Hybridization and stringency
1/8
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
9 Terms
What is hybridization
Hybridization = the binding of a single-stranded molecule of DNA to a complementary single-stranded target DNA molecule
- “sticking together” or “anneal”
- Occurs between PCR primers and the target template
- Usually used to describe binding of special oligonucleotides/probes
What is stringency
Stringency = the combination of conditions under which the target is exposed to the probe
- Conditions of HIGH stringency are more demanding of probe binding (more specific)
- Conditions of LOW stringency are more forgiving (less specific)
What factors affect the stringency of a probe binding to its target
1. Temperature = as temp INCREASES, stringency INCREASES (hotter = only [erfect matches can stay bound)
- Creates a more challenging environment for the probe to bind, only allowing perfectly-matched probes to hybridize
- If temp is too low, the probe will find less-than perfectly matched sequences to bind
- If temp is too high, the probe cannot form its bond and will fail to hybridize
2. Salt concentration = as salt conc. Increases, stringency DECREASES
- Because salt promotes DNA binding (stabilizes DNA) it promotes any kind of binding, even non-specific kinds
- By lowering salt, only the most specific probe target hybrids can stay stable
- Too little salt means no hybridization at all
3. Denaturant in buffer = INCREASE denaturing/destabilizing agents will INCREASE stringency
- Agents = formamide, urea, TMAC (all break up secondary structures)
- Presence of formamide increases stringency because it promotes denaturation even at lower temps (makes it harder for probe to bind => more specific binding)
- When the probe and target are prone to denaturation, only the most specific probe-target hybridization can occur
4. Time
a. Hybridization time = INCREASED time of hybridization (incubation of probe and DNA) DECREASES stringency
- The longer the probe mixes with the DNA, the more likely it will bind non-specifically
b. Wash time = INCREASED time of washing the blot INCREASES stringency
- The longer the wash time, the more likely it will detach weak non-specific bonds, leaving only specific probe target complexes (washing too long can knock off the targets)
5. Length of probe = a LONGER probe will bind under MORE stringent conditions
6. GC content of probe = HIGHER GC probe will bind under MORE stringent conditions
- Presence of formamide will allow GC-rich probes to denature and hybridize at lower temperatures than without formamide
Conditions that are not stringent may be more likely to produce non-specific products for a GC-rich probe than for an AT-rich probe
What is a probe
Probe = a single-stranded fragment of nucleic acid attached to a signal-producing moiety
- Used in molecular methods to identify a sequence of interest within a large mixture of nucleic acid
- Hybridizes specifically with the target DNA/RNA to be analyzed
- Can be denatured DNA/RNA
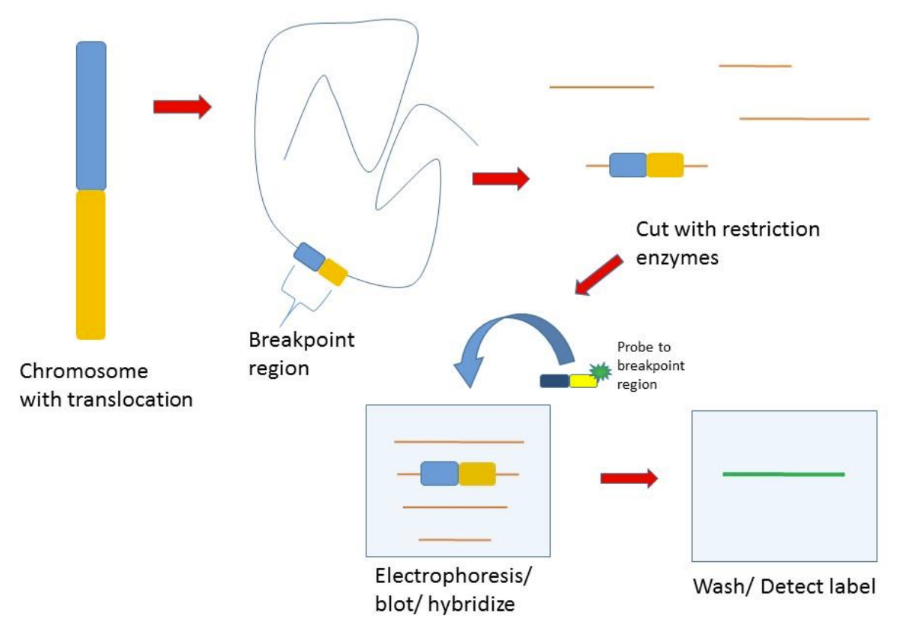
How does the size of the probe determine hybridization
DNA probes:
Size of the probe determines hybridization
- The length of the probe helps determine the specificity of the hybridization reaction
- When probing the entire genome, longer probes are more specific because they must match a longer sequence on the target (unlikely to be found elsewhere in the genome)
o Shorter probes are more likely to be found in multiple locations in the genome
§ Because higher chance to find more identical sequences
- Short probes play an important role in mutational analysis, often with PCR amplification, because shorter probes can be sensitive to single-base mismatches
o Probes will only bind if there is mutation
What are probe labels
Probe labeling: the probe must be labeled and generate a detectable signal (how to detect if probe has bound or not)
- Classically = radioactive labels like 32P
o Introduce nucleotides containing radioactive phosphorus into the probe
- Common = non-radioactive methods (exclusively used in medical laboratories)
o Usually fluorescently tagged
How does the length and location of the probe affect binding/PCR
Probe design
- Length of probe determines specificity (depending on application)
o Genomic DNA (no PCR) = long probes are more specific
o PCR amplified DNA (smaller pieces of DNA) = short probes are better to find mutations
- Location/sequence of the probe can affect binding performance
o Internal complementary sequences will fold and hybridize with itself rather than staying single-stranded and linear
What is the relationship between stringency and PCR
The conditions of PCR determine stringency of PRIMER binding and thus determine the specificity of the PCR reaction (do you produce your target band and ONLY your target band)
IMAGE: Don't need to make all these changes at once. If you had many non-specific products start on the left or if you had no products start on the right
The conditions you use for probe incubation determine stringency of hybridization
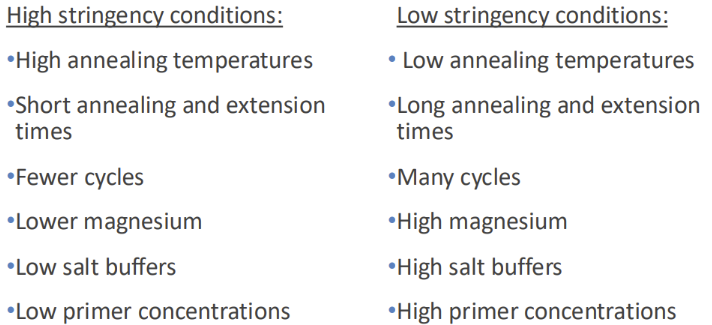
What do different probe results mean (in relation to stringency)
1. If conditions make it TOO HARD for the probe bind, it won’t bind and send fluorescent signal EVEN IF the mutation is present
- Stringency too high
2. If conditions make it TOO EASY for the probe to bind, it will bind and send fluorescent signal EVEN IF the mutation is NOT present
- Stringency is too low
3. Need conditions for probe binding (stringency) to be JUST RIGHT to produce specific probe binding when the mutation is present, and no probe binding when there is no mutation