Cell Bio Making ATP
1/73
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
74 Terms
oxidative phosphorylation overview
- ETC transfers electrons through complexes I-IV
- proton pumping into intermembrane space
- ATP synthase uses gradient to make ATP
pathway integration of making ATP
glycolysis -> TCA -> ETC -> ATP
glycolysis
cytosolic ATP + NADH (Malate-Aspartate Shuttle)
TCA cycle (Krebs cycle) important components
NADH, FADH2, GTP
ETC purpose
oxidizes carriers, pumps protons
ATP synthase function
converts PMF into ATP
electron donors
- NADH donates at Complex I
- FADH2 donates at Complex II
- different entry points -> different ATP yields
glycolysis: ATP yield distribution from glucose in cells
2 ATP gained + 2 NADH
pyruvate oxidation: ATP yield distribution from glucose in cells
2 NADH
TCA cycle: ATP yield distribution from glucose in cells
- 6 NADH
- 2 FADH2
- 2 GTP/ATP
oxidative phosphorylation: ATP yield distribution from glucose in cells
majority of ATP
~26 ATP
theoretical yield from glucose: ATP yield distribution from glucose in cells
36
actual yield from glucose: ATP yield distribution from glucose in cells
~30-32
ETC overview: complexes I-V
- four main complexes in inner membrane
- transfer electrons from NADH/FADH2 to O2 at the end of chain
- proton pumping to IMS at Complexes I, III, IV
Peter Mitchell's Chemiosmotic Hypothesis
- proposed in 1961
- proton gradients drive ATP synthesis
- radical shift from chemical intermediates model (substrate level phosphorylation to generate ATP)
Racker and Stoeckenius experiment
- reconstituted vesicles with bacteriorhodopsin (proton pump) + ATP synthase
- light-driven proton pumping
- ATP synthesized without ETC
Complex I (NADH Dehydrogenase)
- entry point for NADH electrons
- donates 2 electrons to FMN reducing it to FMNH2
-> iron-sulfur clusters
- passes electrons down to ubiquinone (Q), reducing it to ubiquinol (QH2)
- pumps 4 protons into intermembrane space due to conformational changes as electrons move through the enzyme
Complex II (Succinate Dehydrogenase)
- dual role in TCA and ETC
- FAD accepts electrons from Succinate to generate FADH2
- ultimately transfers electrons to ubiquinone (Q) to generate ubiquinol (QH2)
-> iron-sulfur clusters/heme
- does not pump protons
ubiquinone (coenzyme Q10) shuttle
- lipid soluble electron carrier
-> 0.5-3% of the total lipid content of the IMM depending on cell type
- carries protons across/through the IMM
- shuttles 2 electrons as ubiquinol (QH2) between Complexes I/II to Complex III
Complex III: Q Cycle and Cytochrome C
- accepts electrons from QH2
- pumps 4 protons via Q cycle
- passes one electron to cytochrome c at a time
cytochrome c shuttle
- small, soluble heme protein, present in high concentrations around the IMM
- binds the intermembrane side of the IMM directly with cardiolipin, an acidic phospholipid
- transfers 1 electron at a time
- connects Complex III to IV
Complex IV (Cytochrome C Oxidase)
- Complex IV pumps 4 protons per O2 reduced
- reduces O2 -> H2O
1. accepts 2 electrons from 2 cytochrome c attachments
-> one moves to a terminal Cu, one to a Fe after Cu is full
2. reduced Fe and Cu uptake O2 and form a peroxide bridge
3. 2 additional cytochrome c attachments bring 2 more electrons
-> collects 2 H+ from matrix and forms hydroxides
4. 2 more H+ come from the matrix to produce 2 water molecules
electron transfer sequence proceeds energetically "downhill"
stepwise redox transfers release energy
{NADH-CoQ reductase} -> {Succinate-CoQ reductase} -> {CoQH2-cytochrome c reductase} -> {cytochrome c oxidase}
electron transfer sequence proceeds energetically "downhill": NADH
NADH -> Complex I -> Q -> Complex III -> cytochrome c -> Complex IV -> O2
electron transfer sequence proceeds energetically "downhill": FADH2
FADH2 enters at Complex II -> Q -> Complex III -> cytochrome c -> Complex IV -> O2
importance of oxygen as final electron acceptor
- O2 accepts all electrons from ETC and generates H2O
=> 2 O2 -> 2 H2O
- keeps ETC flowing, without it NADH accumulates
- lack of O2 in ischemia stops ATP synthesis
=> clinical oxygen deprivation causes rapid ATP deletion and cell death
cyanide inhibition of Complex IV
- cyanide binds Complex IV
- blocks electron transfer to O2
- collapses PMF -> no ATP
- rapidly fatal: seizures, coma, cardiac arrest appear in minutes due to ATP depletion in the brain and heart
Complex I: proton pumping summary
4 protons
proton pumping summary: Complex II
none
proton pumping summary: Complex III
4 protons
proton pumping summary: Complex IV
2 protons
proton motive force (PMF)
energy stored in gradient
PMF = ΔΨ + ΔpH
-> ΔΨ = electrical potential
-> ΔpH = proton concentration gradient
-> drives ATP synthase and other processes
contribution of membrane potential (ΔΨ) to PMF
- negative charge inside matrix due to charge separation of H+
- electrical force strongly pulls protons back across inner membrane to the matrix
- major contributor to PMF (~70%)
contribution of proton gradient (ΔpH) to PMF
- matrix ~0.5-1 pH unit higher than intermembrane space
- adds chemical component to PMF
- protons flow "downhill" into matrix
-> always towards equilibrium (high to low concentrations)
ΔΨ value in mitochondria
~150-200 mV
ΔpH value in mitochondria
~0.5-1 unit
- contributes ~30-60 mV
total PMF value in mitochondria
~180-220 mV
other uses of PMF in bacteria
- powers flagellar rotation through proton influx
- drives nutrient uptake and efflux pumps
- energizes transport of ions and metabolites
ΔΨ vs ΔpH dominance in mitochondria
ΔΨ dominant (~70%)
ΔΨ vs ΔpH dominance in chloroplasts
ΔpH dominant
ΔΨ vs ΔpH dominance in bacteria
balance varies with environment
ATP synthase overview (F0F1 complex)
- large enzyme spanning inner membrane
- F0 = proton channel
- F1 = catalytic head
- couples proton flow -> ATP synthesis
F0 proton channel (c-ring)
- F0 embedded in membrane
- proton binding drives c-ring rotation
- ring size (stoichiometry of subunits) affects H+/ATP ratio
F1 catalytic head (α3β3 hexamer)
- matrix-facing catalytic domain
- 3 αβ pairs form nucleotide-binding sites
- site of ATP synthesis
γ subunit
central rotating stalk
γ subunit rotation
- interacts with β-subunits of F1 catalytic head
- drives binding-change mechanism
rotary catalysis model
- proton flow rotates c-ring and the γ-subunit
- produces ATP in cyclic manner
- induces conformational changes in β-subunits of F1
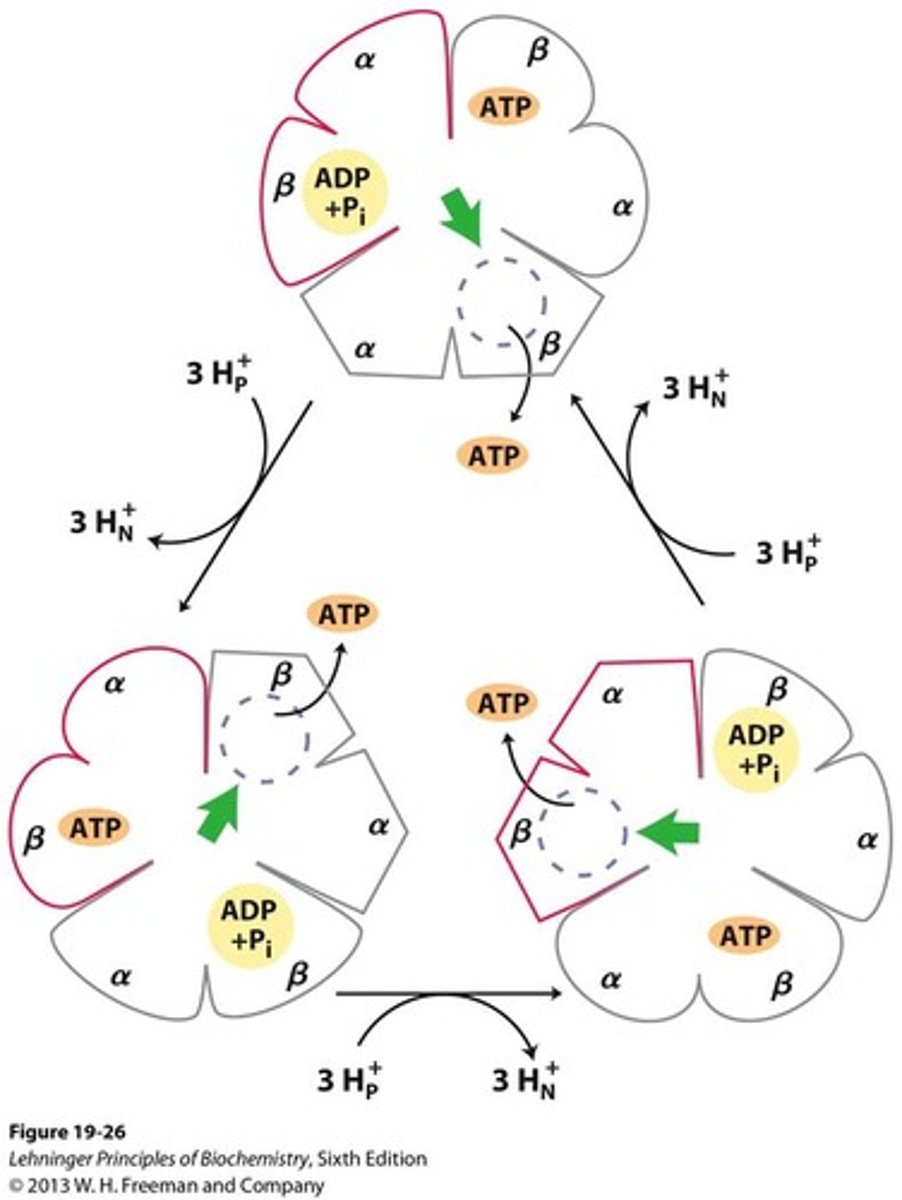
binding-change mechanism
each individual β-subunit of F1 cycles through: open -> loose -> tight
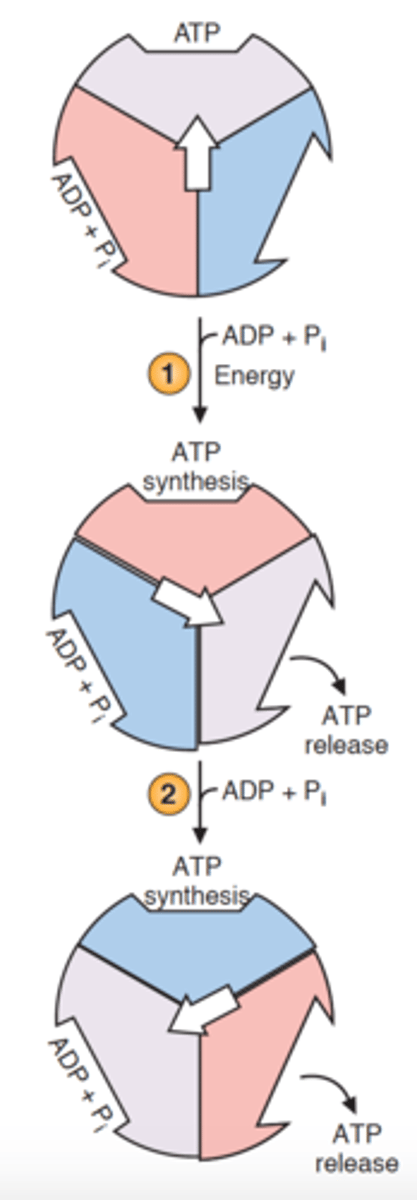
loose
ADP + Pi bond
tight
ATP formed
open
ATP released
experimenal models of rotary motion
- single molecule fluorescence imaging (attach GTP)
- visible rotation of actin filament attached to γ-subunit
- confirmed rotary catalysis model
energetics of proton flow
- ~3-4 protons required per ATP
-> allows γ-subunit to interact with catalytic subunits as it rotates enough to meet the next β subunit of F1
- PMF energy converted into chemical bond energy
ATP synthase as a molecular motor
- proton flux = "fuel"
- rotation of c-ring and γ-subunit = "mechanical motion"
- ATP synthesis = "output work"
uncouplers
collapse the proton gradient
- lipid soluble molecules shuttle protons without ATP synthase
- dissipate PMF -> no ATP made
- energy lost as heat
physiological uncoupling
- brown adipose tissue and UCP1
- energy released as heat (thermogenesis)
brown adipose tissue (fat)
rich in mitochondria
UCP1
uncouples ETC from ATP synthesis (membrane protein)
DNP and obesity
- DNP used as weight loss drug in 1930s
- increased metabolism, but fatal overheating
- illustrates danger of uncoupling
P/O ratios for NADH vs FADH2
- per NADH: 10 protons pumped (~2.5 ATP total)
- per FADH2: 6 protons pumped (~1.5 ATP total)
- difference due to entry points and proton pumping various regulations of ETC
efficiency of oxidative phosphorylation
- ~30-32 ATP per glucose (theoretical yield)
- efficiency impacted by stoichiometry subunits within the c-ring
-> more rings, more H+ per ATP
- overall efficiency ~34% (rest lost as heat)
-> complete glucose oxidation: ~686 kcal/mol
-> ATP hydrolysis: ~7.3 kcal/mol per ATP
- balances energy production and thermoregulation
mitochondrial diseases
- caused by defects in mitochondrial or nuclear genes
- affect high-energy tissues (muscle, brain, heart)
- often multi-systemic and progressive
key neurological clinical features of mitochondrial diseases
- seizures
- stroke-like episodes
- developmental delay
key muscular clinical features of mitochondrial diseases
- weakness
- exercise intolerance
misc. key clinical features of mitochondrial diseases
- hearing loss
- cardiomyopathy
- endocrine dysfunction
classic mitochondrial diseases
- MELAS
- LHON
- MERRF
MELAS
mitochondrial encephalomyopathy, lactic acidosis, stroke-like episodes
LHON
Leber's hereditary optic neuropathy
MERRF
myoclonic epilepsy with ragged red fibers
mechanisms behind pathology of mitochondrial diseases
- reduced ATP production -> energy crisis
- increased ROS generation -> oxidative damage
- defective apoptosis regulation through signaling can lead to neurodegeneration
diagnosis of mitochondrial diseases
- muscle biopsy
- genetic testing
- biochemical assays
supportive treatments of mitochondrial diseases
- exercise
- diet
- cofactor supplements
experimental therapies for mitochondrial diseases
- gene therapy
- mitochondrial replacement