06_Evolutionary Analysis Lecture 6: Population Genetics 2
1/14
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
15 Terms
What is differential genotype selection?
Differential genotype selection is a concept in evolutionary biology and genetics that refers to the process by which certain genotypes (specific genetic compositions) have a higher probability of being passed on to the next generation compared to others. It is a fundamental mechanism driving the process of natural selection.
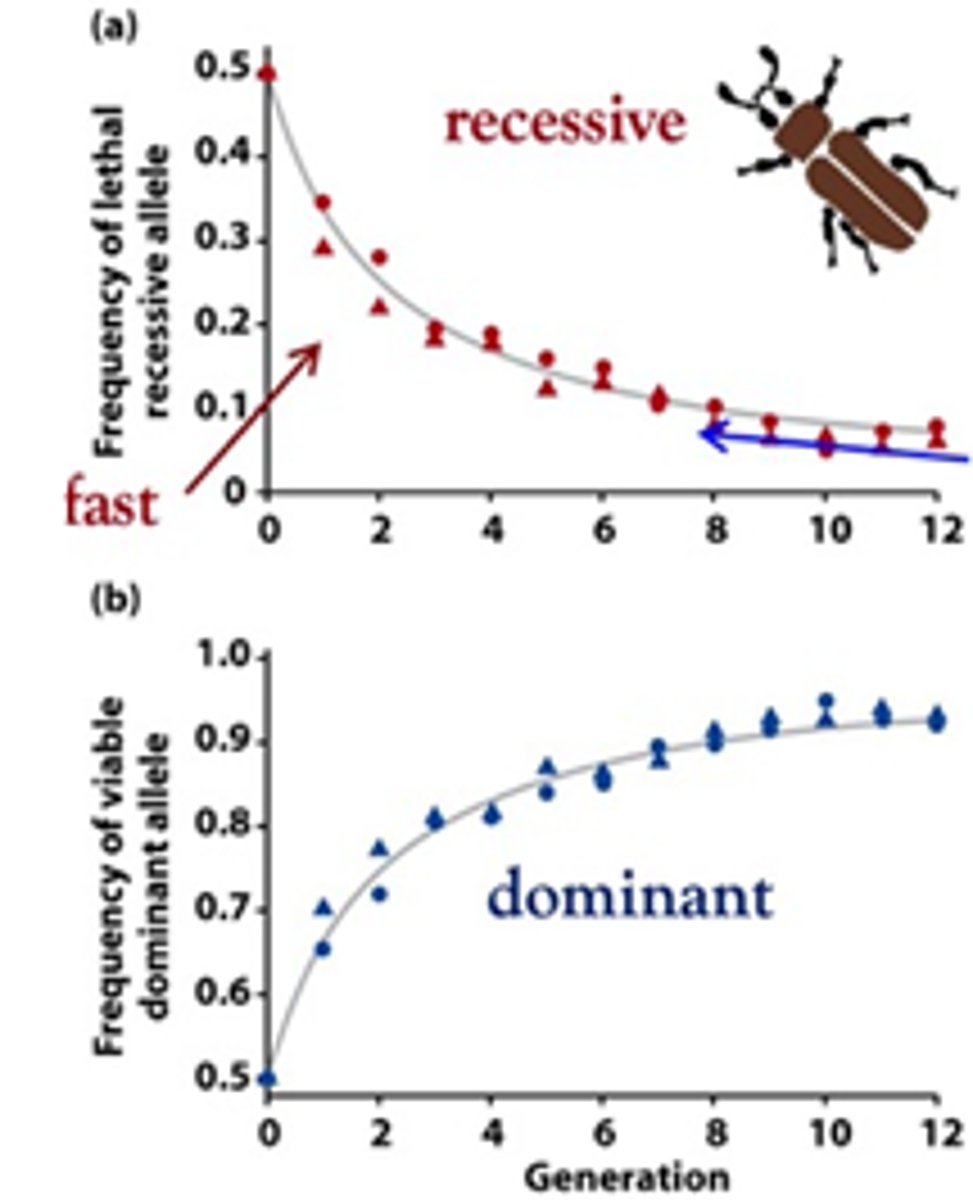
Explain how differential genotype selection acts in a case where recessive allels are disadvantageous.
After several generations the recessive allele frequency is approaching 0%, while the frequency of the dominant allele is approaching 100%. The slowing down is because the number of recessives hidden in heterozygotes increases in time and selection against the lethal allele is less and less efficient. For these reasons, recessive disease alleles are difficult to purge from sexual populations
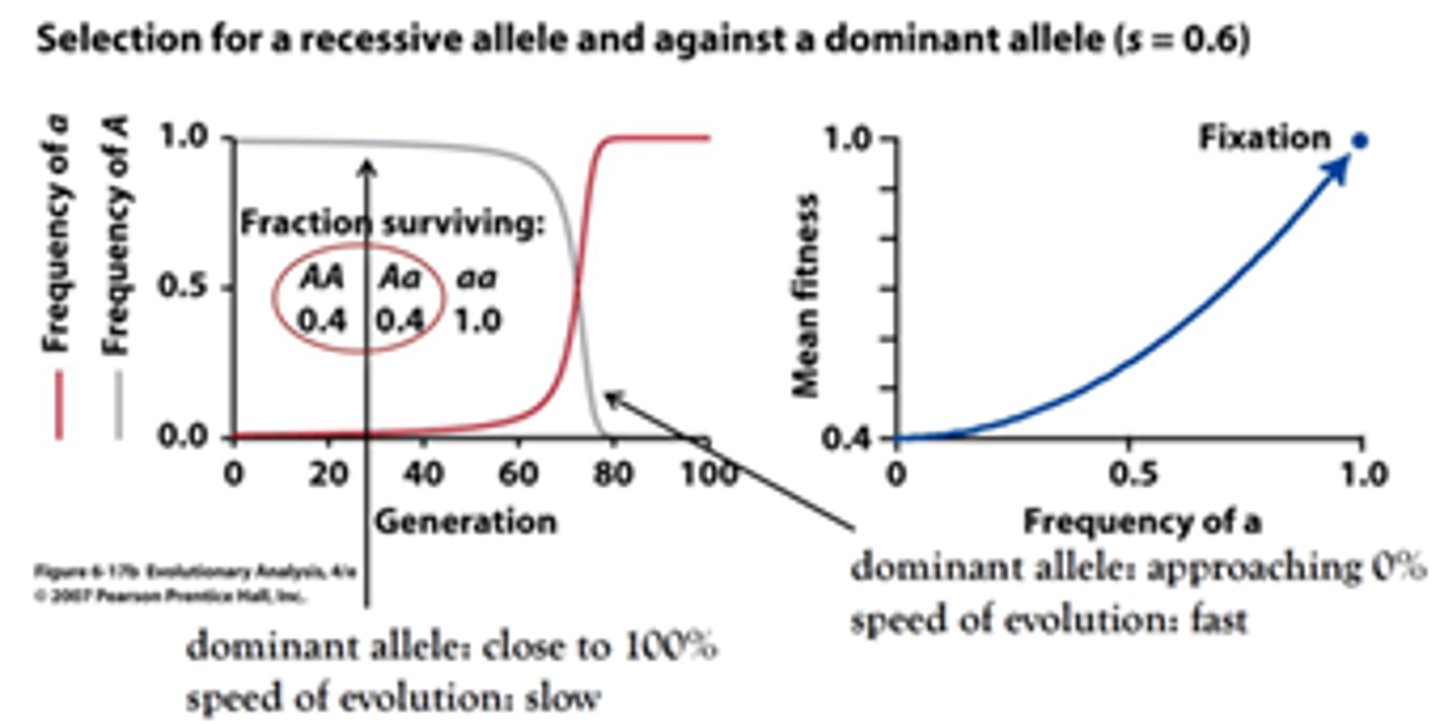
Explain how differential genotype selection acts in a case where dominant allels are disadvantageous.
The evolution in the beginning is slow because at first most recessives are hidden in heterozygotes. Selection for recessives is very efficient only later, when homozygote, recessive frequency increased. Therefore, dominant disease alleles are easy to purge from sexual populations.
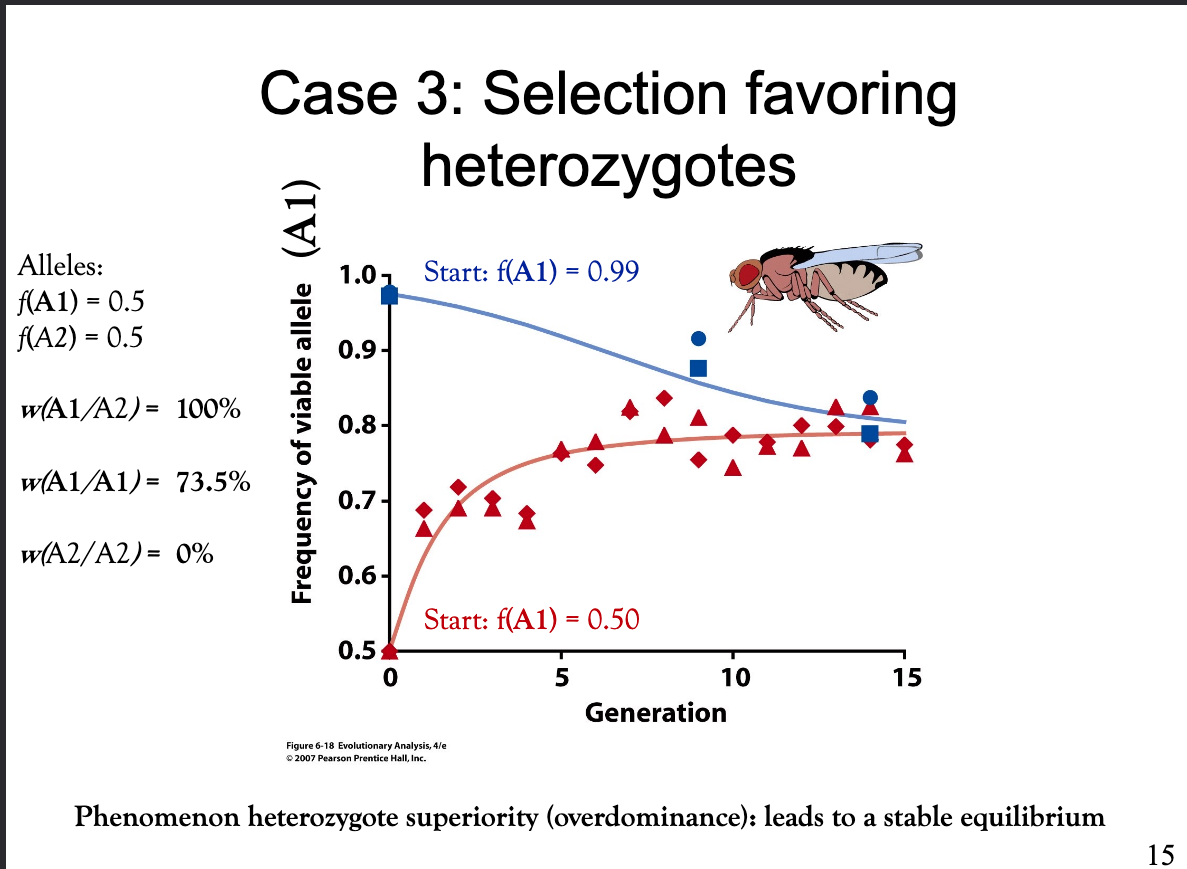
Explain how differential genotype selection acts in a case where heterozygotes hava an advantage.
Heterozygotes have the highest fitness. No matter at which allele frequency you start the experiment, the same equilibrium frequency is reached. This is the phenomenon of heterozygote superiority (overdominance) that leads to a stable equilibrium.
No genetic diversity is lost.
Bild: 1) Dominant, viable allele: A1
2) Recessive, harmful allele: A2
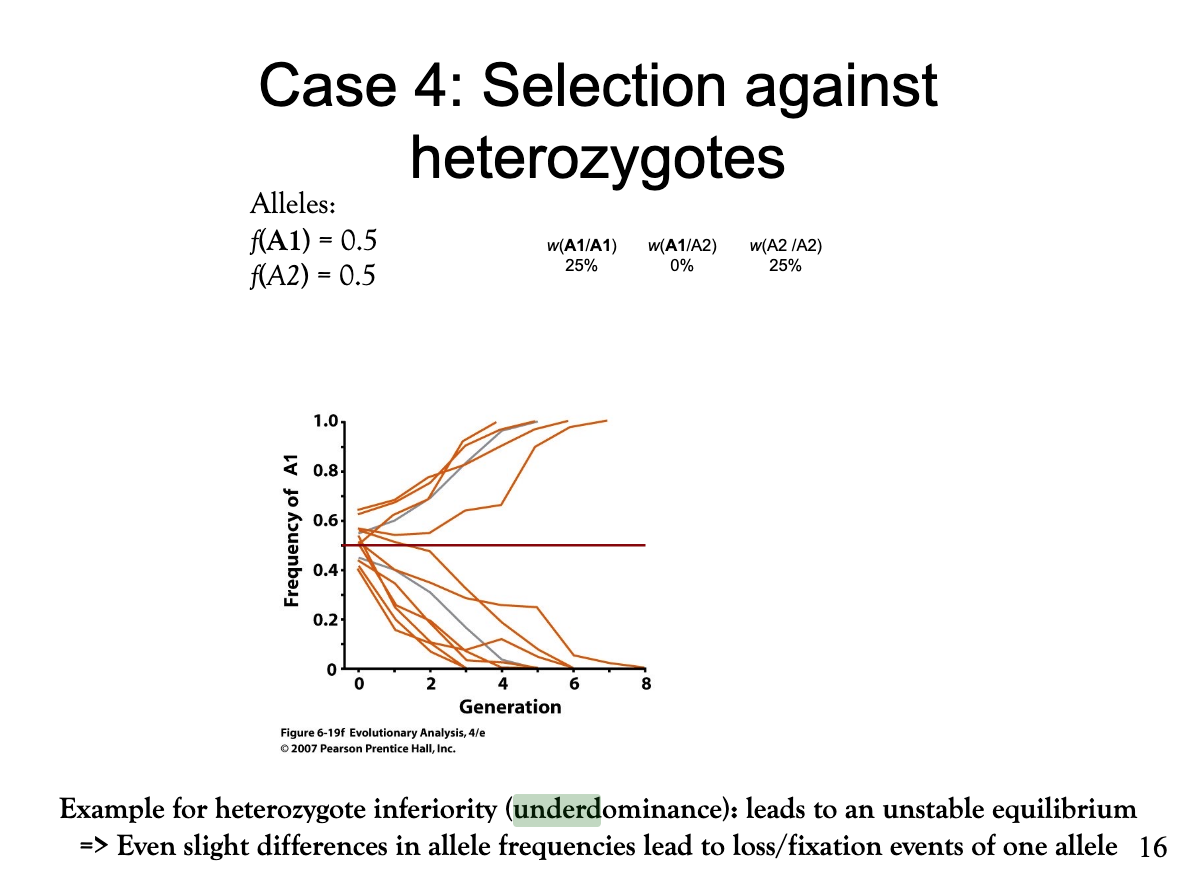
Explain how differential genotype selection acts in a case where heterozygotes hava a disadvantage.
In this case there is a critical threshold. Alleles with frequencies above it are fixed and alleles below it are lost. This threshold is set by the fitness difference of the homozygote phenotypes. In this example, both homozygotes are equally fit, thus the threshold is at a frequency of 50%. If both phenotype frequencies equal to this 50%, they are in an equilibrium. However, the slightest deviations can lead to allele fixation or loss. This is an example of heterozygote inferiority (underdominance) that leads to an unstable equilibrium.
Bild: 1) Dominant, viable allele: A1
2) Recessive, harmful allele: A2
What is genetic drift and what are it's properties?
Genetic drift is differential reproductive success that just so happens (chance process). To compare, selection is differential reproductive success that happens for a reason (deterministic process).
Genetic drift is:
- random survival of alleles due to sampling errors
- most powerful in small populations
- Strongest when natural selection is weakest
- cannot produce adaptations
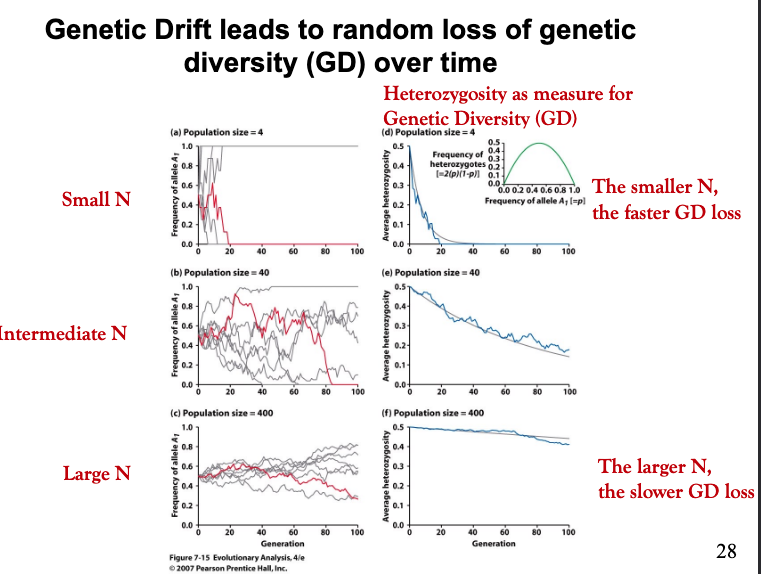
What effect does population size have on genetic drift?
The population size has an important effect on the rate of change by drift. Replicate simulated populations show that each population follows a unique random path and each drifts toward allele fixation or loss. However, evolution by genetic drift occurs more rapidly in small populations but genetic drift can also change frequencies in large populations over long periods of time.
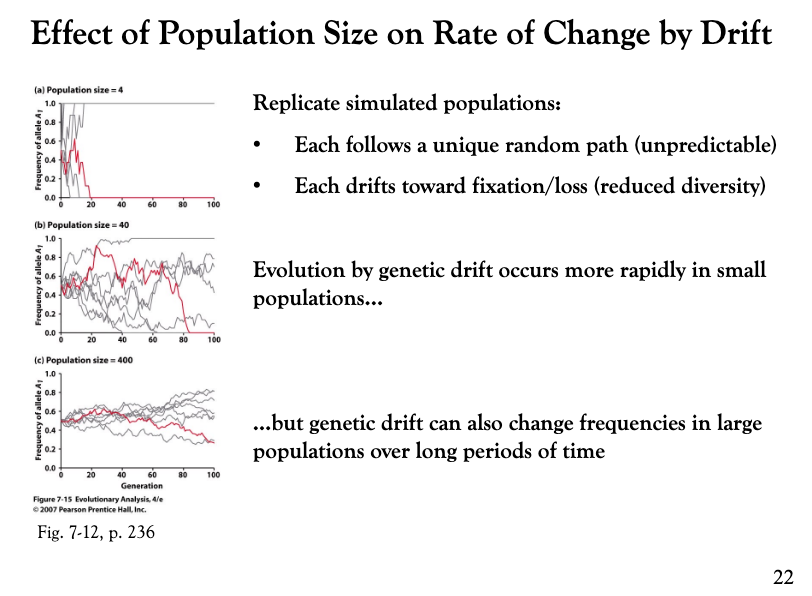
What is the issue with genetic drift?
The issue with genetic drift is that it reduces heterozygosity over time. Heterozygosity is an estimate of genetic variation, that is necessary for efficient natural selection.
How can gene flow be measured?
Differentiation index or FST-value
high FST = great differences between populations
low FST = few differences
Species are divided into subpopulations due to temporary or permanent barriers in space and time. FST measures the degree to which separate populations are genetically distinct due to the absence of gene flow.
Differences in gene flow (i.e. migration) between subpopulations will lead to different distribution of genetic variation across the populations, hence different degrees of population structure (FST).
What is non-random mating? What is the result and in which two extreme forms can it apear?
Non-random mating is evolutionary force that does not change allele frequencies but genotype frequencies determening the fate of variation.
Leads to a deviation from the Hardy Weinberg equilibrium.
There are two extreme forms of non-random mating:
1. inbreeding when relatives (like and like) mate among each other (more common)
2. outbreeding, opposites attract each other -> inbreeding is actively avoided (less common)
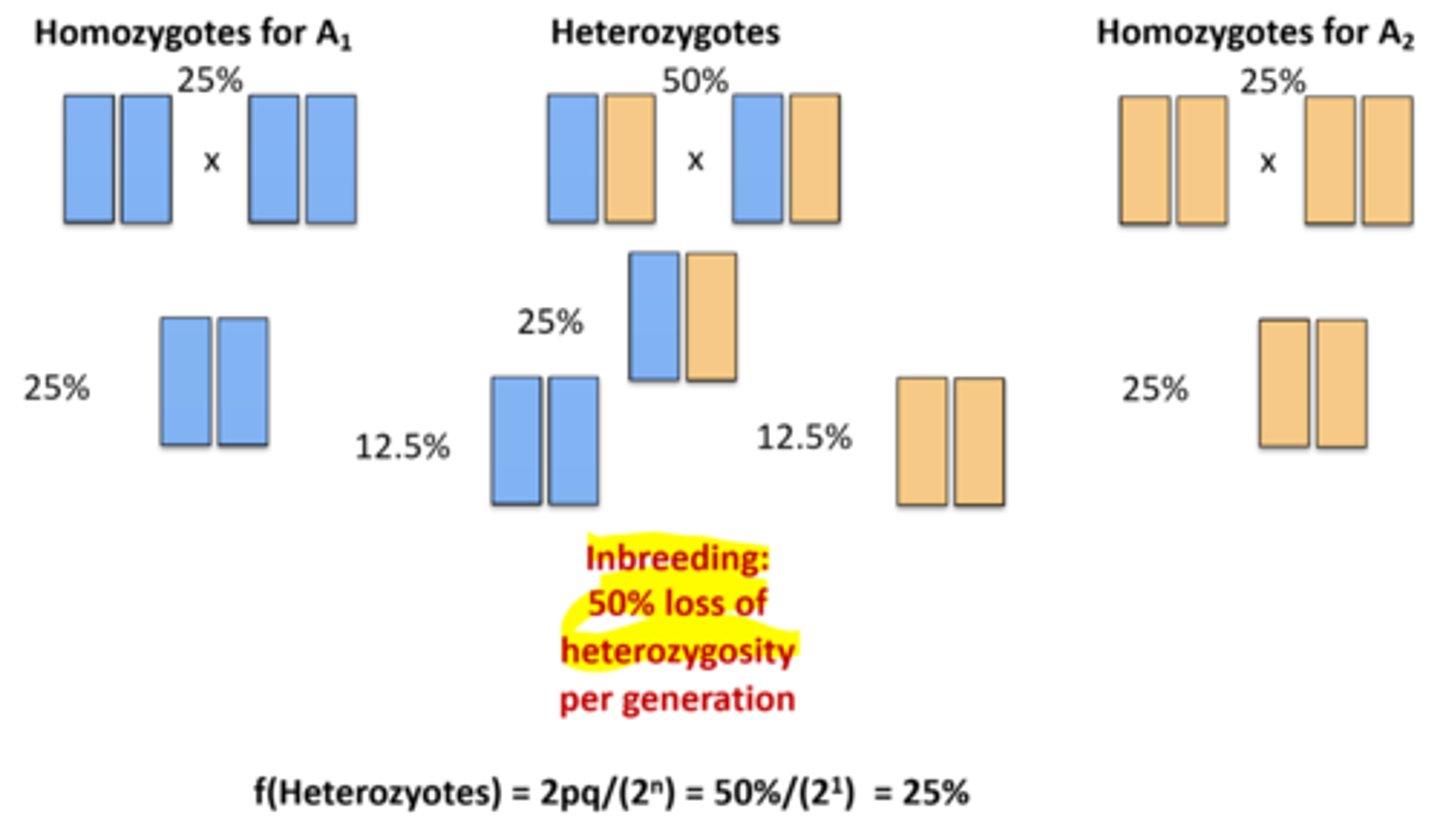
What is selfing and how can the number of heterozygotes after n generations be calculated?
Selfing is the strongest form of inbreeding in which an organism fertilizes itselfe. This leads to a reduction of heterozygosity of 50% each generation.
f(Heterozygotes) = 2pq/2ⁿ
Fitness, Selection coefficient
1 = high fitness
0 = low fitness

* Peter Buri's fruit fly experiment:
** you know which evolutionary force was tested for in this experiment
** you can describe the changes in allele frequency-distribution over time
** you understand the consequences genetic drift on genetic diversity
1. Evolutionary Force Tested:
Genetic drift, showing random changes in allele frequencies over generations.
2. Changes in Allele Frequency:
Started with heterozygous populations (bw/bw75).
Over 20 generations, allele frequencies shifted:
Initially bell-shaped distribution.
Ended with U-shaped distribution (30:28 fixed-to-loss ratio).
3. Consequences on Genetic Diversity:
Random loss of genetic diversity; smaller populations lost diversity faster.
Drift acts randomly, not adaptively, reducing heterozygosity over time.
Chance & Determinism in Evolution
proportionality from drift to selection
Mutational input is random (randomly increases genetic diversity).
Evolution by selection (feeds on mutation and) is deterministic (leads to adaptation).
Evolution by genetic drift is random (does not lead to adaptation).
è The Power of drift is inversely proportional to the power of selection.