BIOL*1090: Cellular Biology
1/243
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
244 Terms
information (plan/recipe)
What does it take to make a cell?
DNA → transcription → RNA → translation → proteins (enzymes, channels, receptors)
Important for differentiation (creating different specific cell types
DNA
The hereditary material of genes; where info is encoded.
RNA
Provides the information necessary to build various proteins.
proteins
The cell’s primary machinery.
Enzymes, channels, receptors
enucleation
Mechanism by which maturing red blood cells (erythrocytes) eject their nucleus (during synthesis).
Features of red blood cells evolved to accommodate maximum hemoglobin carrying capacity
Lack of nuclei and organelles, mature cells do not contain DNA and cannot synthesize RNA
Thus: cannot divide and have limited repair capabilities
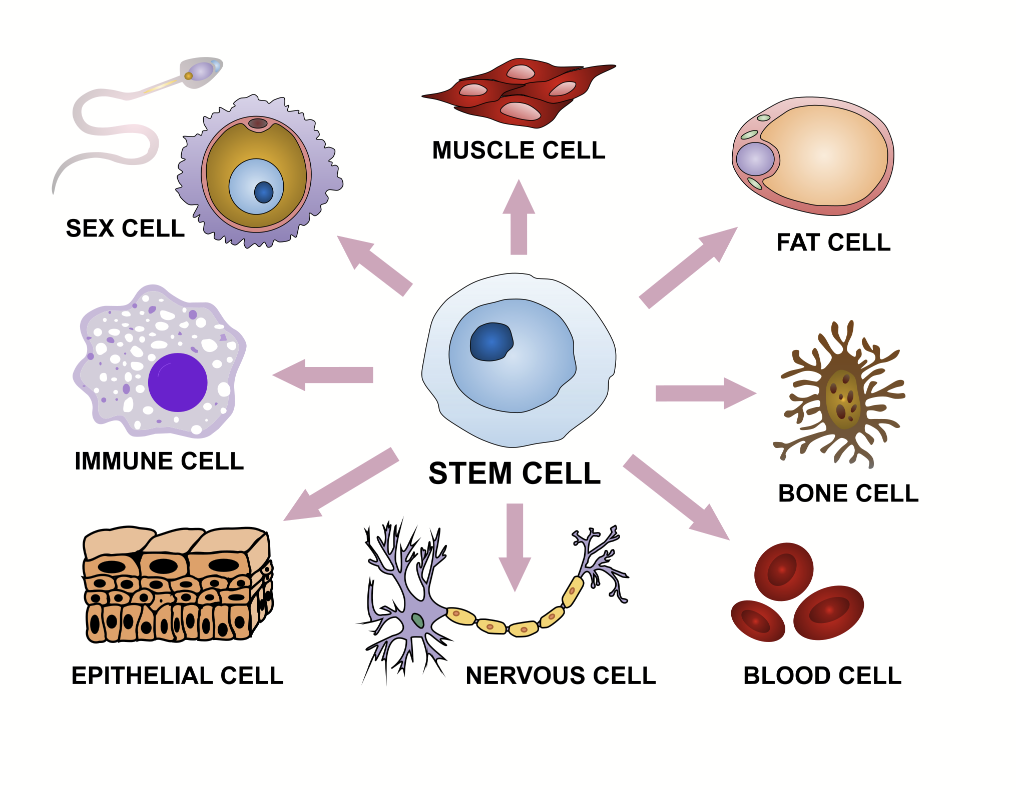
differentiation
During development, most cells in multicellular organisms will become specialized.
Information is important to create different specific cell types.
A stem cell, if in the right environment (specific nutrients, etc.), can become anything
chemistry
What does it take to make a cell?
Amino acids, lipids, nucleic acids → building blocks!
Cells carry out lots of reactions!
ABIOGENESIS
Miller-Urey Experiment
Chemical experiment that simulated the conditions thought to exist on the early Earth and to test the chemical origin of life under those conditions.
AMINO ACIDS CAN BE SYNTHESIZED IN THE LAB UNDER CONDITIONS MIMICKING EARLY EARTH (ANAEROBIC).
Simple precursors + energy = formaldehyde + hydrogen cyanide (first group of intermediate products)
First group + energy = urea + formic acid + AMINO ACIDS
Primordial Soup Hypothesis
Putative conditions on the primitive Earth favoured chemical reactions that synthesized more complex organic compounds from simpler inorganic precursors.
Molecules essential for life: nitrogen, ammonia, methane, carbon dioxide, water, hydrogen
abiogenesis
The chemical origin of life.
Essential molecule + energy = simple organic compound.
compartments
What does it take to make a cell?
Putting everything into a physical space where reactions can happen!
Usually defined by single or double lipid layer membrane
Establish physical boundaries that enable the cell to carry out different metabolic activities
Generate a micro-environment to spatially and temporally regulate biological processes
e.g. mitochondria, chloroplasts, cell nucleus, vesicles, endoplasmic reticulum
The Cell Theory
All living organisms are composed of one or more cells.
The cell is the most basic unit of life.
Omnis cellula e cellula: All cells arise only from pre-existing cells.
Credited to Schleiden and Schwann.
basic properties of cells
Highly complex and organized.
Activity controlled by a genetic program.
Can reproduce - make copies of themselves (exception: red blood cells).
Assimilate and utilize energy.
Carry out many chemical reactions (enzymes).
Engage in mechanical activities.
Respond to stimuli.
Capable of self-regulation.
They evolve.
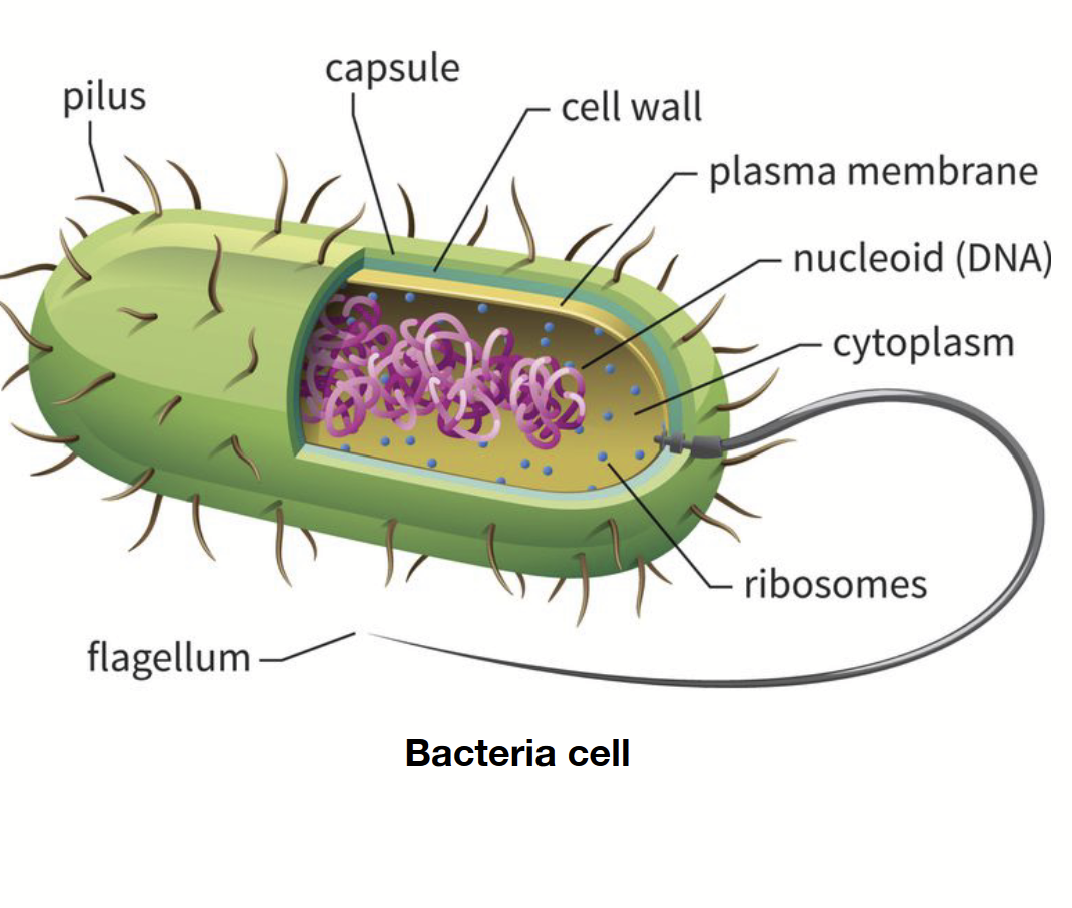
prokaryotes
Single-cell organism. 1-10 micrometres. No nucleus or organelles. Ribosomes are present but small. Reproduce asexually. Genetic material found in nucleoid (not a true chromosome, rather, DNA arranged in a circular fashion: plasmid).
Randomly arranged/spread out.
eukaryotic cells
Found in multicellular organisms, but can be unicellular (protozoa). Typically 10-100 micrometres. Contain nucleus and organelles. Ribosomes are large. Genetic material is found in the nuclear compartment and arranged as chromosomes.
Very condensed.
Include protozoa, fungi, plants, and animals.
plasmid
A small circular DNA molecule found in bacteria and some other microscopic organisms. Reproduce independently.
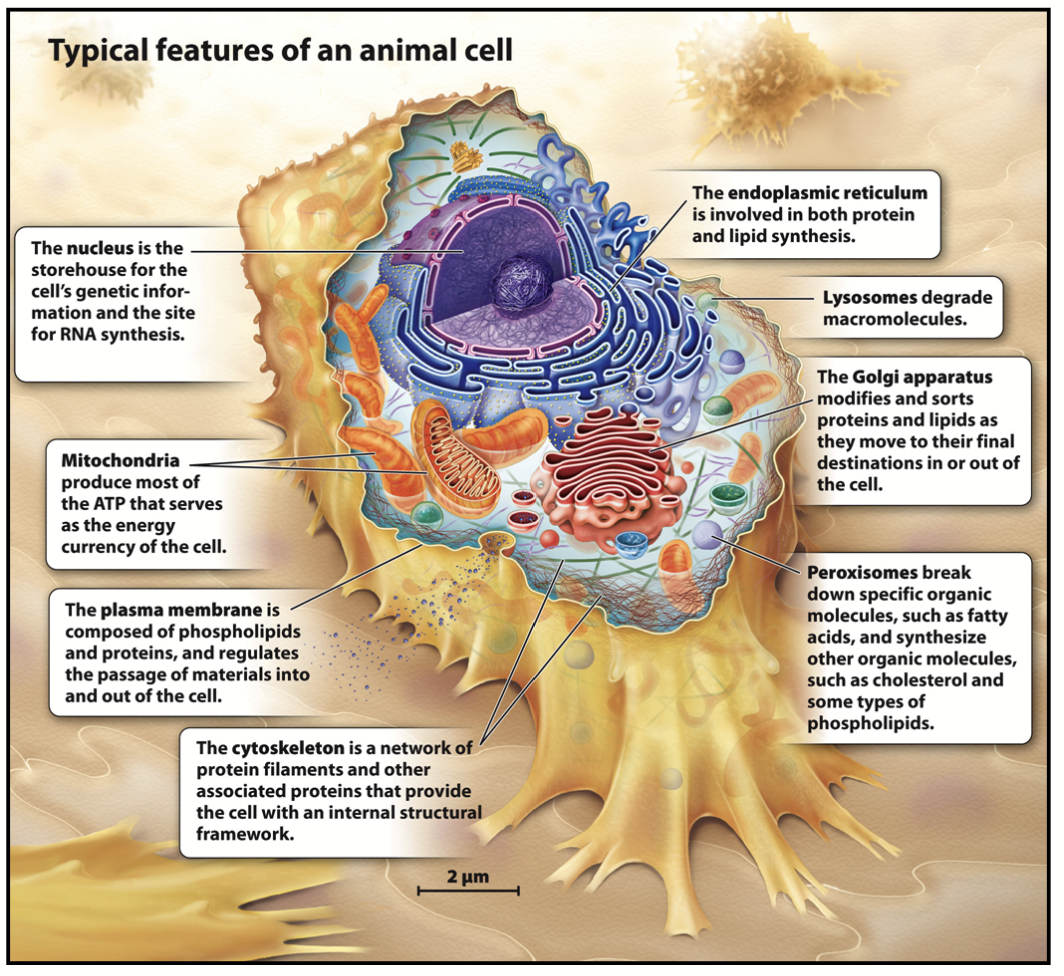
plasma membrane
Controls movement of molecules in and out of the cell and functions in cell-cell signaling and cell adhesion.
mitochondria
Surrounded by a double membrane, generates ATP by oxidation of glucose and fatty acids.
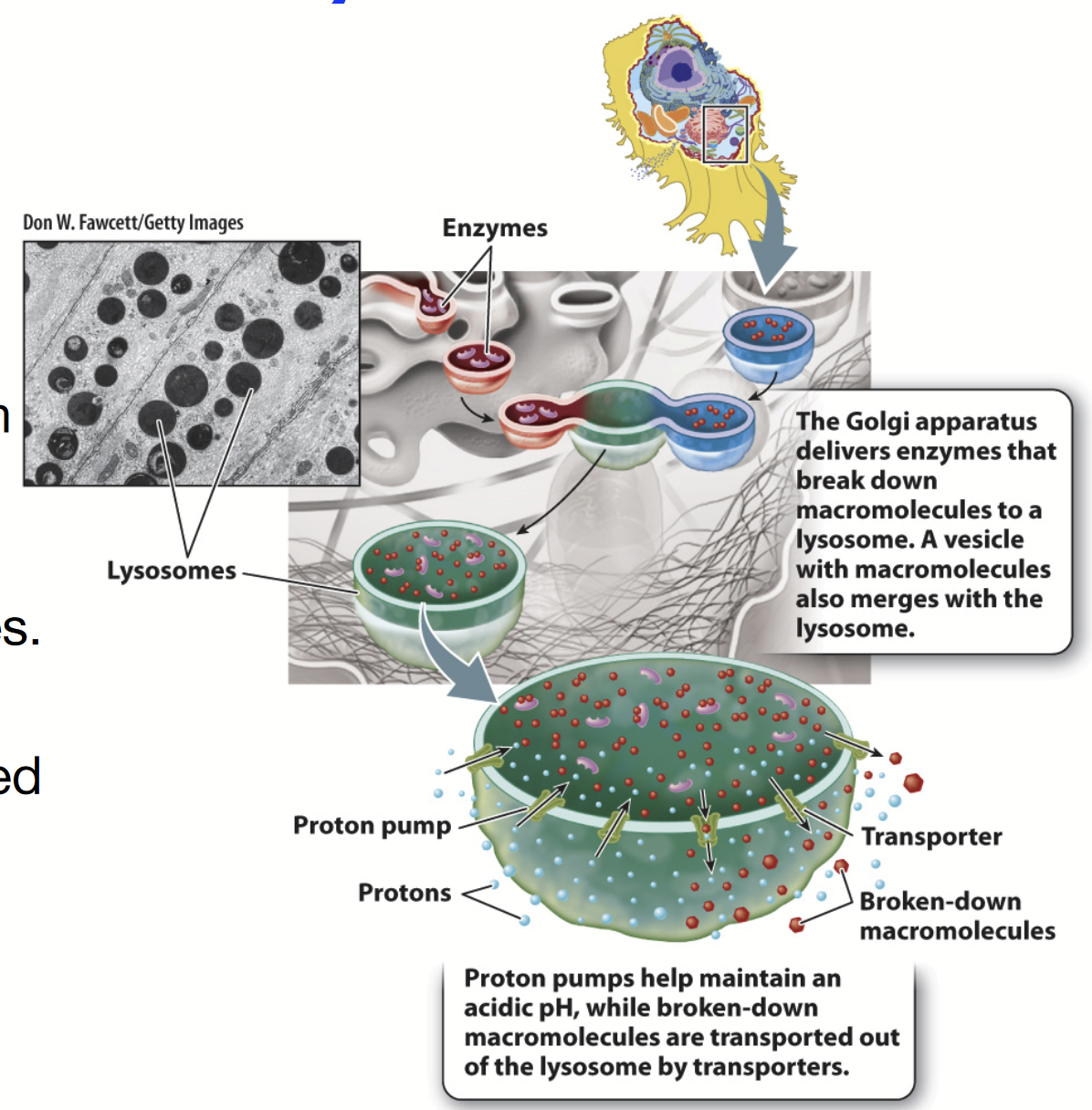
lysosome
Only in animal cells!
Digestive organelle that degrades material internalized by the cell and worn-out cellular membranes and organelles. Contains an acidic lumen.
Degradation of internalized material: recycling of plasma membrane components like receptors and extracellular material, destroy pathogens like bacteria and viruses (only in phagocytic cells).
The Golgi apparatus delivers enzymes that break down macromolecules here. A vesicle with macromolecules also merges with this organelle.
Functions: AUTOPHAGY!!!, degradation of internalized material
Size: 25 nm to 1 micrometre
Internal pH of 4.6 (proton pump or H+-ATPase pumps H+ into lumen to maintain acidity)
Contains hydrolytic enzymes (acid hydrolases)
Membrane is composed of glycosylated proteins that act as a protective lining next to the acidic lumen
When the conditions are right, cargo is degraded and broken down into smaller pieces
Transporters transports broken-down macromolecules out
Rounded shape + VERY DARK INSIDE because of the very acidic interior!!!!
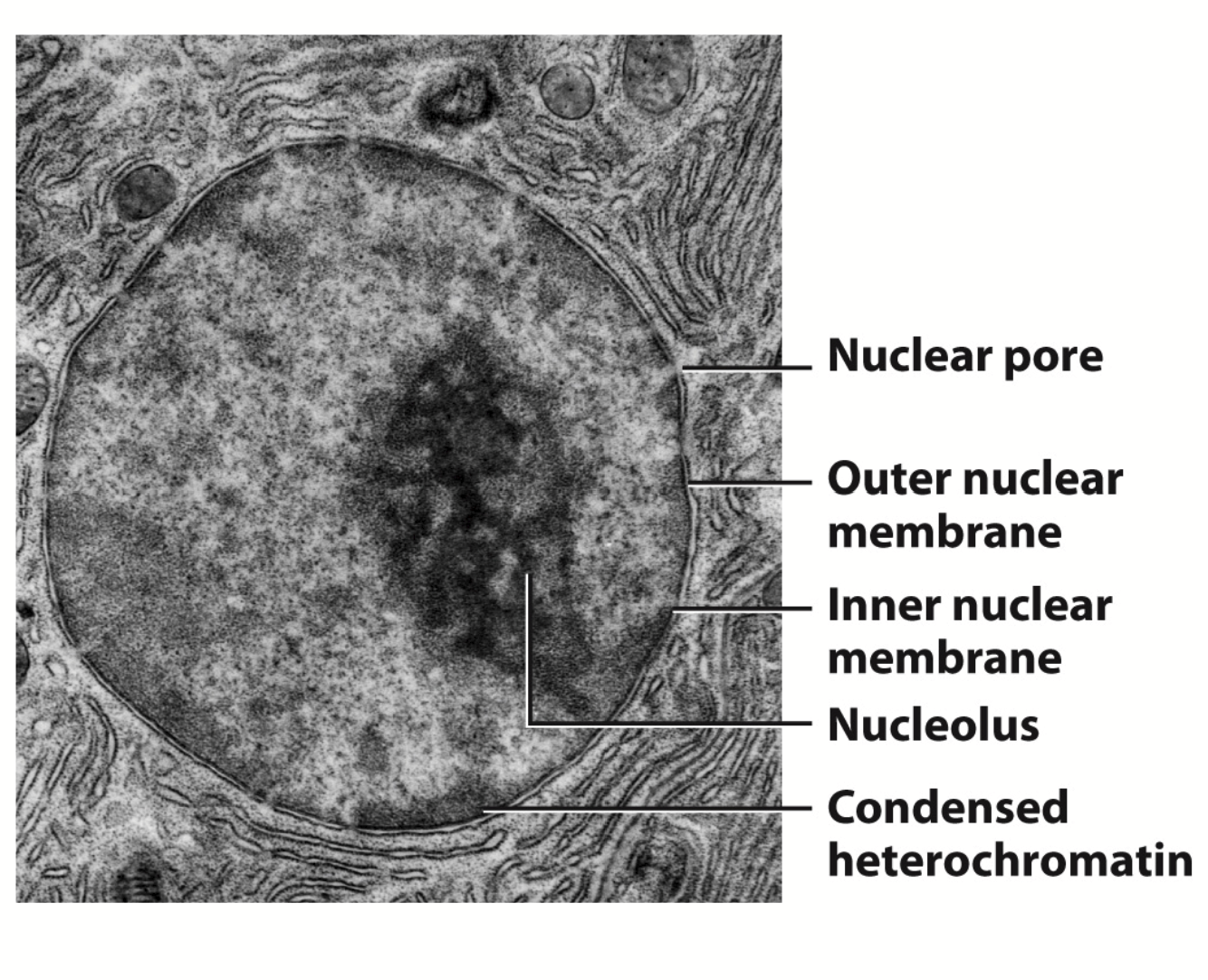
nuclear envelope
A double membrane that encloses the contents of the nucleus. The outer membrane is continuous with the rough endoplasmic reticulum.
nucleolus
A nuclear sub-component where most of the cell’s rRNA is synthesized.
Largest structure inside the nucleus of eukaryotic cells. Primary function is biosynthesis of ribosomes.
KNOW FOR EXAM!!!!!!!!!
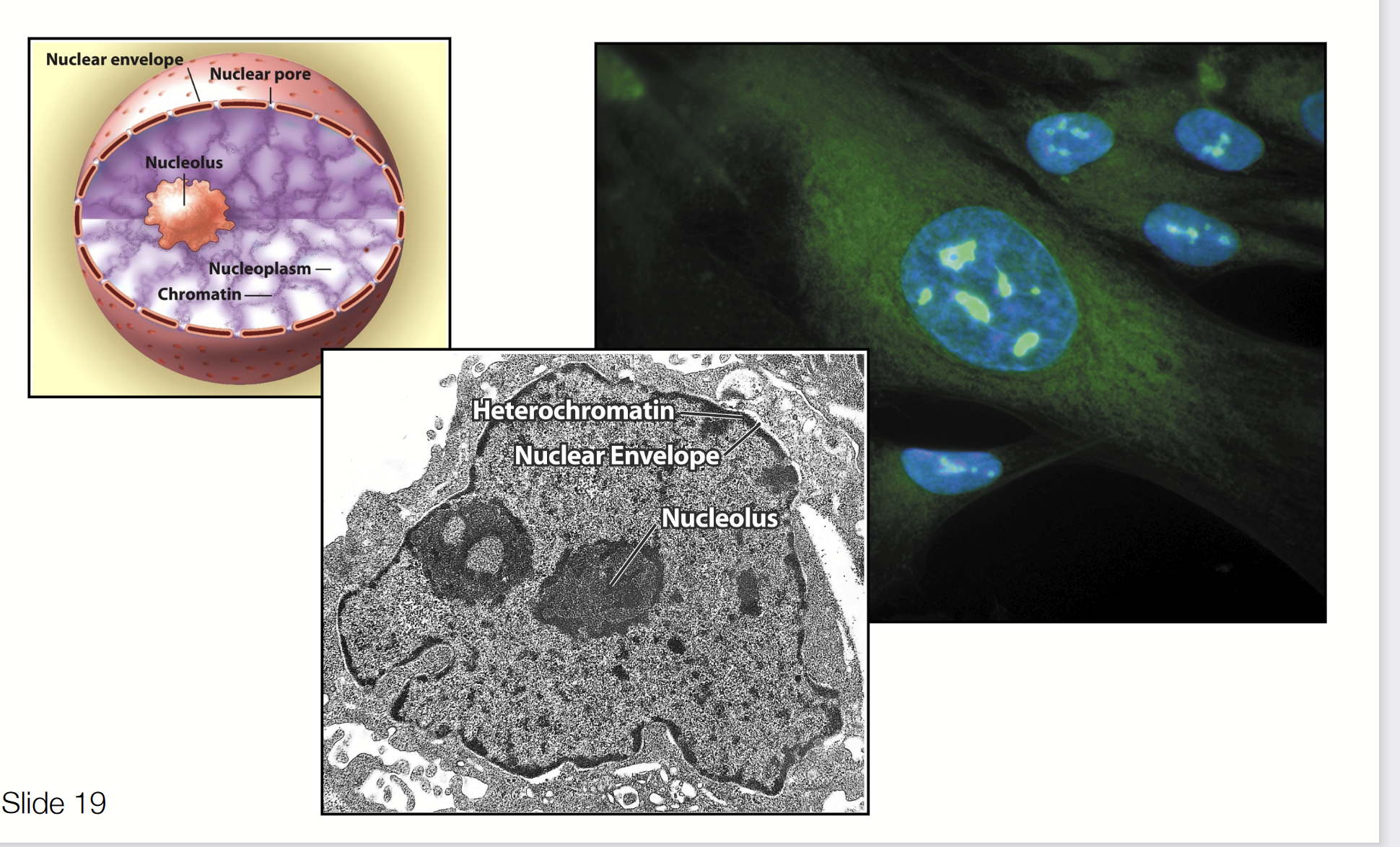
nucleus
Filled with chromatin composed of DNA and proteins. Site of mRNA and tRNA synthesis.
Function:
Storage, replication, and repair of genetic material (contains DNA repair machinery) - very efficient level of compacting
Stage 1: DNA duplex 2 nm in diameter, nucleosome fiber 10 nm in diameter
Stage 2: chromatin fiber 30 nm in diameter
Stage 3: coiled chromatin fiber 300 nm in diameter
Stage 4: coild coil 700 nm in diameter, condensed chromatid 1400 nm in diameter
Expression of genetic material
Transcription: mRNA, tRNA, rRNA → promotor region = contrl of transcription
RNA splicing → occurs before nuclear export of mRNAs
Ribosome biosynthesis → noncoding RNAs do not encode proteins and include tRNA and rRNA; theese RNAs are transcribed from DNA found in the nucleus
Structure:
Nuclear Envelope
Nuclear membrane (inner + outer)
Nuclear pores (small gaps; allow protein to go in and out of membrane)
Nuclear lamina (continuous structure, goes all the way to the rough ER)
Nuclear content
Chromatin, nucleoplasm, nucleolus
smooth endoplasmic reticulum
Contains enzymes that:
Synthesize lipids
Produce steroid hormones
Detoxify certain hydrophobic molecules (liver cells contain enzymes that modify foreign compounds)
Sequestration (storage) of Ca2+
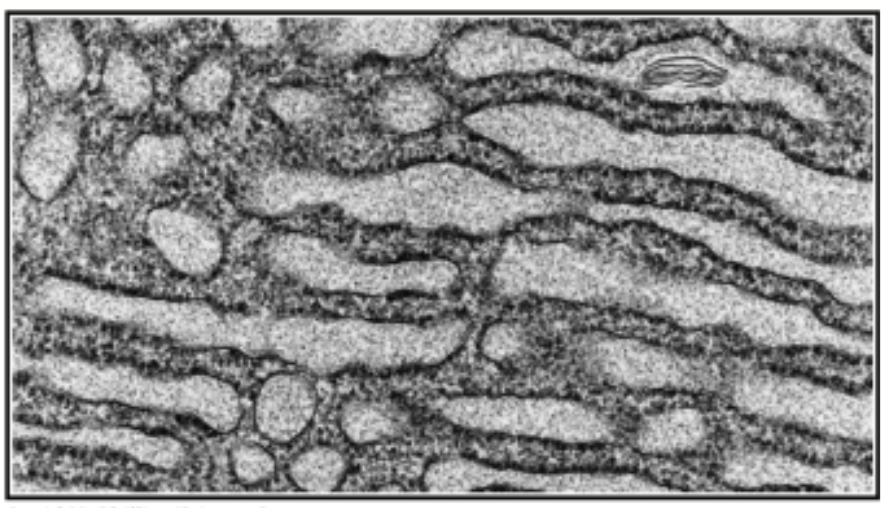
rough endoplasmic reticulum
Functions in the synthesis, processing, and sorting of secreted proteins, lysosomal proteins, and certain membrane proteins. HAS RIBOSOMES ATTACHED!
Synthesis of membrane phospholipids
Glycosylation of proteins - addition of carbohydrate chains to specific proteins
Protein folding / quality control - involve the activity of molecular chaperones, a specific type of protein that assists in the folding proteins
Protein synthesis, modification, and transport - proteins targeted to ER, proteins targeted to other endomembrane compartments (soluble and transmembrane), proteins targeted to plasma membrane (secreted and transmembrane)
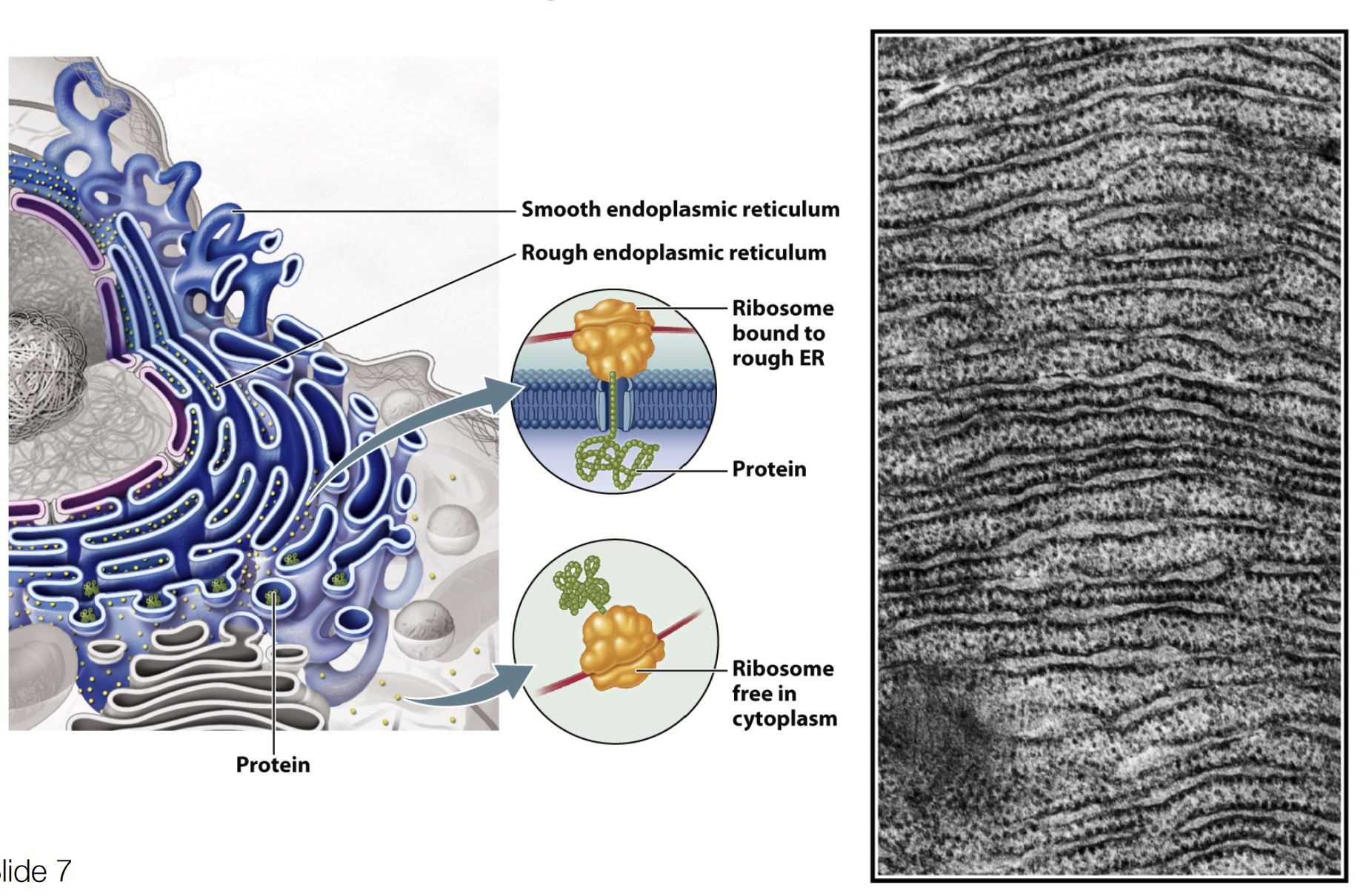
Golgi complex
Processes and sorts secreted proteins, lysosomal proteins, and membrane proteins synthesized on the rough ER. Also involved in the synthesis of polysaccharides and the glycosylation and proteolytic modification of proteins and lipids.
THE PROCESSING PLANT OF THE CELL!
Receives proteins and lipids from the ER and sorts them to other organelles, the plasma membrane, or the cell exterior.
Proteins modified step-wise as they traverse. Different cisternae contain different enzymes that modify proteins; the differential staining of the cisternae reflects their biochemical differences!
Material moves in a PROXIMAL to DISTAL direction (closer to ER → closer to transmembrane)!!!!!
Cis-Golgi Network (CGN) → Medial-Golgi (MG) → Trans-Golgi Network (TGN)
Smooth, flattened, disk-like cisterna (flattened membrane vesicle; ~0.5-1 micron in diameter)
~8 or fewer cisternae/stack - ranges from a few to to several 1000 stacks per cell (layers will be consistent but numbers will differ)
Curved like a shallow bowl
Shows POLARITY: cis-medial-trans cisternae
Cisternae are biochemically unique
Membrane supported by protein skeleton (actin, spectrin)
Scaffold linked to motor proteins that direct movement of vesicles in and out
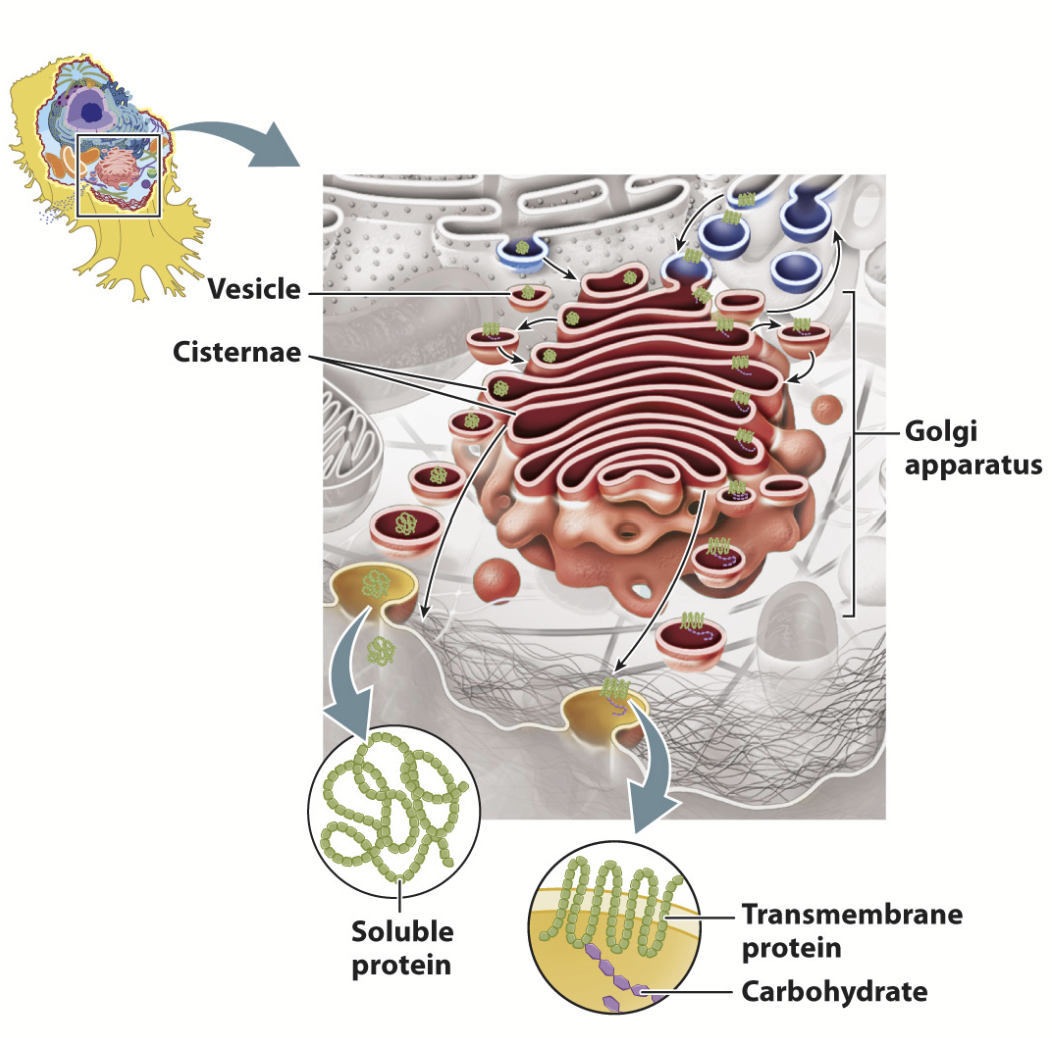
secretory vesicles
Store secreted proteins and fuse with the plasma membrane to release their contents.
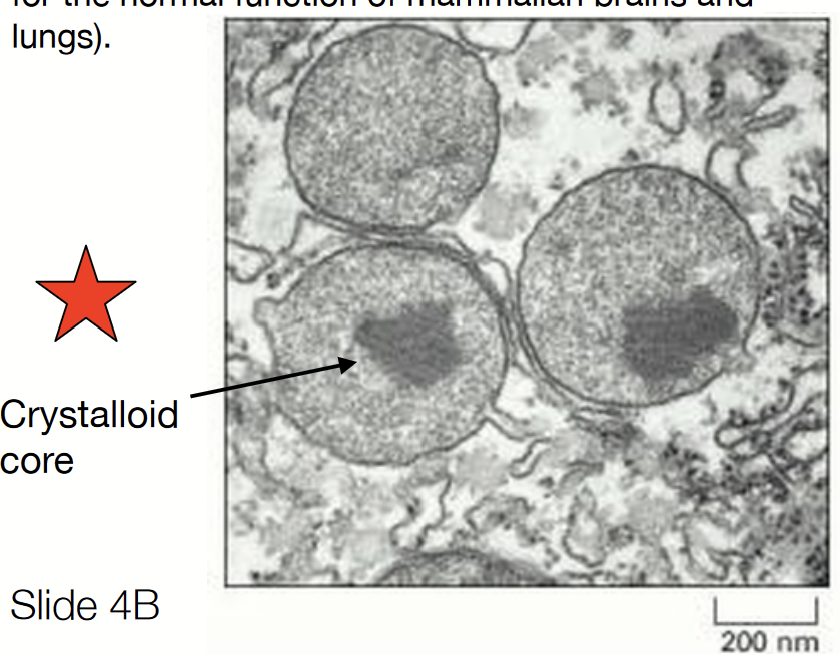
peroxisome
Contains enzymes that break down fatty acids into smaller molecules used for biosynthesis. Detoxifies certain molecules.
Found in virtually all eukaryotic cells.
Catabolism of very long chain fatty acids
Reduction of reactive oxygen species
Biosynthesis plasmalogens (phospholipids critical for the normal function of mammalian brains and lungs)
Critical to bring different species together!
Have a crystalloid core (denser area).
cytoskeleton
Fibers form networks and bundles that support cellular membranes, help organize organelles, and participate in cell movement.
microvilli
Only in animal cells!
Increase surface area for absorption of nutrients from surrounding medium.
cell wall
Only in plant cells!
Composed largely of cellulose. Helps maintain the cell’s shape and provides protection against mechanical stress.
vacuole
Only in plant cells!
Stores, water, ions, and nutrients. Degrades macromolecules. Functions in cell elongation during growth.
chloroplast
Only in plant cells!
Carry out photosynthesis, surrounded by a double membrane, contain a network of internal membrane-bound sacs.
plasmodesmata
Only in plant cells!
Tubelike cell junctions that span the cell wall and connect the cytoplasms of adjacent plant cells.
genome
Dynamic information system. The complete set of genes or genetic material present in a cell or organism.
slime molds
Originally classified as part of the Fungi branch, but now grouped with the kingdom Protista - not a mold.
Eukaryotic single cell organism.
Use a primitive form of sexual reproduction (spores).
Size: less than 1 cm to several square meters.
Multinucleated (1 cell has thousands of nuclei).
They have an amazing capacity to move around and
respond to their environment.
They have no neurons and no brain, but can navigate complex decisions such as which direction to move.

virus
NOT CELLS! Macromolecular packages that can function and reproduce only within living cells. VERY SMALL!
Basic structures: nucleic acid genome and protein capsid that covers the genome.
Main factor that determines what cell type a virus can infect: surface expression of a specific surface protein.
Highly complex and organized
Activity controlled by a genetic program
Needs to hijack a host/body in order to reproduce
Cannot assimilate or use energy, must randomly hijack other bodies
Do not carry out many chemical reactions (but sometimes enzymes are added into genetic material, found on surface in order to penetrate into host cell)
Do not engage in mechanical activities; are randomly spread out and penetrate host cells only through random contact (don’t actively chase hosts)
Do not respond to stimuli
Not capable of self-regulation
Can evolve
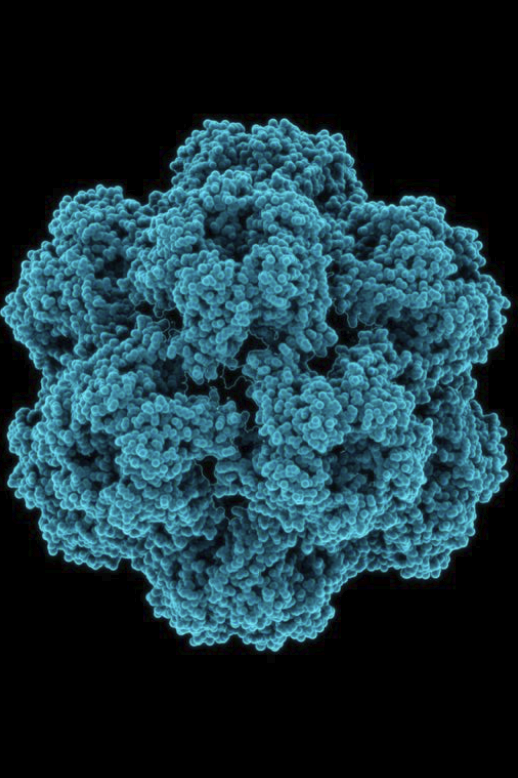
virion
The inanimate particle that viruses exist as outside of cells.
Like dust!
Made out of:
Small amount of DNA (DNA virus) or RNA (RNA virus) that encodes hundreds of genes
A protein capsule called capsid
capsid
The protein shell of a virus, enclosing its genetic material.
Baltimore classification
Classification system of viruses that categorizes them based on type of genome (RNA/DNA) and their method of replication.
retrovirus
RNA virus that can insert a copy of its genome into the DNA of a host cell.
HIV and AIDS
hepadnavirus
DNA virus that affects human liver and causes serious infections.
Hepatitis B
filovirus
Virus that encodes their genome in the form of single-stranded negative-sense RNA. Causes hemorrhagic fever (makes many copies and then bursts, killing host and destroying integrity of organism). VERY DEADLY, gets into cell.
adenovirus
DNA virus that can cause respiratory illnesses (bronchitis, pneumonia, etc.) or conjunctivitis (an infection in the eye).
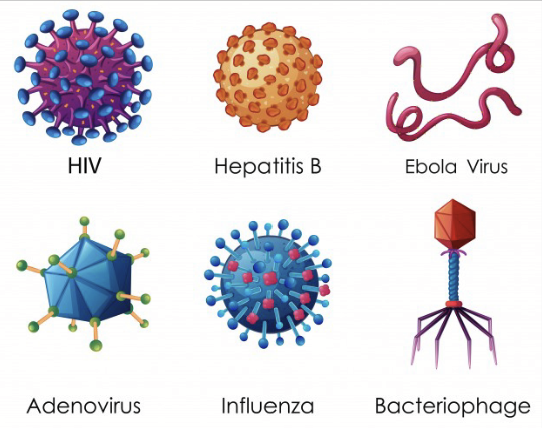
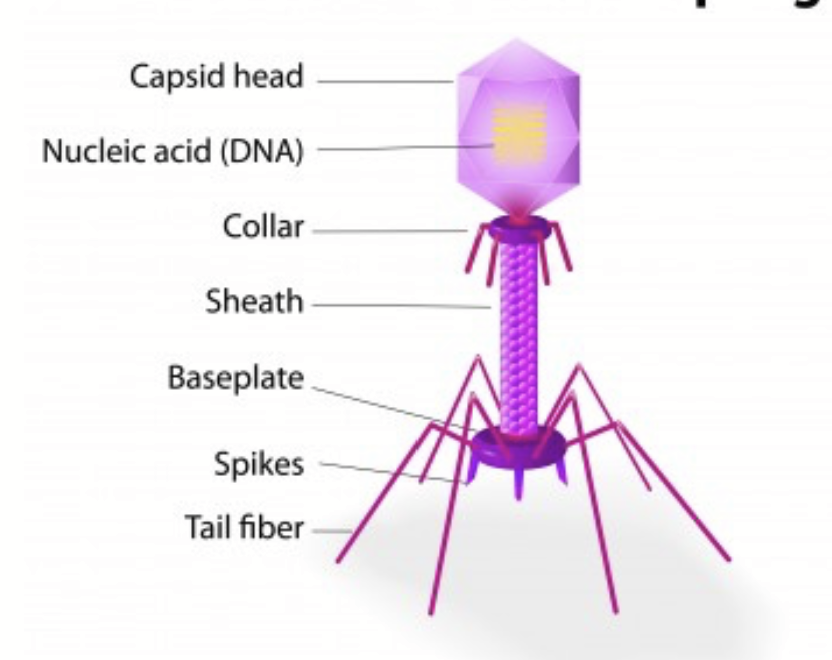
bacteriophage
Virus that infects and replicates within bacteria and archaea (prokaryotes).
“Holds” cell to transfer DNA inside
Made up of capsid head, nucleic acid (DNA), collar, sheath, baseplate, spikes, tail fiber
virosphere
Each microbe/cell type is almost certainly associated with at least one virus, there are probably billions of distinct viruses. So far, just 4958 virus species have been formally described.
CRISPR-Cas
An immune-like system that can battle continuous attacks from bacteriophages.
virus host range
Viruses bind to a cell surface via specific proteins and then enter into the cell.
narrow host range
Viruses that can infect one species/host.
Human cold, influenza infect epithelial cells of human respiratory system
wide host range
Viruses that can infect cells in any species/host.
Rabies can infect cells in dogs, foxes, bats, raccoons, humans
viral infection
Once inside a cell, the virus hijacks cellular machinery to synthesize nucleic acids (DNA/RNA) and proteins.
Two types: lytic and non-lytic
lytic viral infection
Viral infection where the production of virus particles ruptures and kills cell (e.g. influenza, rabies).
non-lytic viral infection
Viral infection where viral DNA is inserted in host genome (PROVIRUS). The infected cell can survive, often with impaired function, and can pass on the virus from one cell generation to the next.
e.g. HIV, Chicken Pox
Also known as integrative or lysogenic
papovavirus
A large family of viruses that includes the human papillomavirus (HPV).
rabies virus
Virus that has an enveloped single stranded RNA genome.
The RNA genome encodes five genes: nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and the viral RNA polymerase (L).
Evolved within 1500 years.
Currently 7 genotypes known.
Zika virus
Associated with microcephaly (a birth defect where a baby's head is smaller than expected when compared to babies of the same sex and age).
Positive-sense RNA genome directly translated into viral proteins.
Transmission: mosquitoes, sexual contact, blood transfusion
Infection of adults often asymptomatic.
Vertical transmission from mother to fetus → crosses the placental barrier by targeting trophoblasts and macrophages.
If infection of the fetus occurs during the period of neurogenesis, the survival of the neural progenitor cells (NPCs) can be affected → a lot of stem cells will die.
microcephaly
A condition where the head (circumference) is smaller than normal. This is typically associated with defect during early brain development.
Results in delayed motor and speech functions, intellectual disability, seizures, facial distortions, hyperactivity, balance and coordination problems, and other brain-related or neurological problems.
Possible causes: genetic abnormalities, alcohol, viruses
RNA vaccine
Vaccines that work by tricking the body’s cells into producing a fragment of a virus, an antigen, from an RNA template (there is no actual virus inside, only the template).
To make them more efficient, instructions for assembling a replicase, which can make lots of copies of the RNA template for producing antigens.
The immune system can then target the RNA template proteins.
antigen
A toxin or other foreign substance which induces an immune response in the body, especially the production of antibodies.
antibody
A blood protein produced in response to and counteracting a specific antigen. Antibodies combine chemically with substances which the body recognizes as alien, such as bacteria, viruses, and foreign substances in the blood.
DNA virus
Double-stranded virus. Travels to the host’s nucleus, where it is transcribed by the host’s infrastructure. Stays as a separate module and does not fuse with the host’s DNA.
Generally bigger than RNA viruses and thus can store more information - thus, viruses with these large genomes are less dependent on their hosts.
However, this replication process is much slower than RNA replication.
Contains repair mechanisms as this type of genome is meant for long-term storage of genetic information.
RNA virus
Virus with a single-stranded genome. Can remain in the host’s cytoplasm and either functions as mRNA and is translated into a protein OR it is transformed into mRNA. These viruses require their own RNA polymerase to replicate their genomes, as the host cell cannot replicate the RNA.
As host cells can’t use single-stranded DNA for replication or transcription, these single-stranded viruses also transport proteins required for converting their DNA into double-stranded DNA.
Negative-sense (complementary to mRNA) must be converted to positive-sense (similar to mRNA) by RNA polymerase before translation into a protein.
Short-lived molecule, likely to be degraded rather than repaired upon damage.
provirus
Viral DNA inserted in host genome.
biological membranes
Functions:
Define cell boundary
Define enclose compartments
Control movement of material into and out of cell
Allow response to external stimuli
Enable interactions between cells
Provide scaffold for biochemical activities
ASYMMETRICAL: two leaflets have distinct lipid composition in many plasma membranes; outer leaflet contains glycolipids and glycoproteins (lipids and proteins with carbohydrates attached to them).
plasma membrane
The most studied cell membrane - trilaminar structure made of a phospholipid bilayer.
~ 6 nm thick, very consistent between cell types.
Makes up the outermost layer of cells.
sarcoplasmic reticulum
A complex network of specialized smooth endoplasmic reticulum that is important in transmitting the electrical impulse as well as in the storage of calcium ions.
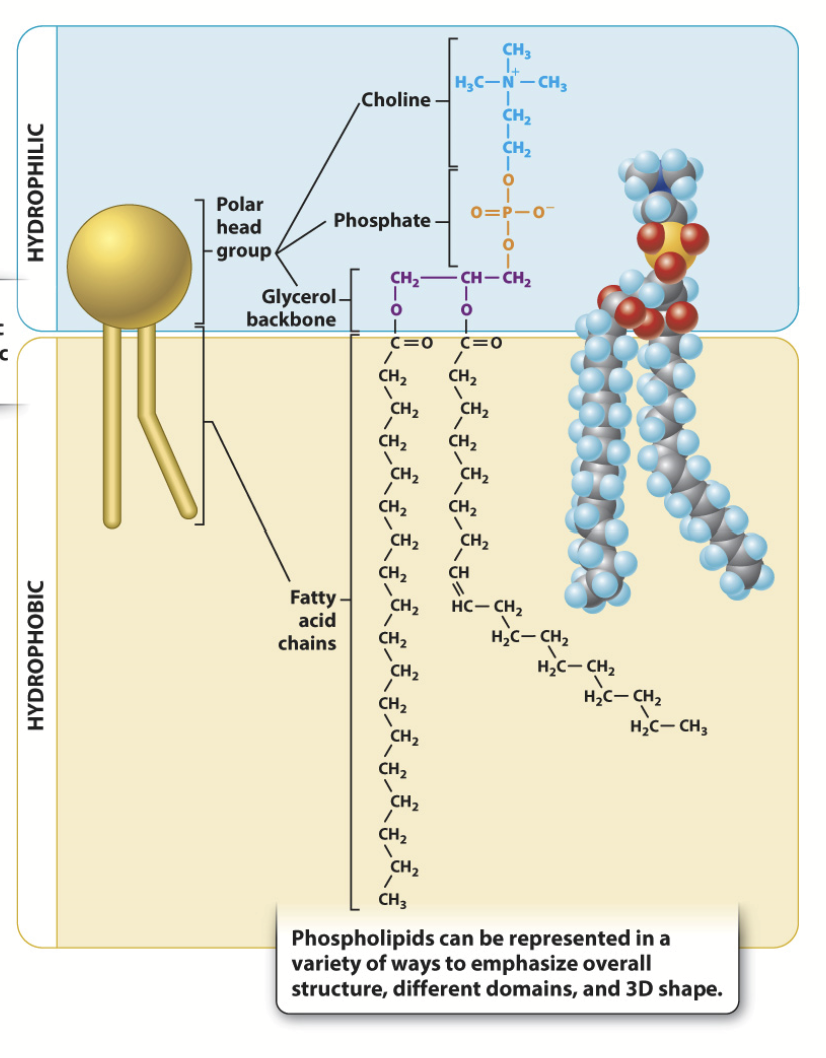
phospholipids
Made up of choline + phosphate (polar) + glycerol backbone + two fatty acid chains (nonpolar). Major lipid found in cell membranes.
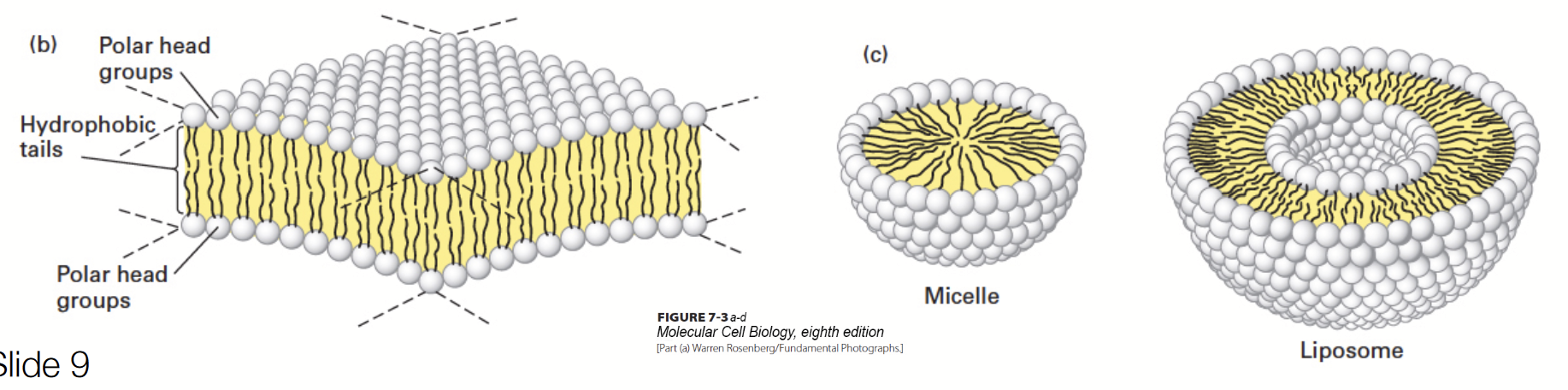
Lipids molecules spontaneously aggregate to bury their hydrophobic tails in the interior and expose their hydrophilic heads to water.
A molecule will always be in a conformation in which it is the most stable. Micelles are usually formed by fatty acids with only one hydrophobic chain.
Amphipathic molecules - they have both hydrophobic (non-polar) and hydrophilic (polar) regions.
Head groups are variable → lots of diversity!
amphipatic
Having both hydrophobic (non-polar) and hydrophilic (polar) regions.
e.g. phospholipids
hydrophobic
A molecule not attracted by water; repelled by water.
hydrophilic
A molecule attracted to water molecules and tends to be dissolved by water.
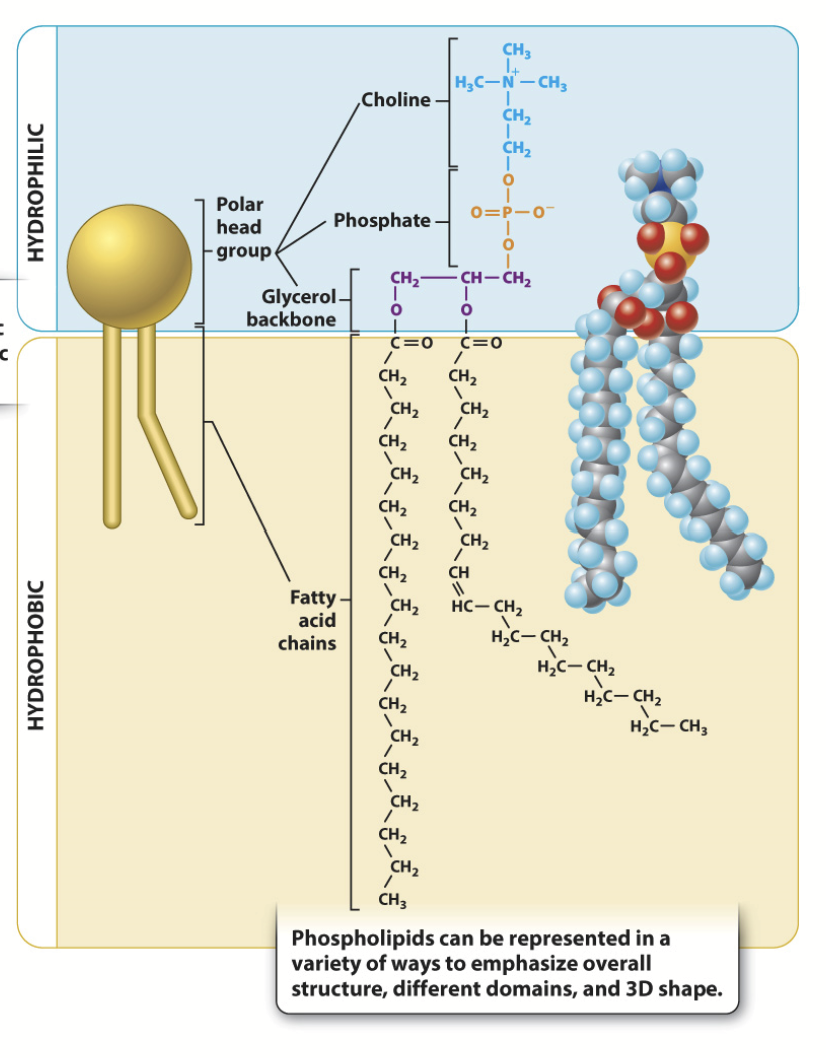
saturated phospholipid
The terms saturated and unsaturated refer to whether or not double bonds are present between the carbons in the fatty acid tails. Saturated tails have no double bonds and as a result have straight, unkinked tails.
Will pack together tightly, less membrane fluidity.
unsaturated phospholipid
The terms saturated and unsaturated refer to whether or not double bonds are present between the carbons in the fatty acid tails. Unsaturated tails have double bonds and, as a result, have crooked, kinked tails.
Will not pack together as tightly, resulting in higher membrane fluidity.
phospholipid synthesis
Occurs at the interface of the cytosol and outer endoplasmic reticulum membrane, which has all the molecular machinery (enzymes) for synthesis and distribution.
A multistep process, requiring the activity of many specialized effectors.
IN CYTOSOL: fatty acids (derived from carbohydrates via the glycolytic pathway) activated by attachment of a CoA molecule.
Activated fatty acids bond to glycerol-phosphate and are inserted into the cytosolic leaflet of the ER membrane via acyl transferase.
The phosphate is removed by a phosphatase enzyme.
A choline already linked to phosphate is attached via choline phosphotransferase.
Flippases transfer some of the phospholipids to the other leaflet.
Eventually a vesicle will come off from the ER containing phospholipids destined for the cytoplasmic cellular membrane on its exterior leaflet and phospholipids destined for the exoplasmic cellular membrane on its inner leaflet.
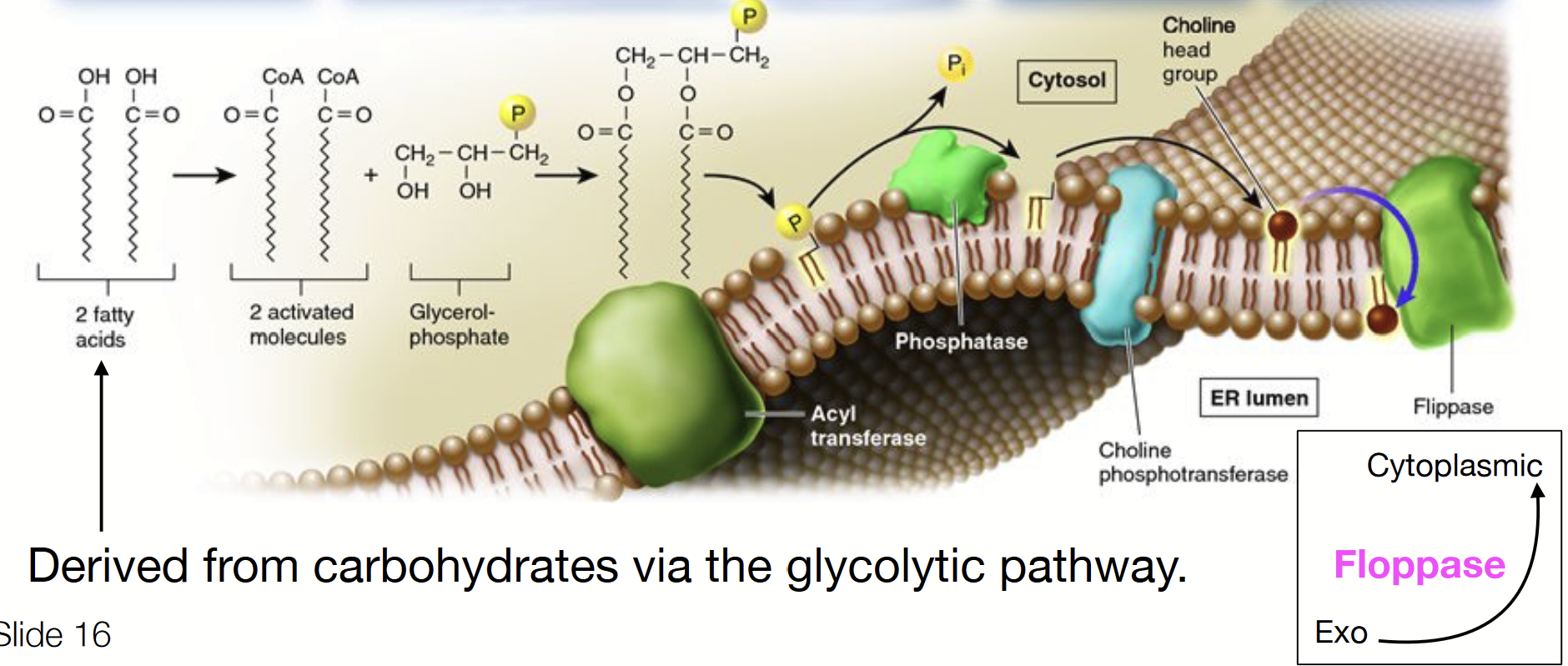
flippase
Flips a phospholipid from the cytoplasmic side to the ER lumen (exo) side.
floppase
Flips a phospholipid from the ER lumen (exo) side to the cytoplasmic side.
fluid
Individual lipid molecules move (unless they’re attached to the cytoskeleton).
mosaic
Diverse “particles” like proteins, carbohydrates, and cholesterol penetrate the lipid layer.
Fluid Mosaic Model
Proposed by Jonathan Singer and Garth Nicolson in 1972.
Plasma membrane = two-dimensional liquid that restricts the diffusion of membrane components.
Different proteins are embedded in the phospholipid bilayer.
Components are mobile.
Components can interact.
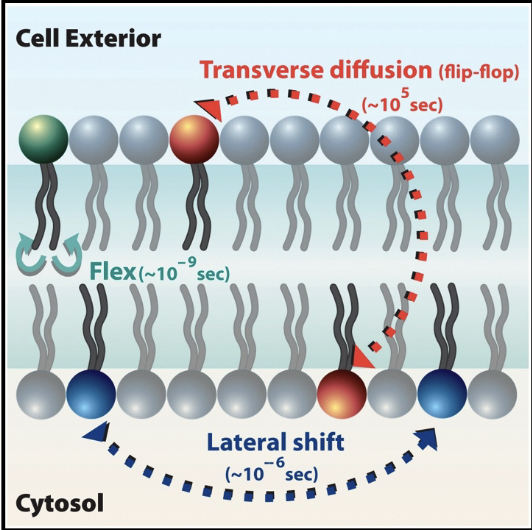
lipid movement in plasma membrane
Lipids: move easily, laterally, within leaflet; movement to other leaflet is difficult and slow.
protein diffusion in plasma membrane
Membrane proteins: diffuse within the bilayer.
Movement of proteins is restricted
Rapid movement is spatially limited
Long range diffusion is slow
Biochemical modification can alter protein mobility in the membrane (important for signal transduction!)
lipid rafts
Areas of lipids that are more fluid and allow proteins to move through faster.
membranes
Approximately 6 nm thick (with water), stable, flexible, capable of self assembly.
Islands of different phospholipids in different regions with different properties.
Different ones contain different types of lipids and proteins, giving them different functions. There are differences between cells as well as within an individual cell (e.g. inner mitochondria membrane v.s. myelin sheaths).
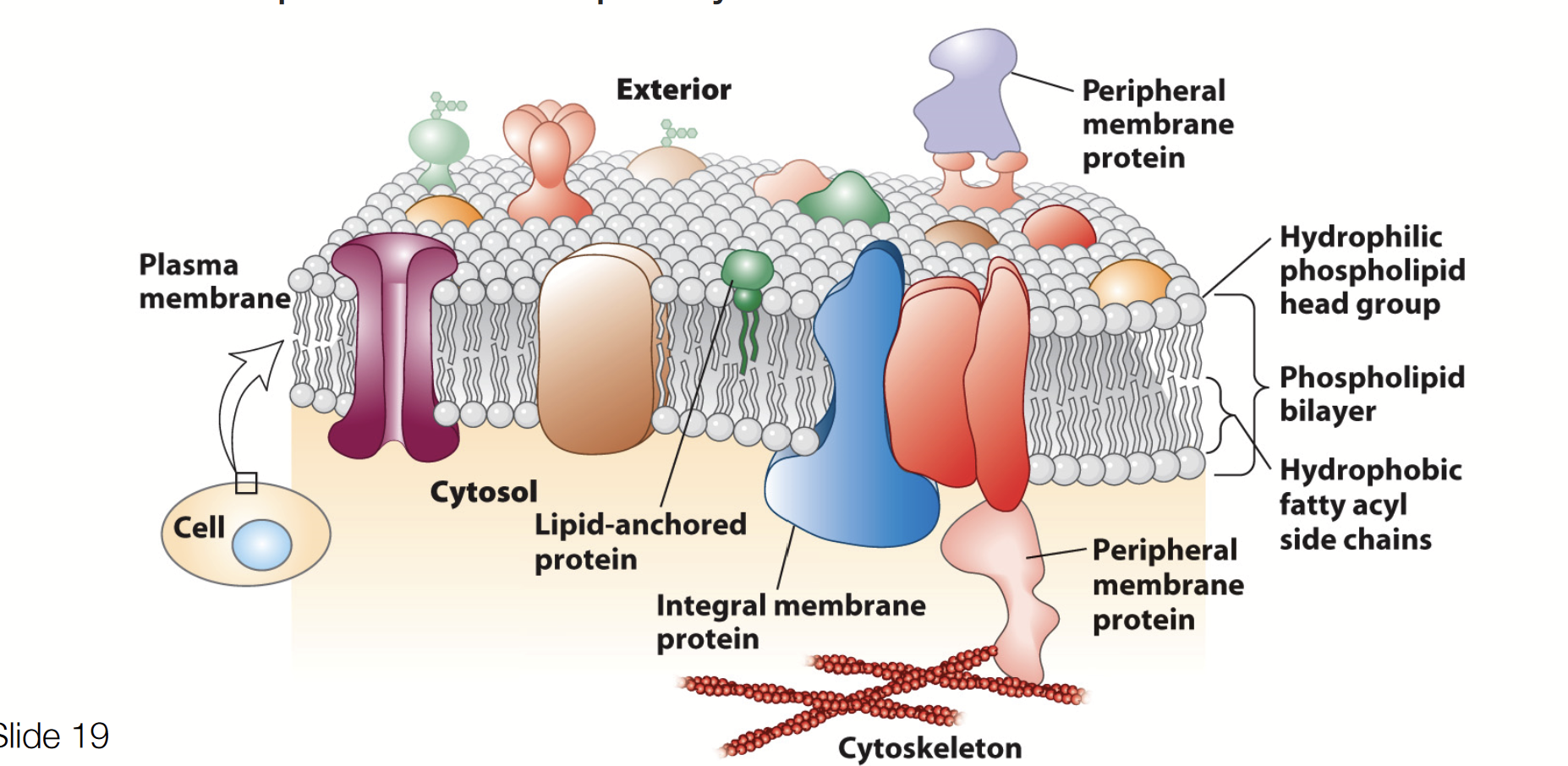
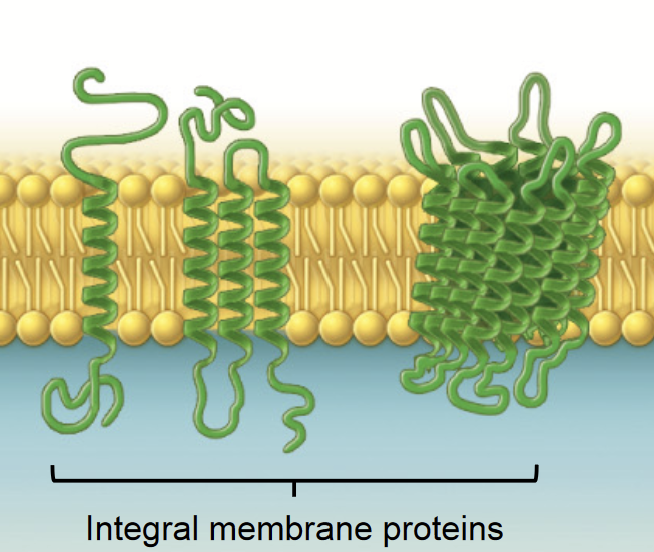
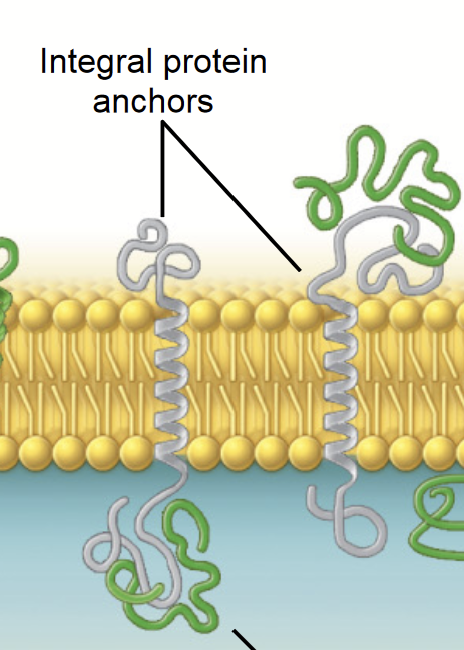
integral membrane protein
THREE CLASSES OF MEMBRANE PROTEINS
Membrane proteins that span the lipid bilayer - they are permanently embedded within the plasma membrane.
Transport of nutrients and ions
Cell-cell communication (gap junction)
Attachment
peripheral membrane proteins
THREE CLASSES OF MEMBRANE PROTEINS
Membrane proteins associated with the surfaces of the lipid bilayer. (a protein that is found temporarily attached to the cell or mitochondrial membrane).
Support, communication, enzymes, and molecule transfer in the cell
lipid-anchored proteins
THREE CLASSES OF MEMBRANE PROTEINS
Proteins that attach to a lipid in the bilayer, assume place alongside similar fatty acid tails.
target and stabilize the membrane association of proteins
fluidity
TEMPERATURE affects the fluidity (Iike butter!):
Warming INCREASES fluidity → liquid crystal
Cooling DECREASES fluidity → crystalline gel
FLUIDITY IS CRUCIAL TO CELL FUNCTION:
Determined by lipids! Unsaturated lipids increase fluidity (kinks that reduce tightness), saturated lipids reduce fluidity.
Lipid composition can be changed by: desaturation of lipids AND/OR exchange of lipid chains.
BALANCE BETWEEN ORDERED (rigid) STRUCTURE AND DISORDERED STRUCTURE ALLOWS:
Mechanical support and flexibility
Membrane assembly and modification
Dynamics interactions between membrane components (e.g. proteins can come together reversibly)
CHOLESTEROL modulates membrane fluidity:
Acts as a bidirectional regulator of membrane fluidity
At high temps, stabilizes the membrane and raises its melting point; at low temps, intercalates between the phospholipids and prevents them from clustering together and stiffening
cholesterol
Acts as a bidirectional regulator of membrane fluidity. At high temps, stabilizes the membrane and raises its melting point; at low temps, intercalates between the phospholipids and prevents them from clustering together and stiffening. Makes up ~30% of lipid membranes.
Its amphipathic structure allows it to pack tightly with phospholipids.
Alters packing and flexibility of lipids
If added to a liquid crystal (high temps) membrane, fluidity will DECREASE
If added to a crystalline gel (low temps) membrane, fluidity will INCREASE
(regulator, chooses the equilibrium/midpoint)
membrane
Physically separate cells from their external environment and define spaces within many cells that allow them to carry out their diverse functions.
Main components: lipids embedded with proteins and carbohydrates attached to lipids and proteins.
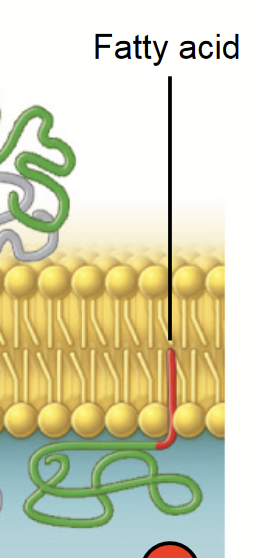
transmembrane protein
A peptide sequence that is largely hydrophobic (uncharged) and spans across the plasma membrane. This sequence permanently attaches the protein to the plasma membrane.
alpha helice
The most common protein structure element crossing biological membranes, twisted into a coil.
Transmembrane domain made of hydrophobic amino acids.
Most frequently used for sequence-specific interactions in protein–DNA interfaces.
glycine (Gly)
One of nine amino acids with hydrophobic side chains.
Gly
alanine (Ala)
One of nine amino acids with hydrophobic side chains.
Ala
valine (Val)
One of nine amino acids with hydrophobic side chains.
Val
leucine (Leu)
One of nine amino acids with hydrophobic side chains.
Leu
isoleucine (Ile)
One of nine amino acids with hydrophobic side chains.
Ile
proline (Pro)
One of nine amino acids with hydrophobic side chains.
Pro
phenylaline (Phe)
One of nine amino acids with hydrophobic side chains.
Phe
methionine (Met)
One of nine amino acids with hydrophobic side chains.
Met
tryptophan (Trp)
One of nine amino acids with hydrophobic side chains.
Trp
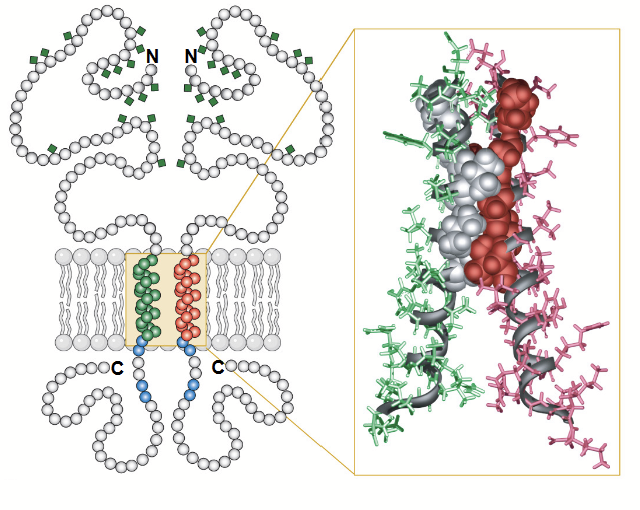
tetraspanins (TM4SFs)
A family of membrane proteins found in all multicellular eukaryotes. They have four transmembrane alpha-helices and two extracellular domains: one short (EC1) and one longer (EC2). Contains an N-terminus on the EC1 side and a C-terminus on the EC2 side.
Some can be glycosylated (attachment of a carbohydrate molecule) on the long extracellular loop (EC2).
Play a role in cell adhesion, motility, proliferation, and more.
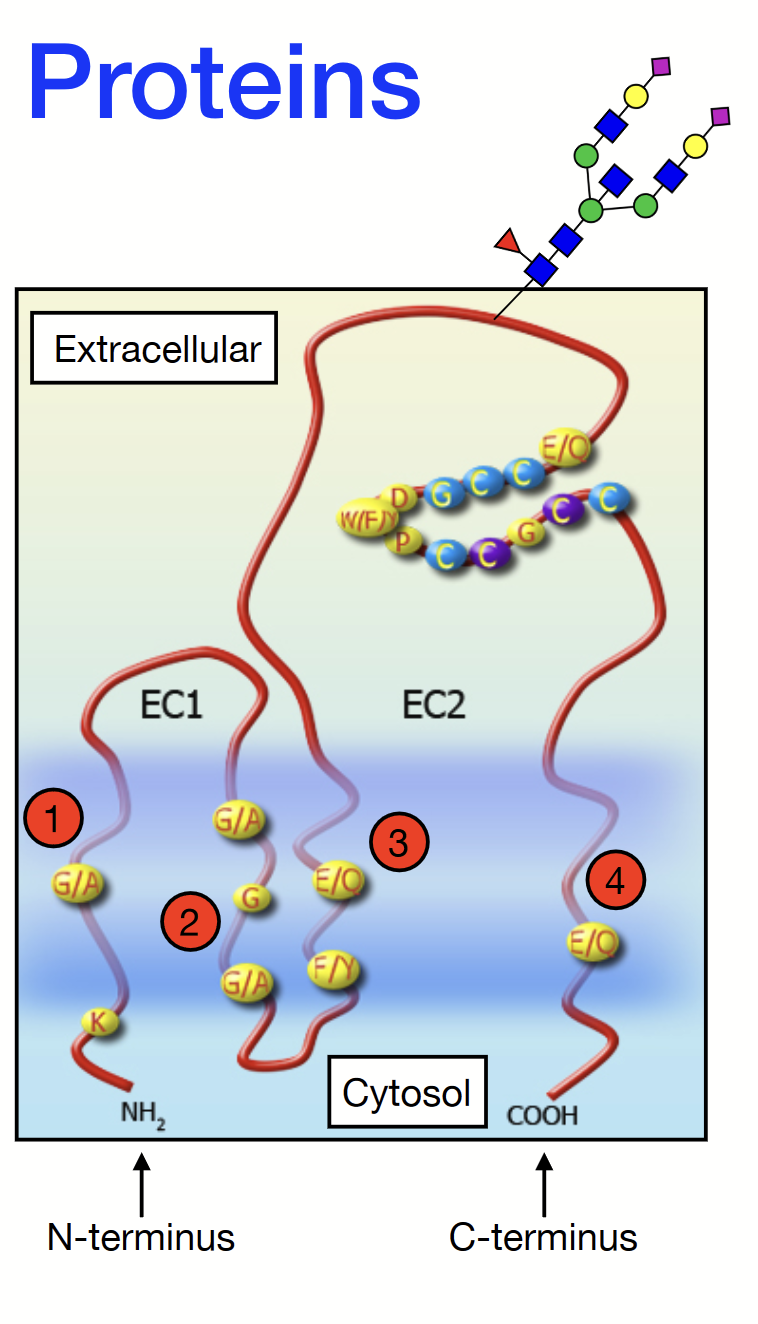
glycosylation
The process by which a carbohydrate is covalently attached to a target macromolecule, typically proteins and lipids.