct its folding into native conformation, according to the surrounding environment (in aqueous solution) - Hydrophobic Effect
Aliphatic side chains interact with each other releasing water.
Decrease in entropy of protein and increase in entropy of water.
What is Levinthal’s Paradox?
Random attempts of every protein conformation for folding are mathematically impossible until the lowest-energy one is found.
The search for the fold with minimum energy is not random. The native structure is thermodynamically the most favorable
Folding Funnel Hypothesis
Nucleation can begin at multiple points, and these partially folded structures can be guided towards the final state by energy minimisation
i.e. they ‘fall’ down a funnel into their lowest energy state as their native structure
Measuring Protein Unfolding
When 50% of the protein is unfolded this is called the melting temperature (Tm)
Can be measured by heating the proteins and then determining the percent that is unfolded
Mutations in proteins can change the melting temperature - can be used to determine if the protein will be active or not/cause disease or not
Protein Folding and Disease
Cells use a lot of ATP to fold proteins correctly, preventing aggregation or removing misfolded ones.
In some cases, ordered aggregates in prion and amyloid fibers can cause severe diseases.
Prions
Misfolded proteins that can induce misfolding in normal proteins.
Accumulation of Prions
PrPsc acts as templates for transforming normal proteins into prions (Prion hypothesis).
They strongly associate and form insoluble protease-resistant aggregates called fibrils.
Deposits of these aggregates in neural cells cause neuronal loss, which can be infectious, genetic, or sporadic.
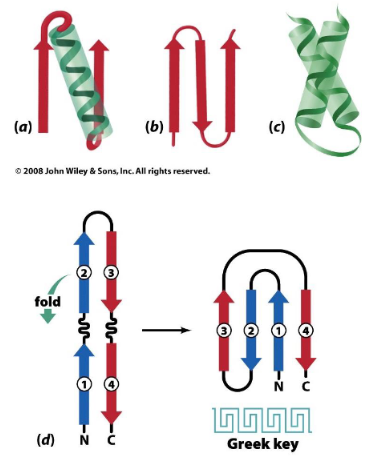
Motifs
A specific arrangement of secondary structure elements including:
β-⍺-β motif
β hairpin motif
⍺-⍺ motif
Greek key motif (β hairpin folded over to form a 4 stranded antiparallel sheet).
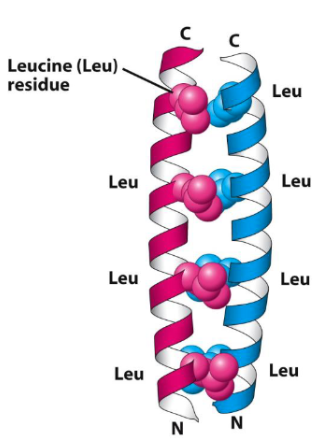
Coiled-Coil Motif
A bundle of right-handed alpha - helices forms a left-handed superhelix resembling a rope.
Each right-handed alpha-helix consists of 7- amino acid repeats with hydrophobic side chains (isoleucine, leucine or valine) on positions 1, 4 and 7
Pairs of helices intertwine in a left-handed coil, burying the hydrophobic residues and stabilising the structure.
Domains and Families
Domains are larger, independently folding units within a protein, often associated with a particular function, such as the SH3 domain involved in protein-protein interactions.
Domains become a family when they are seen in many other proteins are perform a very similar function
Burying Residues
It is worse to bury a charged residue than expose a hydrophobic one, as charged and polar residues are usually found on the surface in contact with water. However, they may be buried if H-bonding and charge balance are satisfied.
Fibrous Proteins
Separate themselves from aqueous solution and are elongated and fibrous in nature or have a sheet like structure.
Ex. collagen and elastin
Abundant in non-bulky amino acids (glycine, alanine, serine, glutamate, and glutamine). Exception: high proline in collagen and elastin.
Alpha Keratin
A fibrous protein that is mechanically durable and is chemically nonreactive. It has two main types: hard alpha keratin, found in shells, fingernails, and claws, and soft alpha keratin, found in skin, hair, and wool. Keratin fibres cross-link through disulphide bonds between cysteine residues on adjacent chains, forming an alpha-helical rod with non-helical N- and C-termini caps.
Silk Fibroin
Fibrous protein that is made of anti-parallel beta-sheets. Alternating sequence: Gly-Ala-Gly-Ala (Ser).... Gly on one sheet interacts with Gly on an adjacent sheet (same for Ala/Ser).
Spider Dragline Silk
Comprised of 2 proteins: fibroin which gives structure and seracin acts like glue or matrix
Overall composite material like carbon fibre – higher tensile strength than stee
Elastin Overview
Fibrous protein that makes up the main part of connective tissue in lungs and arteries, allowing them to regain their shape after stretching or contracting. It’s hydrophobic, insoluble, and forms a 3D elastic network made of loose, unstructured polypeptide chains.
Elastin Structure
This random coil or disorder conformation allows the protein to stretch and recoil. There are many possible random coil conformations. It can stretch in any direction, making it more elastic than rubber
Protein Sequencing
Actual sequence generally determined from DNA sequence using Edman Degradation (Classical method) or Mass Spectrometry (Modern method).
Edman Degradation
Successive rounds of N-terminal modification, cleavage, and amino acid identification
Can be used to identify protein with known sequence
Polypeptides > 40 to 100 cannot be directly sequenced and requires chemical or enzymatic cleavage into smaller fragments
Mass Spectrometry
MALDI MS and ESI MS can precisely identify the mass of a peptide and amino acid sequence
Can be used to determine post-translational modifications
Chemical Peptide Synthesis
The artificial synthesis of peptides using insoluble polystyrene beads
In a few days with an excellent yield of 96% per step a peptide containing 100 amino acids will have a final yield of 1.8% (from 100g starting you will get 1.8g).
In vivo using E.coli to biologically synthesise the same peptide will take 5 seconds with 100% yield
Proteostasis
The degradation of proteins keeps pace with synthesis.
Peptide Exiting from Ribosomes
It will start exiting at 18 amino acids in length and once it exits it can start folding. As it emerges from the ribosome, the signal sequence is bound by signal recognition particle (SRP).
ER Targeting Sequence
Found at the N-terminus and is a core of 6-12 hydrophobic amino acids, often preceded by one or more basic amino acids (Arg, Lys). The sequence is removed.
Mitochondrion Targeting Sequence
Found at the N-terminus and is an amphipathic helix, 20-50 residues inlength, with Arg and Lys resideues on one side and hydrophobic residues on the other. The sequence is removed.
Chloroplast Targeting Sequence
Found at the N-terminus and has no common motifs. However, it’s generally rich in Ser, Thr, and small hydrophobic residues and poor in Glu and Asp. The sequence is removed.
Peroxisome Targeting Sequence
PTS1 signal (SER-Lys-Leu) at extreme C-terminus (few proteins) or PTS2 signal at N-terminus (most proteins).
Nucleus Targeting Sequence
Multiple different kinds, a common motif includes a short segment rich in Lys and Arg residues.
Directing Proteins After Synthesis
Step 1 – Synthesis of proteins is completed on free ribosomes
Step 2 – Proteins with no targeting sequence remain in the cytosol.
Steps 3–6 – Proteins with an organelle-specific targeting sequence (pink) get imported into mitochondria, chloroplasts, peroxisomes, or the nucleus
Secretory Pathway
Step 1 – Ribosomes initiate protein synthesis in the cytosol.
Step 2 – ER signal sequence directs ribosome docking onto rough endoplasmic reticulum (ER). Nascent proteins translocated into the ER lumen or embedded in the ER membrane.
Step 3 – Proteins in ER membrane or lumen can move via transport vesicles to the Golgi complex.
Step 4a or 4b – Further sorting of proteins to the plasma membrane or to lysosomes in vesicles
Structure of Sec61α Complex (Translocon)
The channel is shaped like an hourglass with a narrow constriction
The pore is formed by a ring of isoleucine residues
When there is no translocation the pore is blocked by a helical plug.
When hydrophobic sequence containing an Anchoring Signal the purple helix in the translocon slides sideways to insert the peptide sequence into the membrane.
Collagen
A fibrous protein that is the most abundant protein in the body. It is a crucual component of many connective tissues and is made up of amino acids that arggane themselves in a unique triple helix structure.
Collagen Triple Superhelix
3 intertwined helical (1000 amino acids each) more extended than alpha helix
3.3 residues per turn – LEFT HANDED
Repeating triplet: -(Gly-Pro-(Pro/HyP))-
Procollagen cleaved to tropocollagen
4-Hydroxyproline in Collagen
The proline ring adopts into a favorable conformation.
Increases hydrogen bonding between collagen strands.
The post-translation modification is catalyzed by prolyl-hydroxylase, requiring α-ketoglutarate, molecular oxygen, and Ascorbate (Vitamin C – deficiency causes Scurvy).
Lysine Cross-Linking in Collagen
Collagen lacks cysteine, hence no disulfide cross-links.
These cross-links are formed by covalent bonds between modified side chains.
One type involves oxidized lysine side-chains, formed by lysyl oxidase
Sterol
Steroid nucleus: four fused rings (almost planar)
Hydroxyl group (polar head) in the A-ring
Cholesterol and sterols function in membranes
Modulate fluidity and permeability and thicken the membrane.
Packing of different types of FAs
Saturated fatty acids pack in an orderly way - extensive favorable interactions (high melting point)
Unsaturated cis fatty acids pack less orderly due to the kink - less-extensive favorable interactions (lower melting point)
Unsaturated trans fatty acids adopt an extended conformation - extensive favorable interactions (highest melting point)
How can the charge of membranes be modified?
By adding a modifer such as cardiolipin or phosphatidic acid to the X group of lipoproteins.
Phosphatidylcholine
The major component of most eukaryotic cell membranes
Sphingolipids Structure
The backbone of sphingolipids is a long-chain amino alcohol sphingosine
A fatty acid is joined to sphingosine via an amide linkage rather than an ester linkage as usually seen in lipids
A polar head group is connected to sphingosine at C1 by a glycosidic or phosphodiester linkage
Sphingolipis Location
In the outer face of plasma membranes because they are quite neutral in charge and are more stable than glycerophospholipids.
Difference between a vesicle and a micelle
A vesicle has two layers with an aqueous cavity in the centre whereas a micelle just has one layer of lipoproteins.
Gel or Fluid Phase of Membranes
Depending on their composition and the temperature lipid bilayers can be in
Gel phase: individual molecules do not move around, or
Fluid phase: individual molecules can move around
Whatdetermines membrane fluidity?
By the fatty acid composition.
Shorter and unsaturated fatty acids increase membrane fluidity.
Movement of Molecules in Membranes
Individual lipids undergo fast lateral diffusion (move side to side) within the leaflet
Spontaneous flips from one leaflet to another are rare