Applied Lectures #4-6: Theme 2 Module 3 - Theme 3 Module 4
1/135
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
136 Terms
Development of eukaryotic organisms depend on
Gene regulation and signal sequences
Gene regulation significance
Allows for the creation of various specialized cells derived from a single-celled zygote
Transcription factors
Proteins that bind to specific DNA sequences to control transcription. Work together with other proteins that cause changes in gene expression in successive cell divisions
Transcription factors significance
Determine the pathway that a specific cell will follow and determine the final mature/differentiated cell type
Eukaryotic vs Prokaryotic transcription
Similar:
RNA Pol
Activating/repressing proteins
Different:
Compartmentalization
Each gene controlled by its own promoters and enhancers (E)
Compact chromatin (E)
Chromatin remodeling
Unraveling of chromatin to initiate transcription
Why do we need chromatin remodeling
DNA in chromatin form is tightly wound, and thus genes are not expressed, need chromatin remodelling for transcription to occur
Chromatin structure
DNA wound around histones to form nucleosomes. 1 Nucleosome = 8 histones = 150 BP’s. DNA & histones bonded through + charged histone tails and - charged phosphates in DNA
Chromatin remodelling process
Transcription factor binds to enhancer site and recruits coactivator enzyme HAT
HAT attaches acetyl groups to lysine a.a’s along + charged histone tail to reduce + charge
Weakened + charge causes bond to weaken, allowing DNA to be unwound and transcribed
Chemical modifications
Acetylation of lysine
Methylation of lysine & arginine
Phosphorylation of serine & threonine
Alter charges of histone tails to weaken DNA bond and allow space for transcriptional proteins. # of groups correspond to different functions (Ex. 1 methyl = transcriptional activation, 3 methyl’s = repression)
Eukaryotic transcription factors
Bind based on structural and chemical complementarity. Have alpha-helical domains that bind to DNA through hydrogen bonds between a.a’s in alpha helix and nitrogenous bases in DNA
Types of eukaryotic transcription factors
Hair-loop-hair, helix-turn-helix, zinc finger, leucine zipper
Cis-sequences
Eukaryotic promoter regions required to initiate transcription
Core promoter
Binding site required for RNA Pol and transcription factors (Ex. TATA box, BRE region). Close proximity to initiation site
What is TATA box sequence recognized by
TATA-binding protein (TBP), subunit of the transcription factor TFIID
What is BRE region recognized by
TFIID general transcription factor
Enhancer sequences
Located upstream, bind specific transcription factors that interact with promoter to form transcriptional complex and enhance transcription of a gene. Distant from promoter but due to flexible nature of DNA via looping, far-away regions are brought closer. Adaptor/mediator proteins connect proteins from enhancer region to promoter region.
Transcriptional repressors
Bind to silencer regions positioned upstream of target gene to activate, causing the interference of general transcription factor assembly, and subsequent transcription
Blood cell progenitor differentiation
Progenitor cells activate transcription of globin proteins.
1st half of hemoglobin: 2 alpha-globin
2nd half (fetus): 2 gamma-globin
2nd half (adult): 2 beta-globin
Blood cells & transcriptional regulation
While fetuses require gamma globin to bind oxygen better and garner enough oxygen in the womb, this is not required into adulthood. Thus, progenitor cells halt the transcription of gamma-globin and increases the transcription of beta-globin subunits as the fetus matures into adulthood
How is beta-globin transcription repressed in fetuses
In fetal blood cell progenitors, chromatin is tightly wound around the beta-globin gene to inhibit transcription, whereas the gamma-globin gene site is open to allow transcription
Chemical modification of cystine bases
Addition of methyl group to CpG islands (string of C and G) located near/in promoter regions represses transcription by altering shape of DNA binding site to prevent RNA Pol binding.
Epigenetic mechanism
Process influenced by external factors (ex. diet) that alters gene expression, causing functionally relevant and heritable changes to the genome (Ex. methylation is heritable, maintains repressed state)
How do transcriptionally repressed methylated promoter further repress transcription
Though RNA Pol can no longer bind, other proteins can ONLY bind to CpG methylated sequence, further inhibiting transcription. Ex. HDAC binding removes acetyl groups from nearby histones, allowing nucleosomes to reassemble to mask promoter/enhancer sequences
Prokaryotic vs Eukaryotic regulation
Prokaryotic:
Genes are default “on” for transcription
Eukaryotes
Genes are default “off” for transcription, must be remodelled to expose promoter sequence
When can genes be regulated
Transcription initiation
RNA processing
Protein synthesis
Protein modification & transport
Protein degradation
What is the benefit of multi-level regulation
A cell can rapidly alter the levels of active proteins in response to internal and external signals
How can examine patterns of gene expression
Analyze where the corresponding mRNA that is transcribed from that gene is found during development in situ.
in-situ hybridization
Detects site of mRNA transcription with fluorescently tagged complementary probe composed of RNA/DNA that binds/hybridizes to the target mRNA molecule. Limited by the number of genes it can examine.
DNA microarraying
in-vitro, examines the expression of thousands of genes at once. Use glass slides containing tiny spots of known DNA sequences/genes. These probes detect gene expression
Benefits of DNA microarraying
Allow visualization of gene expression variation across different stages of development, cell types, and in response to cell signals. Identify differences in gene expression levels between normal and cancerous cells.
DNA microarraying & breast cancer
Grow healthy and cancerous cell cultures and isolate genes/mRNA transcribed by both cell types
Isolated mRNA serves as template for synthesis of complementary DNA molecules using reverse transcriptase enzyme
Fluorescent nucleotides are synthesized as part of cDNA
Combine equal amounts of mRNA from normal cells (green) and cancer cells (red) to microarray chip and measure fluorescence
GREEN = EXPRESSED MORE IN NORMAL CELLS
RED = EXPRESSED MORE IN CANCER CELLS
Intensity of light determines relative gene activity
mRNA degradation
mRNA must be degraded to ensure halting of gene expression. Ex. length of polyA tail regulates stability, thus must be degraded
Regulation at Translation
Short, non-coding regulatory double-stranded RNA molecules activate the RNA interfering machinery, which ensures that some mRNA are not translated. Ex. MicroRNA synthesized from RNA Pol forms hairpin loops due to complementary base pairing within itself that fragment to activate RNA machinery
RISC Complex
RNA-induced silencing complex, composed of microRNA that contain complementary sequences for mRNA sequences requiring regulation. Bind to mRNA in a non-specific manner to inhibit translation
siRNA’s
Exact complements of target mRNA sequence, bind to the complementary sequence via RISC complex and induce the cutting of target mRNA
Post-translational modifications
Cleavage
Disulphide bond formation
Acetylation
Phosphorylation
Methylation
Selective degradation
Dictate how long a protein functions in a cell. Proteasome’s help regulate the concentrations of specific proteins by breaking long polypeptides into small fragments of a.a’s that are further degraded and used in subsequent rounds of translation
Proteasomes
Large protein complexes that break peptide bonds and degrade unneeded/damaged proteins.
How are proteins destined for degradation identified
Target proteins are tagged through enzymatic cascade with small ubiquitin proteins in an ATP-dependent process facilitated by ubiquitin activation, conjugation to target protein, and ligation. Multiple ubiquitin’s attached forms a polyubiquitin chain that attaches to tagged protein, allowing proteasomes to degrade.
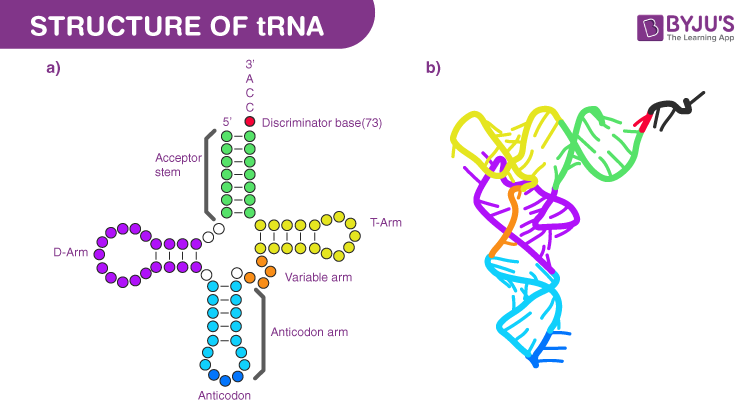
tRNA structure
70-90 nucleotides long, hydrogen bonds between complementary bases allows for 4 double-helical segments and 3 loops. Can also fold on itself into an L shape. a.a attached to 3’ end’s CCA nucleotide sequence, terminal Adenine acts as actual attachment for a.a during tRNA molecule activation
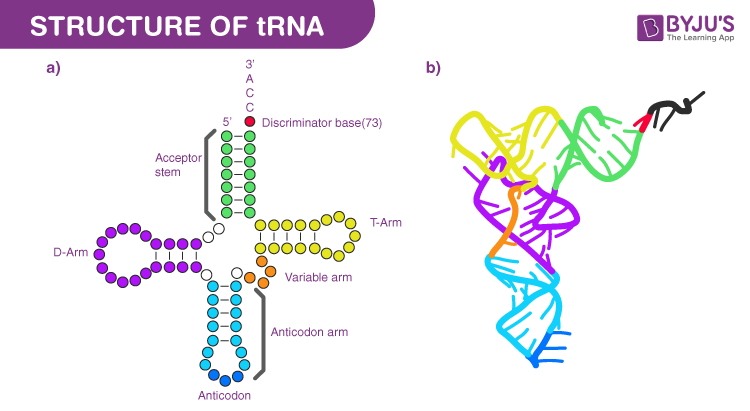
tRNA anticodon region
Nucleotide triplet that forms complementary base-pairs with specific mRNA codon. Written 3’ to 5’ and align with mRNA codons in 5’ to 3’
Aminoacyl tRNA syntheases
Facilitates activation of tRNA molecule with specific a.a. Specific to each type of tRNA and corresponding a.a, 20 for each a.a. Active site recognizes the anticodon of tRNA and a.a attachment site, and catalyze covalent attachment through ATP hydrolysis, creating a charged tRNA molecule
How many codons can mRNA code for
64
How many tRNA molecules are involved in translation
45, <60 so some can bind to more than one codon due to the wobble theory
Where does translation occur
Cytoplasm
Translation initiation in Eukaryotes
Translation initiation complex forms near the 5’ cap, scans sequence until AUG start codon found
Translation initiation in Prokaryotes
Due to lack of 5’ cap, translation initiation complex forms at Shine-Dalgarno ribosome binding sites, located few bases upstream of AUG
Polycistronic
mRNA codes for multiple proteins
What is beneficial about translation initiation in prokaryotes
It can occur along multiple regions of the sequence, allowing it to have specific open reading frames for more than one protein along a single mRNA strand (polycistronic). Works because prokaryotes have functionally related genes grouped together, and thus transcribed as a single unit from one promoter
Ribosomal initiation complex assembly
Translation factors bind to 5’ cap, allowing for assembly of small ribosomal unit
Other initiation factors bind to charged tRNA
Unit moves along mRNA in 5’ to 3’ direction until AUG is encountered
Allows for large ribosomal unit to bind to initiation complex through energy from GTP hydrolysis
Translation Elongation
As each subsequent tRNA enters and binds to A (aminoacyl) site, they each attach to a GTP-bound elongation factor which is hydrolyzed when the correct codon-anticodon pairing has been made. The deacylated tRNA move from P-site to E-site, and the subsequent tRNA enters the A-site, allowing for the release of the E-site tRNA
Peptidyl-transferase reaction
Facilitates formation of peptide bonds, catalyzed by the large ribosomal subunit
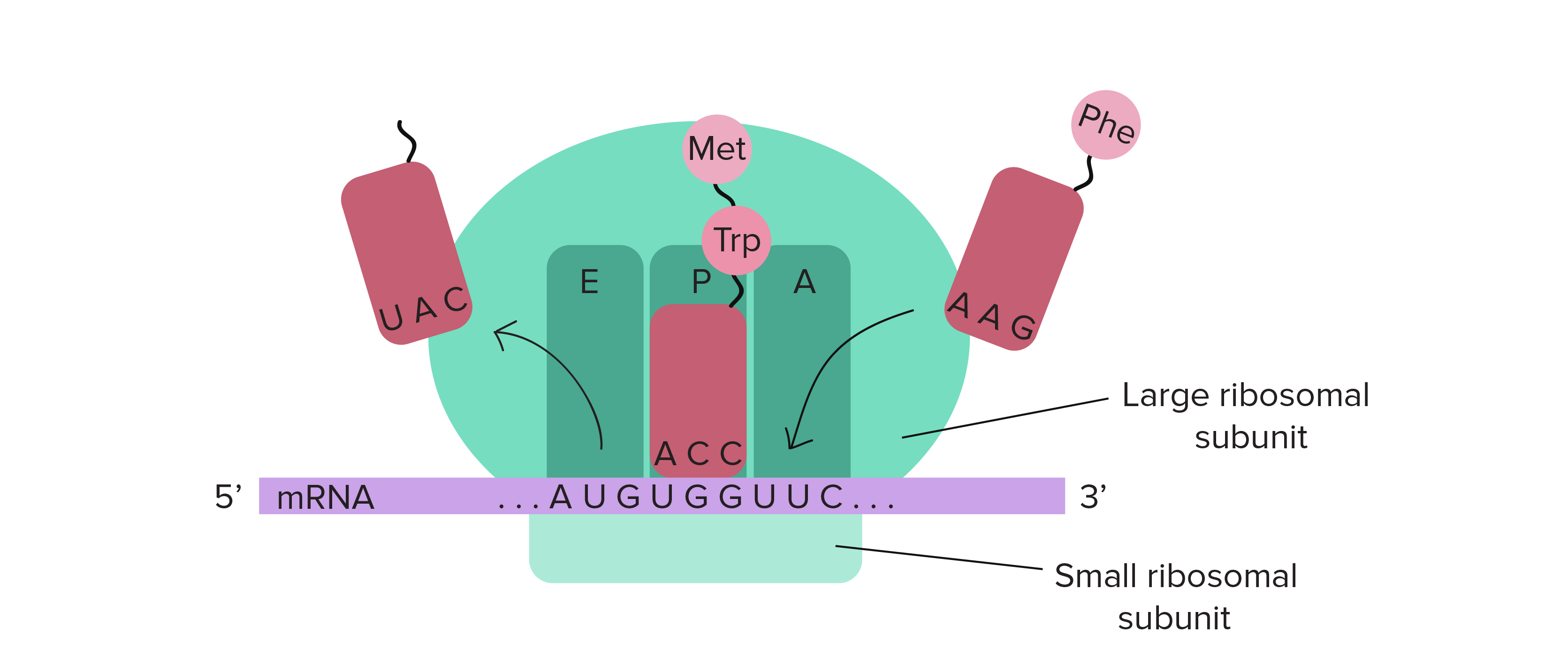
Translation Termination
Detection of stop codon allows GTP-bound release factors to bind to the A-site and catalyze hydrolsis of bond between terminal amino acid and tRNA in P site, and causes the dissociation of ribosomal subunits
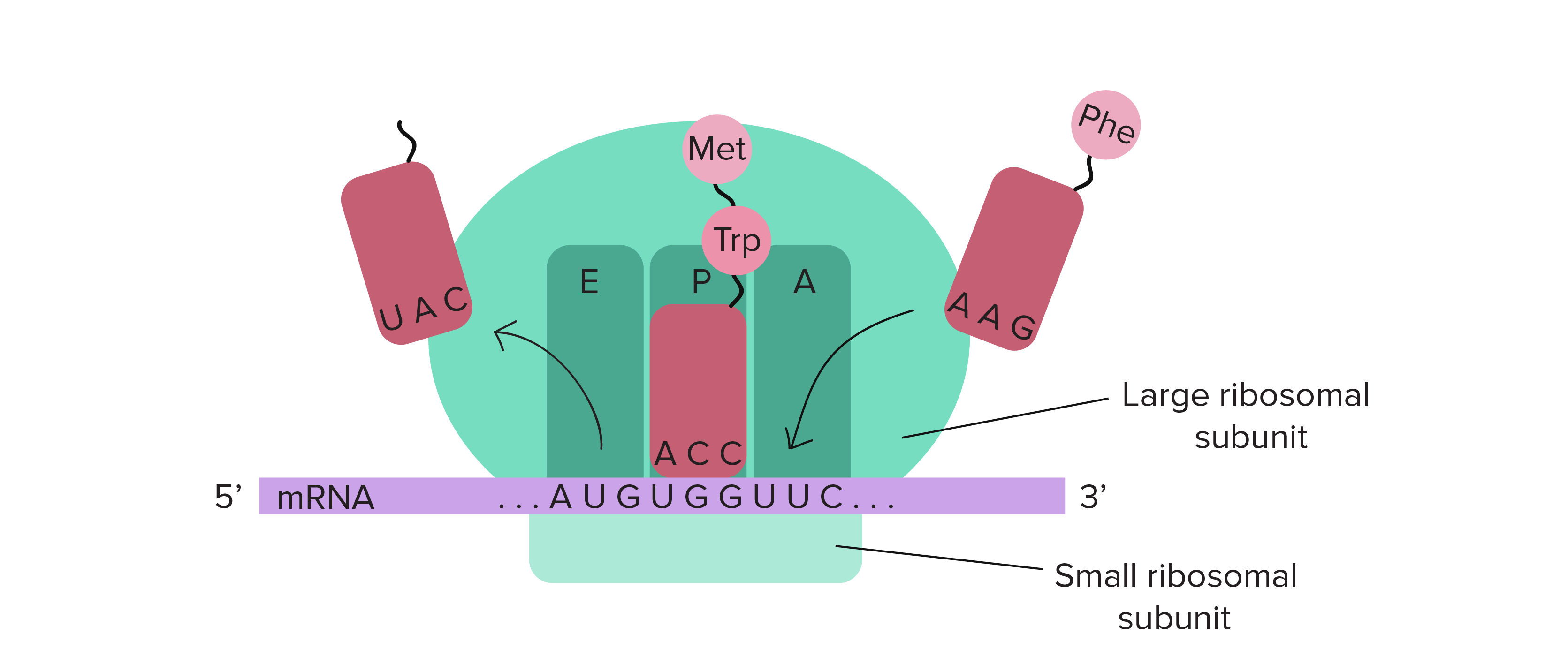
What machinery is required in translation (prokaryotes and eukaryotes)
Large and small ribosomal subunits, mRNA molecule, and charged tRNA a.a’s, initiation, elongation, and termination factors
Beadle & Tatum Experiment Significance
one-gene-one-enzyme hypothesis established the relationship between genes and proteins using the idea that Neurospora can grow well on minimal medium because it has some enzymes that can convert simple substances into arginine, as Neurospora can synthesize its own arginine through metabolic pathways (Ornithine - Citrulline - Arginine)
Srb and Horowitz
Modified it to the one-gene-one-polypeptide hypothesis. Performed study on radiation treated Neurospora to determine whether there are specific genes that produced the enzymes in the metabolic cascade for the synthesis of arginine. To prevent radiation from causing mutations in the DNA Neurospora was raised on medium containing nothing apart from the 3 enzymes
Srb and Horowitz conclusion
Mutations in specific genes block individual steps in metabolic pathways, preventing the organism from making certain nutrients. By adding the missing intermediate or final product to the growth medium, they could rescue the mutants, demonstrating that each gene directs the production of a specific enzyme
Exceptions to the one-gene-one-polypeptide hypothesis
The human genome has 20-25,000 protein-coding genes, proving that more than one protein can be produced from one gene. Post-translational modifications of proteins allow for diverse proteins
Human proteome
Full number of proteins expressed by genome
How are glucose levels regulated
Sensory responses in Beta islet cells of the pancreas lead control glucose levels. High levels = pancreas modulates synthesis and secretion of insulin to regulate
Insulin
Small effector protein produced by pancreatic beta cells that facilitates a response on target cells, thus reducing blood glucose levels
Where is glucose absorbed
In the mouth through thin epithelial surfaces in close contact to capillaries and blood vessels & in the microvilli of the small intestine also in close contact to capillaries and small blood vessels
Insulin biosynthesis
Regulated at the transcriptional and translational levels.
Insulin structure
P.p translated from gene is 110 a.a’s long, but the functional protein itself is made of 51 a.a’s.
Why does insulin decrease in length
Post-translational modifications cause decrease in length
Preproinsulin
Original a.a chain length as coded by the gene. Synthesized by bound ribosomes and transported to lumen where it is processed through cleavage of the signal sequence, creating a proinsulin molecule.
Proinsulin
Undergoes folding in ER along with the addition of 3 disulphide bonds after which it is cleaved in golgi to form an insulin dimer (A & B chains) and release a small C chain that previously linked the A & B chains
Protein folding
Occurs in the ER through assistance of chaperone proteins, then transported to golgi
Why does preproinsulin need post-translational modification
Allows the N and C-terminal a.a from the A and B chains to bind to the insulin receptors on the target cells
Why do we need post-translational modifications
Increase functional diversity of proteome
Types of post-translational modifications
Cleavage, folding & disulphide bridge formation, covalent attachment of other molecules, degradation
Types of covalent attachment modifications
Phosphorylation, Acetylation, Methylation (PAM)
Phosphorylation
Reversible post-translational modification, attachment of phosphate group to threonine, serine or tyrosine via kinases
Methylation
Attachment of methyl group
Acetylation
Attachment of acetyl group
Receptor Kinases
Type of monomeric receptor embedded in cell membrane that dimerizes when bound to signal, causing the activation of the cytoplasmic region of the receptor, which behave like kinase proteins to phosphorylate at many regions on the receptor tail to trigger a cellular response
Dimerization
2 Monomers pairing up
How is glucose absorbed into cells
Insulin signal activates receptor kinases, causing a series of cytoplasmic proteins to activate and lead to an intracellular response that activates the glucose transported proteins at the cell surface
Intracellular signal amplification
Amplifier proteins downstream from receptor activate, follow a positive-feedback loop for amplification. Double negative-feedback loops inhibit the inhibitor of a signal, providing fine control to extracellular responses
Where is glucose stored
Fat cells in adipose tissues convert to fatty acids and store as triglycerides. Liver and muscle cells store as glycogen
Alternative splicing of pre-mRNA’s
Allows pre-mRNA to be spliced at different junctions to create mature mRNA molecules that contain a different combination of exons
Isoforms
Formed during alternative splicing when some exons are removed like introns due to the spliceosome recognizing an exon as an intron. Can be from the same or different cell.
How does alternative splicing help in the regulation of gene expression
The same primary transcript can be spliced in different ways to produce mature mRNA isoforms that can produce different but related proteins, promoting genetic diversity
Alternative splicing example
Primary transcript of gene that codes for insulin receptor.
Insulin receptor gene = 22 exons
Exon 11 is removed from the mature mRNA in skeleton muscle cells and translated into a higher affinity version of the insulin receptor in muscle cells
This allows them to have a higher glucose uptake in response to insulin signals
Liver cells retain exon 11 leading to a lower affinity for glucose uptake
Liver cells vs Skeletal cell insulin receptor
Lower vs higher affinity.
Insulin feedback system
Negative, drop in blood glucose is detected, causing subsequent decrease in insulin secretion
What if insulin/insulin receptor was not processed correctly
Lack of ability to bind to insulin receptors. Insulin receptor error = cannot activate glucose transport proteins = inability to take up glucose = hyperglycemia & diabetes
What do prokaryotes require for survival
Amino acids, vitamins, nucleotides, carbs, favoured temperatures
Housekeeping genes
Genes required 24/7 for normal functions, alway being transcribed and translated (Ex. structural proteins, RNA/DNA Pol)
Regulated genes
Can be turned on/off as needed
How do bacterial cells respond to changing environments
Alter the expression pattern of some regulated genes
E. Coli and altering gene expression patterns
Glucose is preferred carb source, once used up bacterial growth is arrested
Unique gene expression mechanism allows them to switch to an alternate fuel source
If E.coli is grown with glucose and lactose, the glucose will still be used up before lactose
The products of glucose breakdown activate the switch to lactose use
How does E.Coli breakdown lactose
Using B-galactosidase to produce glucose and galactose. Enzyme is produced by transcription of the B-galactosidase gene, which only occurs when lactose is present and glucose is unavaliable
What does transcriptional modification target
Amount of mRNA produced, controls the binding of proteins to the promoter to inhbit/activate transcription
What does translational modification control
Translation of mRNA to proteins
What do post-translational modifications control
Modification and activation of proteins
What 2 factors do protein synthesis depend on
Rate of translation and mRNA stability (quickly degraded = less protein)
Which level of regulation is fastest
Post translational, enzyme-driven chemical changes that occur much faster than synthesizing new proteins, respond quickly to external stimuli. Allows cell to have a stock of inactive proteins that can later be activated
Which level of regulation is slowest
Transcriptional, needs cell to start from scratch, only used for drastic environmental changes (Ex. E.coli glucose depletion). MOST EFFICIENT, no energy/resource is wasted making a mRNA/pp unless it needs to