Lecture 11: Linkage and Mapping
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
21 Terms
Describe an experiment to test whether two genes assort independently.
Question: Do the genes for flower color and pollen shape in sweet peas assort independently?
Cross homozygous strains of purple flowers/long pollen with red flowers/round pollen to generate F1 individuals with purple flowers/long pollen
We expect to see the 9:3:3:1 Mendelian ratios in the F2 generation, due to the results of a dihybrid cross where the two genes in question assort independently
However, we observe drastically different ratios:
284 purple/long
21 purple/round
21 red/long
55 red/round
Conclusion: The genes for flower color and pollen shape do not assort independently because the F2 generation does not display the expected 9:3:3:1 Mendelian phenotypic ratios.
Genetic linkage
DNA sequences that are close together on a chromosome tend to be inherited together during the meiosis phase of sexual reproduction
The closer two genes are in physical space on a chromosome, the more linked they are → linked genes do not assort independently during meiosis
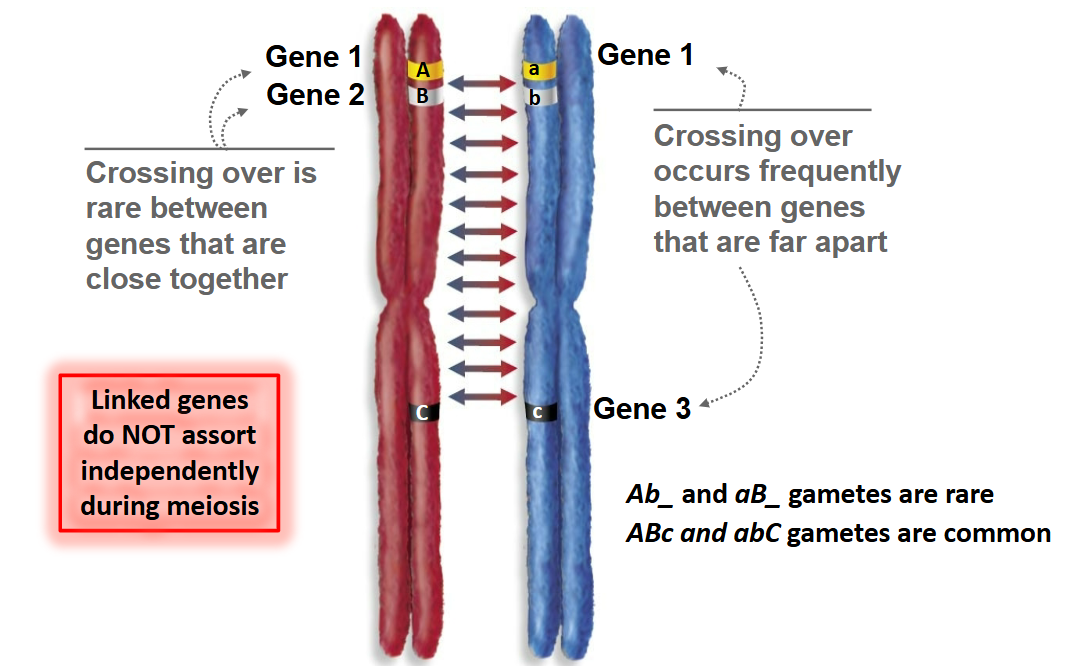
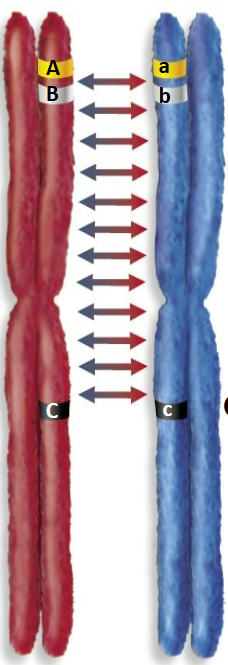
Which gametes are common and which are rare during crossing over?
ABc and abC gametes are common
Ab_ and aB_ gametes are rare
Crossing over is rare…
between genes that are close together.
Crossing over occurs frequently…
between genes that are far apart.
Compare unlinked and completely linked genes.
If genes are completely linked with each other, there is no crossing over
If genes are unlinked, they assort independently and crossing over occurs
However, it’s important to remember that linkage is a spectrum, not a binary concept
Independent assortment → the random orientation of homologous chromosomes during Anaphase I, which leads to different combinations of chromosomes in each gamete
Crossing over → the exchange of genetic material between homologous chromosomes, creating new combinations of alleles on a single chromosome
Crossing over does not occur between sister chromatids! Because sister chromatids are genetically identical, there is no cellular or genetic purpose to do so
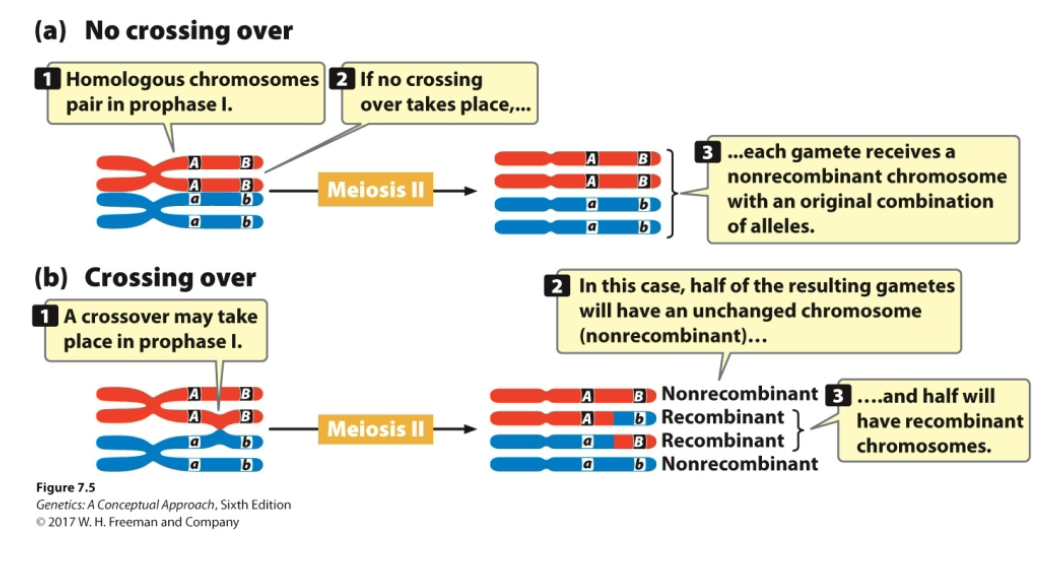
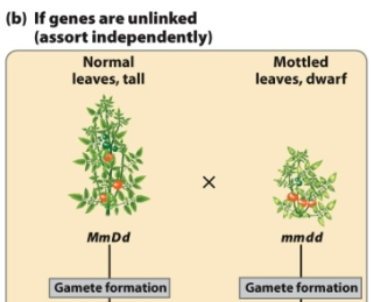
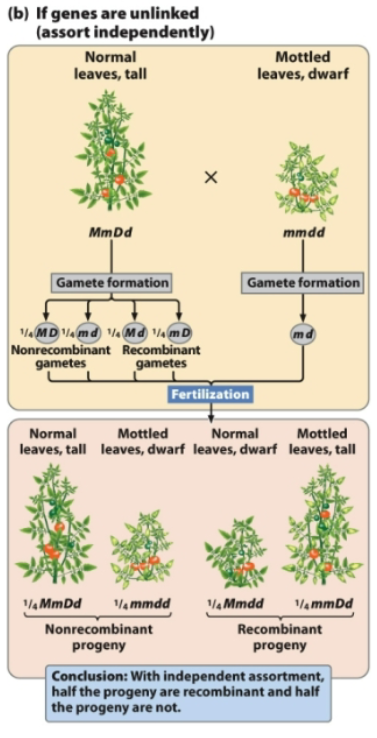
If a plant with genotype MmDd has unlinked genes that assort independently, what gametes does it form?
From MmDd, 4 combinations of gametes are possible
MD and md (nonrecombinant gametes)
Md and mD (recombinant gametes; crossing over has occurred)
With independent assortment, half the progeny are recombinant and half the progeny are not
When crossing over happens in unlinked genes, there is an equal probability of producing all four types of gametes (equal probabilities of recombinant and nonrecombinant offspring)
When crossing over happens in linked genes, there is an unequal probability of producing all four types of gametes (unequal probabilities of recombinant and nonrecombinant offspring)
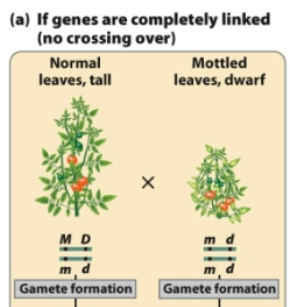
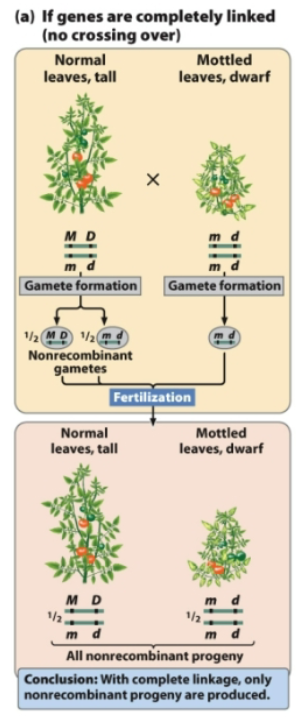
If a plant with genotype MmDd has linked genes that do not assort independently, what gametes does it form?
From MmDd, 2 combinations of gametes are possible
MD and md (nonrecombinant gametes)
With complete linkage, only nonrecombinant progeny are produced
Crossing over
The exchange of genetic material between homologous chromosomes → creates recombinant chromosomes
Occurs during Prophase I (the beginning of Meiosis I)
Coupling (cis) configuration of linked genes
One chromosome contains both wild-type alleles, one chromosome contains both mutant alleles
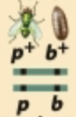
Repulsion (trans) configuration of linked genes
Wild-type allele and mutant allele are found on the same chromosome
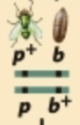

Predict the progeny from this test cross and identify the recombinant and nonrecombinant progeny.
Nonrecombinant gametes: (p+b+) and (pb)
Recombinant gametes: (p+b) and (pb+)
Fertilization with (pb) gametes produces:
p+pb+b (NR)
ppbb (NR)
p+pbb (R)
ppb+b (R)
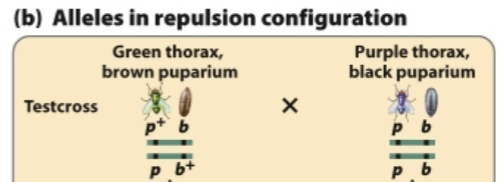
Predict the progeny from this test cross and identify the recombinant and nonrecombinant progeny.
Nonrecombinant gametes: (p+b) and (pb+)
Recombinant gametes: (p+b+) and (pb)
Fertilization with (pb) gametes produces:
p+pbb (NR)
ppb+b (NR)
p+pb+b (R)
ppbb (R)
Describe the steps involved in recombination mapping.
1) Start with an organism that is heterozygous for alleles in two or more genes
2) Do a testcross, crossing the heterozygote to a homozygous recessive individual
3) Count the number of recombinant and nonrecombinant (parental) genotypes and phenotypes among the offspring from the testcross
4) Calculate the map distance between genes based on the observed recombination frequency
Map distance (% or cM or map units) = (# recombinants / total progeny) x 100
1 centiMorgan (cM) = 1% recombination observed = 1 map unit
The following testcross produces the progeny shown: AaBb x aabb → 40 Aabb, 40 aaBb, 10 AaBb, 10 aabb. What is the percent recombination between the A and B loci?
Since there are less AaBb and aabb, these are the recombinant offspring
% recombination = (10+10) / (10+10+40+40) = 20/100 = 0.2 × 100 = 20% recombination
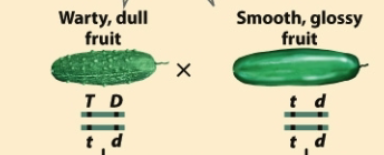
The recombination frequency between T and D alleles is 5%. How many Ttdd (warty glossy cucumber plants) will you get out of 1000 plants?
5% RF = 0.05 = x / 1000, so x = 50 total recombinants
The two possible recombinant gametes are tD and Td, creating ttDd and Ttdd
Number of Ttdd plants = 50/2 = 25 expected Ttdd plants
Describe the three types of crossovers possible with three linked loci, A B C.
A B C
a b c
A single crossover between A and B
a B CA b c
A single crossover between B and C
A B ca b C
A double crossover
A b Ca B c
Three-point testcross
A genetic mapping technique that establishes the order of three loci on a chromosome
Provides more accurate map distances than a regular testcross
Can detect double crossovers
Describe how to identify the order of genes on a chromosome.
Identify the double-crossover (DCO) offspring — there should be two groups, and the double recombinant progeny are always the fewest in number.
Identify the non-recombinant (NR) offspring — there should be two groups, and the non-recombinant offspring are the greatest in number.
Compare the DCO progeny with the NR progeny → the allele that is different in the DCO progeny is the middle allele on the chromosome. Note that you cannot tell which gene is on which end, so the sequence could be A-B-C or C-B-A!
Describe how to calculate the recombination frequencies for a three-point testcross.
Identify the DCO progeny first → there should be two groups of DCO progeny, and they should be the smallest out of all the groups.
Identify the SCO progeny → there should be four groups of SCO progeny, and they should contain more individuals than the DCO groups, but less than the nonrecombinant groups.
Identify the NR progeny → there should be two groups remaining, and these groups should be the largest out of all the groups.
Compare the DCO and NR progeny to determine the middle allele. Then, use the formula for recombination frequency: (# of SCO + # of DCO) / total progeny x 100.
Describe how to calculate the number of progeny from recombination frequencies.
If you are given a linkage map A — 5cM — B — 12cM — C, that means the A-B recombination frequency is 5%, and therefore the probability of a crossover in the A-B region (including single crossovers and double crossovers) is 5%.
First calculate the number of DCO progeny: SCO (A-B) x SCO (B-C) x total progeny / 2. This formula works because the probability of a double crossover is the probability of a crossover in the first region multiplied by the probability of a crossover in the second region, because both the crossovers are independent and must occur simultaneously. This formula gives the number of progeny in each DCO group.
Then use the formula for recombination frequency to find the number of SCO progeny: (# of SCO + # of DCO) / total progeny x 100. Make sure to calculate the number of DCO progeny first to avoid double-counting them!
Lastly, subtract the number of recombinants (SCO + DCO) from the total progeny and divide by 2 to find the number of nonrecombinants in each category.