BIOL 3000 Exam 3
1/409
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
410 Terms
cell division
process by which a parental cell divides into two daughter cells
why is cell size limited?
the ability to transport food and oxygen into the interior becomes harder, as well as the outside cannot keep up with the quick growth of the inside
cell cycle
G0, G1, S, G2, M
G0 phase
resting phase, cell senescence
cell senescence
the period in a cell's lifespan when it loses the ability to divide and grow; cell aging
G1 phase
growth phase, cell increases in size and prepares for DNA synthesis
S phase
DNA is replicated
G2 phase
second growth phase, cell gets larger and prepares for mitosis and cell division
M phase
cell growth stops and cell divides into two daughter cells
why was frederick griffith's transforming principle important?
it was the first suggestion of DNA as a heritable material
why was the avery-macleod & mccarty experiment important?
it confirmed griffith's work and isolated DNA as the heritable material
why was the hershey-chase experiment important?
it confirmed that DNA was the genetic material
image B51
x-ray diffraction image of DNA, used later to determine its double helical structure
semi-conservative model
double stranded DNA contains one parental and one daughter strand following replication
DNA replication is a ____ process
semi-conservative
conservative model
both parental strands stay together after DNA replication and the daughter molecules contain all new nucleotides
dispersive model
parental and daughter DNA are interspersed in both strands; consists of old and new strands
meselson-stahl experiment
used isotope of nitrogen to change the weight of DNA N15 and N14, demonstrated that the semi-conservative model is the best description of replication
DNA replication
use of an existing strand of DNA as a template for the synthesis of new, identical strand
how common are errors in DNA replication?
one error occurring every 1 billion nucleotides
origin of replication
a particular sequence in a genome at which replication is initiated
origin of replication in bacteria
OriC
what is responsible for the timing of DNA replication?
GATC methylation sites
DnaA boxes
AT rich region that is the site for the binding of DnaA protein
initiator proteins
proteins that bind to DNA and mark the point where strand separation will occur
DnaA is an ____
initiator protein
replication bubbles
sections of DNA that look like a bubble where the two strands separate in order to enable replication to occur rapidly
replication occurs.....
bidirectionally
replication fork
a y-shaped region on a replicating DNA molecule where new strands are growing
DNA replicates from ____ to ____
5' to 3'
topoisomerase (gyrase)
reduce torsional strain causing unwinding of double helix
helicase
breaks hydrogen bonds between complementary nucleotides
unwinds DNA to form transcription bubble
single-strand binding proteins
stabilize ssDNA until elongation begins
primase
RNA polymerase that adds a ribonucleotide primer to ssDNA
how long are primers?
typically 10-12 bases in length
when are DNA primers removed?
following elongation. they are replaced with DNA nucleotides in this step
DNA polymerase (prokaryotic)
enzyme that catalyzes attachment of nucleotides to make new DNA during replication
DNA polymerase I
removes RNA primer and starts synthesis
DNA polymerase III
responsible for most of replication process
DNA polymerase II, IV, V
repairs DNA
T/F: prokaryotes contain the enzymes DNA polymerase alpha, sigma, beta
false, prokaryotes have DNA polymerase I, II, III, IV, V
polymerase alpha
synthesizes primer
polymerase sigma
synthesizes leading strands
polymerase epsilon
synthesizes lagging strand
T/F: DNA polymerase can initiate DNA by itself given the right conditions
false, DNA polymerase requires primers in order to initiate synthesis
DNA polymerase can only synthesize in the _ to _ direction
5' to 3'
are the sugars on the outside of the double helix hydrophilic or hydrophobic?
hydrophilic
1st step of elongation
new DNA is synthesized from deoxyribonucleic triphosphates (dNTPs)
2nd step of elongation
in replication, the 3' OH group on last nucleotide "attacks" the 5' phosphate group of incoming dNTP
3rd step of elongation
two phosphates are cleaved off of incoming dNTP
4th step of elongation
a phosphodiester bond form between the two nucleotides
5th step of elongation
phosphate ions are released
t/f: in DNA replication, both parental strands are replicated at the same time
true
DNA ligase
joins together newly synthesized DNA strands by creating a covalent phosphodiester bond
what does high fidelity mean in terms of DNA replication?
mistakes are very rare
exonuclease activity
located in DNA pol III, backs up in 3' to 5' direction to fix mistakes
what is unique about prokaryotic DNA replication?
- single circular plasmid DNA
- one origin (oriC)
- DNA is free floating in cytoplasm
what is unique about eukaryotic DNA replication?
- multiple large linear chromosomes
- multiple origin sites
- DNA tightly packed around histone proteins
- telomeres
telomerase
an enzyme that catalyzes the lengthening of telomeres in eukaryotic cells
how does telomerase work?
1. binds to overhand using complementary RNA primer
2. synthesizes DNA to make it double stranded
3. DNA pol binds to double stranded DNA and fills in the gap
4. ligase joins strands together
cancer
uncontrolled cell division or replication
aging
cell senescence or lack of cell division (werner syndrome)
disease
especially related to DNA repair defects (bloom syndrome)
site of replication
replicon
t/f: replication only occurs on ssDNA
true
chromatin
DNA and protein complex that compacts DNA
t/f: chromatin is only found in eukaryotes
true
histones
proteins found in chromatin, provides structure, shape and gene regulation of chromosomes
euchromatin
lightly packed chromatin rich in gene concentration. is most often under active transcription
heterochromatin
tightly packed chromatin consisting mainly of genetically inactive sequences
in chromosomes, when gene transcription occurs, what is likely the chromatin formation?
euchromatin
facultative heterochromatin
gene silencing; barr bodies
the condensed, inactive X chromosomes found in females is an example of.....
barr bodies
constitutive heterochromatin
VERY gene poor; centromeres and telomeres
folded-fiber model
DNA and protein model that proposed a single, long chromatin fiber makes up each chromosome
folded fiber model: type A fiber
chromatin fiber 1-10 nm. DNA packing ratio is 6:1
folded fiber model: type B fiber
chromatin fiber 20-25 nm. DNA packing ratio 10:1
according to the folded fiber model, what type of fiber is folded more tightly?
type B
according to the folded fiber model, extensive folding of type ____ forms a chromatid
B
nucleosome model
most accepted DNA packing model. proposes nucleosomes to be the packing unit of eukaryotic chromatin, made up of a double DNA strand wrapped core histones
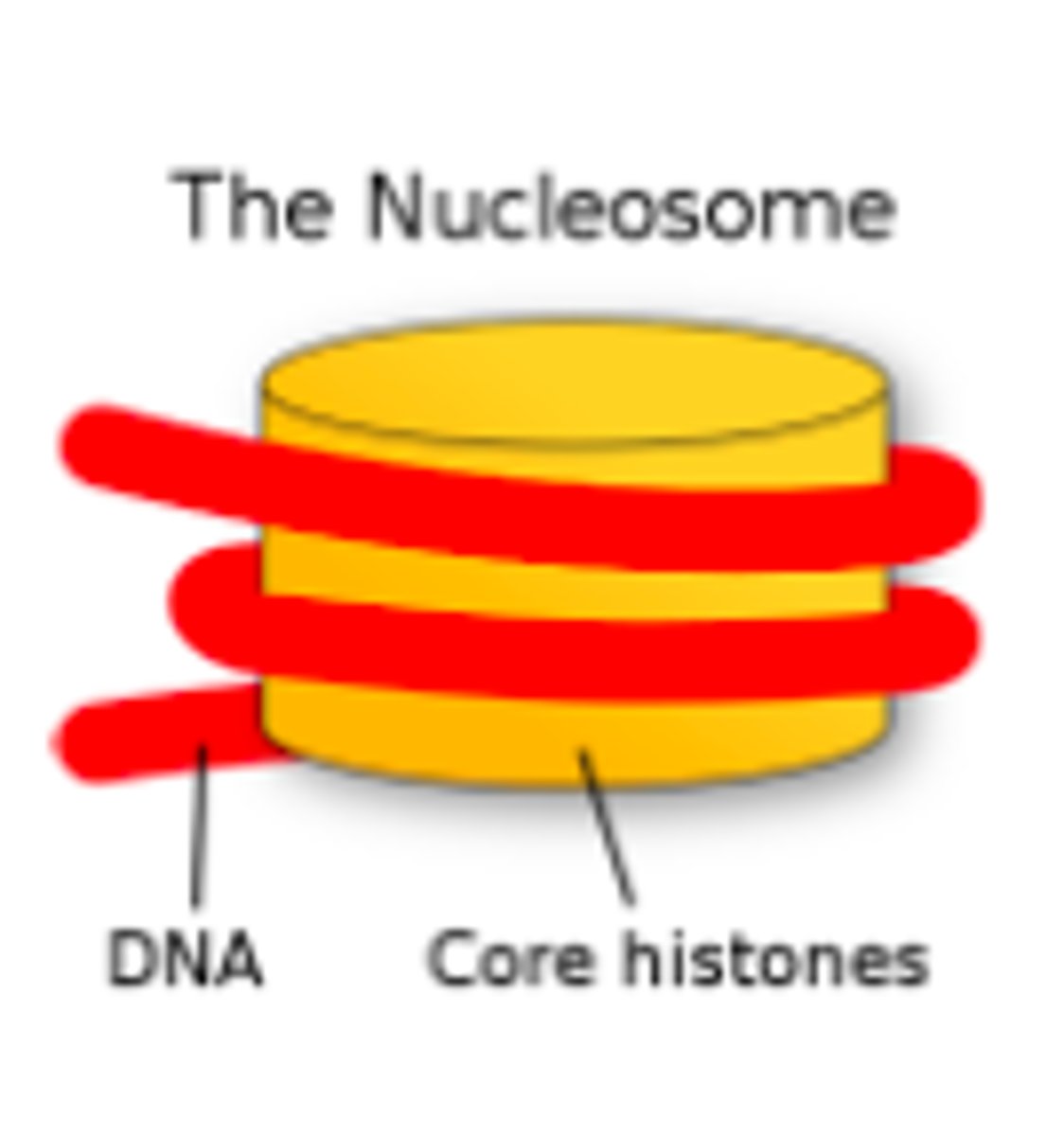
which DNA packing model is better fit for protein biosynthesis?
nucleosome model
which of the following statements is true regarding core histones?
I. H2A, H2B, H3, and H4 are core histones
II. they were highly conserved during evolution
III. have a very basic charge due to being made of cystine and arginine
IV. consist of approximately 120 amino acids each
I, II, and IV
which of the following statements is true regarding linker histones?
I. consists of 200 amino acids
II. they were highly conserved during evolution
III. tissue specific expression
IV. closely associated with core particle
H1 is in what histone class?
linker histones
how many core histones are in a nucleosome?
8, two of each type
how many linker histones are in a nucleosome?
1
what is the importance of 10 nm fiber in nucleosome formation?
they are the primary packing of chromatin
solenoid formation
helical coiling of 10 nm
fibers consisting of 6 nucleosomes
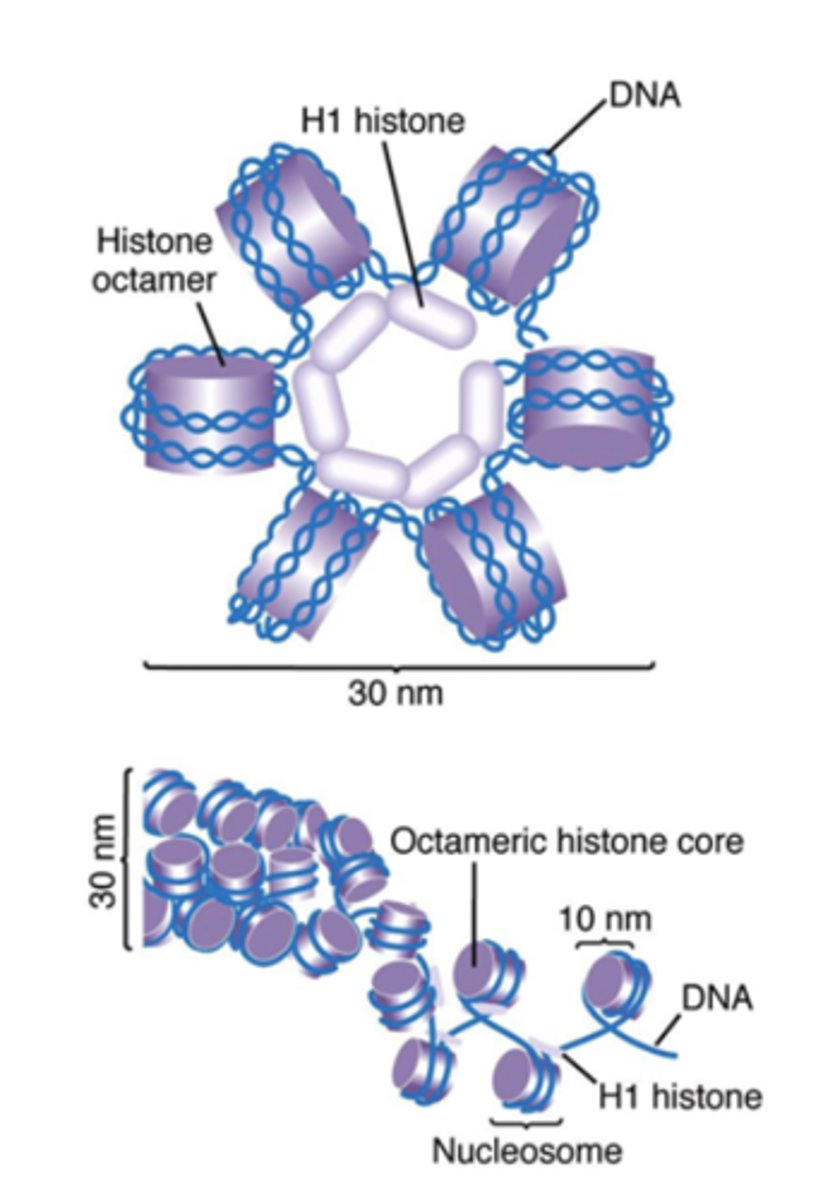
what histone is found in the center of a solenoid, and what is it used for?
H1, used for packing
how does H1 histone work?
binds linker DNA and 146 bp portion of central core DNA, compacts up to 40x!
zig zag model formation
due to the limited flexibility of DNA, straight linker DNA connects opposite nucleosomes
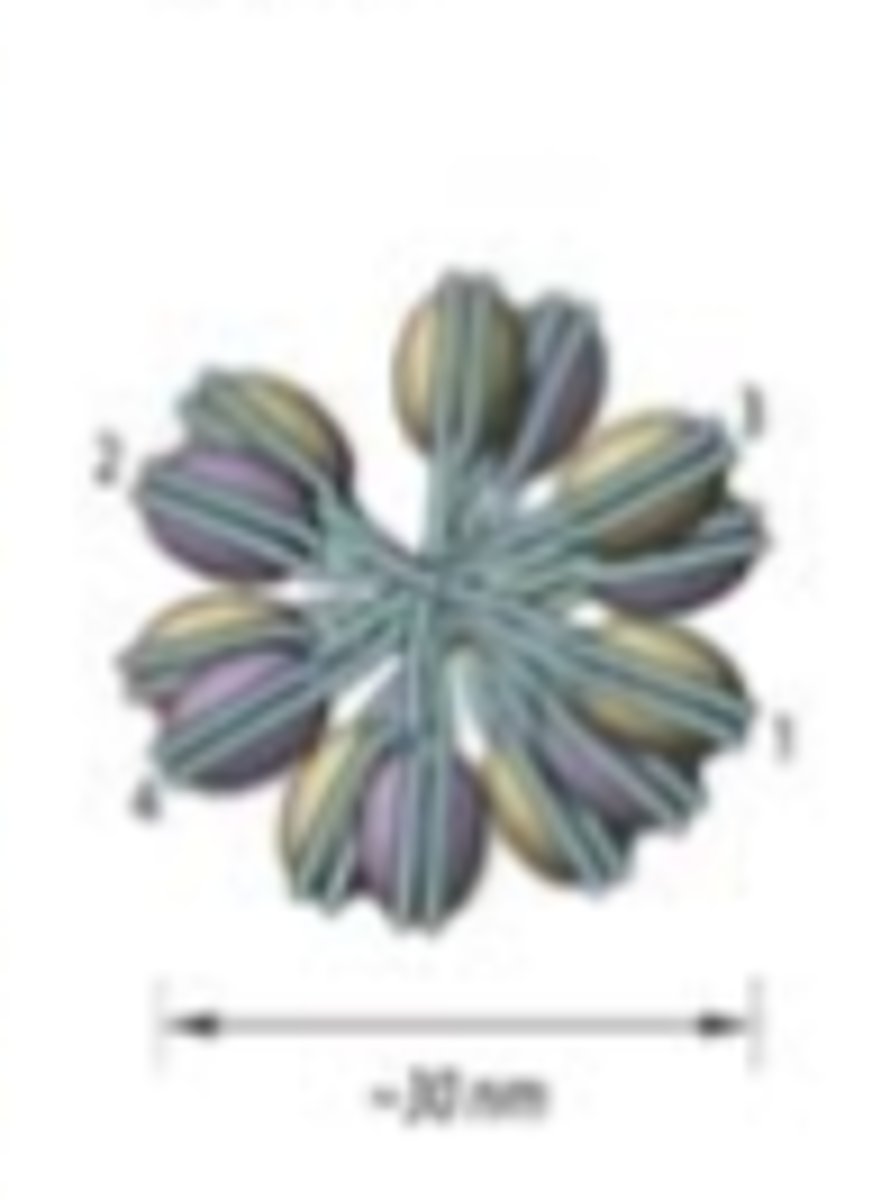
t/f: both solenoid and zig zag typologies may both simultaneously be present in chromatin fiber
true
chromatin loops
higher order of coiling with 300 nm chromatin fiber built around scaffold of topoisomerase II
chromatin loops share the same level of compaction as....
euchromatin
metaphase chromosome
condensed chromatin loops looped around spiral scaffold composed of topoisomerase II and about 15 non-histone proteins
metaphase chromosomes have the same level of compaction as...
heterochromatin
2 nm - 10 nm fibers are known as _____ and are found in the _____ phase
DNA and nucleosomes; G1
30 nm fibers are known as _____ and are found in the _____ phase
chromatin; early G2
300 nm fibers are known as ____ and are found in the _____ phase
chromatin loops; late G2
700 nm fibers are known as _____ and are found in the _____ phase
condensed chromatin loops; beginning prophase