07 - Proteins and Translation
1/57
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
58 Terms
Polypeptides are linear sequences of ____
amino acids
amino acids are linked by _____
peptide bonds
peptide bonds join the _________end of one amino acid to the _________ end of another.
join the carboxyl end of one amino acid to the amino end of another
structure of amino acids
amino end
carboxyl end
side chain (r group)
hydrogen atom
amino terminus is also known as
n- terminus
carboxyl terminus is also known as
c-terminus
Polypeptides:
Linear chains of amino acids linked by peptide bonds
subunits
Individual polypeptide chains in a multi-chain protein; can fold independently and contribute to overall function.
Proteins
One or more folded polypeptide subunits forming a functional 3D structure,
To be functional, a protein must do what
fold into the correct 3-D shape, include the correct cofactors/subunits, and contain any required post-translational modifications
Folding is promoted by
non-covalent interactions
Enzymes involved in the folding process often have
weak affinity for the ribosome and stay near the exit tunnel
Protein domains
the structural units of proteins
usually folds independently and has a particular function
Different proteins can contain the same domain
How could the same DNA-binding domain be found in multiple different proteins?
exon shuffling by transposition
gene duplication and divergance
protein domain relation to evolution
Modular and reusable across different proteins
Key to protein function diversity
Products of gene evolution (domain shuffling and duplication)
Different proteins can vary greatly in size and number of polypeptide
___
subunits
1 polypeptide could contain _____ domain
more than 1
1 protein often contains _____ domains with _____ functions
many
different
is the translation of genetic code conserved or no
One of the highly conserved and most complex process in both eukaryotes and prokaryotes
overall process of Translation of the genetic code
1. Initiation - Ribosome attaches to mRNA and begins translating at the initiation/start codon (AUG → methionine) → puts the ribosome in the correct reading frame
2. Elongation – Polypeptide chain elongation → series of steps repeated over and over until a stop codon is reached (UAA, UAG or UGA)
3. Termination – Stop codon signals to stop further elongation and release the polypeptide
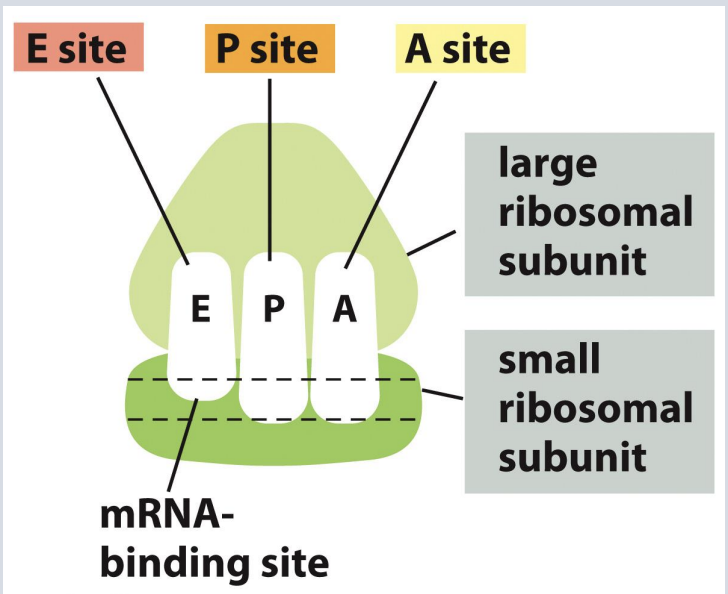
Structure of ribosome
Each ribosome has three sites for association with tRNAs:
1. A (aminoacyl) site
2. P (peptidyl) site
3. E (exit) site
Peptide bond formation during elongation
A new peptide bond forms between amino group of incoming amino acid and C-terminus of the growing chain
translation - elongation
Step 1: AA-tRNA binds to an empty A site
Step 2: A new peptide bond is formed between the growing chain and the new amino acid (peptidyl transferase activity)
Step 3: Large subunit translocates
Step 4: Small subunit translocates by 3 nucleotides (codon), resulting in empty A site that can accept another AA-tRNA
The process repeats until the ribosome reaches a stop codon on mRNA
Translation Elongation Factors
are additional proteins that improve the efficiency and accuracy of translation (EF -Tu and EF -G in bacteria)
EF -G
Hydrolyze GTP to drive transitions in the ribosome subunits
accuracy check as Translation Elongation Factors
Accuracy checks:
• Small subunit rRNA hydrogen bonds with the codon -anticodon
• A tight (correct) codon/anti -codon pairing triggers a conformational change in the ribosome and hydrolysis of GTP by EF -Tu (in this image). EF -Tu then releases the AA -tRNA, freeing it for addition of the AA to the growing chain
how is the reading frame set
Beginning from the AUG start codon allows the ribosome to correctly set the reading frame
directionality of ribosome movement
The ribosome moves 5’ to 3’ along the mRNA
directionality of protein synthesis
the polypeptide is built N-terminus to C-terminus (and in prokaryotes, translation can begin at the 5’ end while RNA polymerase is still synthesizing)
The start codon is at the ____ end of the protein-coding sequence in the RNA
5’
AUG codes for
for methionine, so the initial amino acid at the amino (N) terminus of the polypeptide is always Met (but this can be removed later)
initiator factors in eukaryotes and prokaryotes
In bacteria called IFs and in eukaryotes called eIFs.
Translation initiation in prokaryotes
In prokaryotes, the small ribosomal subunit binds to the first AUG codon guided by a specific sequence: The Shine-Dalgarno Sequence (5’-AGGAGG-3’)
Shine-Dalgarno sequence is complementary to a sequence near 3’ end of 16S rRNA → positions the ribosome at the correct spot
Initiation factors are attached to the small subunit
In bacteria the initial AUG methionine is a modified version called N-formylmethionine (fMet)
Initiator tRNA (carrying fMet) interacts with AUG at what will be the ‘P’ site of the ribosome
IFs are then released and large subunit can bind
Shine-Dalgarno sequence
5’-AGGAGG-3’
where is the Shine-Dalgarno sequences located and why is tgis imp
Shine-Dalgarno sequences can be located anywhere along the mRNA
Therefore, prokaryotic ribosomes can synthesize multiple proteins from a single RNA
IF1:
Helps with attachment to mRNA
IF2:
GTP-binding protein that is required for attachment of first AAtRNA
IF3:
Prevents premature attachment of large subunit
Translation Initiation in Eukaryotes
Processed 5’ and 3’ ends are important for translation initiation and for nuclear export • helps ensure only completed mRNA are translated
Eukaryotes have larger ribosomes and require additional proteins (12 eIFs or more) → initiation is more complex than prokaryotes
eIFs bind to the small subunit and are important for:
Initiator tRNA (Met) binding the ‘P’ site with GTP-bound eIF2
Small ribosomal subunit (along with eIFs and initiator tRNA) finds 5’ end of mRNA → scans along until reaches a Kozak sequence (5’ -CCACCAUG C -3’)
GTP bound to eIF2 is hydrolyzed and is released along with other eIFs •
Dissociation of initiation factors allows the large subunit to attach
eIF1s:
Conformational change to allow binding of mRNA
eIF2
Initiator tRNA (Met) binding the ‘P’ site with GTP-bound
eIF3:
interaction with eIF4G on mRNA complex
mRNA has its own set of eIFs:
eIF4E
eIF4A
eIF4G
eIF4E
binds to 5’ cap
eIF4A
has helicase activity that uses ATP hydrolysis to unwind any double stranded regions in mRNA
eIF4G
links 5’ cap and 3’ poly(A) tail. This converts mRNA into a circular message and interacts with eIF3 on small subunit
The 5’ to 3’ scanning activity searches for the what seq in eukaryotes
Kozak sequence
Kozak sequence
(5’-CCACCAUGC-3’)
consensus sequence (we know which nucleotides are found most commonly at each position)
“leaky scanning” for Kozak sequences
Leaky scanning occurs when the ribosome skips a weak Kozak sequence and initiates at a downstream AUG.
The actual sequence can vary slightly, but the more different it is, the less efficient initiation of translation at that AUG will be
What effect do you think Leaky scanning could have on the proteins produced from an mRNA?
this could create proteins with different amino acid sequences at the n-terminus
Translation Termination
Translation continues until ribosome reaches a stop codon for which no corresponding tRNA is available
Release factors (which resemble tRNA and recognize stop codons) bind in the vacant A site and catalyze the addition of water instead of an amino acid, which frees the C-terminus (no longer attached to a tRNA)
Polyribosomes
Multiple translation initiations typically take place on the same mRNA
Multiple ribosomes associated with an mRNA → Polyribosome (or Polysome)
Attach at AUG, start translating and once AUG is free, another ribosome can assemble
Prokaryotes vs Eukaryotes:Initiation Signal
Prokaryotes - Shine-Dalgarno sequence
Eukaryotes - Kozak sequence (surrounding AUG)
Prokaryotes vs Eukaryotes: Initiator tRNA
Prokaryotes - fMet-tRNA
Eukaryotes - Met-tRNA
Prokaryotes vs Eukaryotes: mRNA Structure
Prokaryotes - Often polycistronic
Eukaryotes - Monocistronic
Prokaryotes vs Eukaryotes: Polysomes
Prokaryotes - Form during transcription
Eukaryotes - Form after mRNA processing/export
Prokaryotes vs Eukaryotes: Initiation Factors
Prokaryotes - IF1, IF2, IF3
Eukaryotes - eIF1–eIF4 (more complex)
Prokaryotes vs Eukaryotes: Ribosome Binding
Prokaryotes - Small subunit binds Shine-Dalgarno via rRNA
Eukaryotes - Small subunit scans from 5′ cap