L19 - Non-coding RNAs and mechanisms controlling pervasive transcription in eukaryotic cells
1/55
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
56 Terms
What is the only mRNA which is not polyadenylated?
Histones
What % do coding regions in higher eukaryotes make up?
Coding regions in higher eukaryotes make up 1-2% of the total genome
What is non-coding RNA?
“The term non-coding RNA (ncRNA) is commonly employed for RNA that does not encode a protein, but this does not mean that such RNAs do not contain information nor have function”
Where are most ncRNAs located?
Many ncRNAs retain in the nucleus, they are exported to the cytoplasm, they are very quickly degraded or stabilised
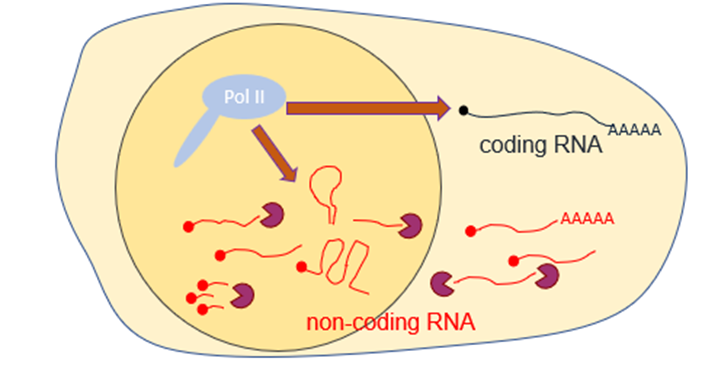
Which ncRNAs are transcribed by RNA Polymerase II (Pol II)?
• small nuclear and small nucleolar RNAs (snRNA and snoRNAs)
• long intergenic RNAs (lincRNAs)
• enhancer RNAs (eRNAs)
• promoter-associated ncRNAs (PROMPTs, paRNAs, TSS-ncRNAs)
• many more short classes: miRNAs, piRNAs, telsRNAs, siRNAs
Which ncRNAs are long ncRNAs (lncRNAs)?
Over 200 nts
eg lincRNAs, eRNAs, PROMPTs
Which ncRNAs are transcribed by RNA Polymerase I and III?
• rRNA
• tRNA
How are ncRNAs separated?
In general, we separate ncRNAs into those which are dependently synthesised (co-expressed with mRNA) and independently synthesized (expressed from their own promoters)
What are the different types of ncRNAs associated with protein-coding genes (dependent ncRNAs)?
ncRNAs originating from nucleosome free regions
ncRNAs processed from pre-mRNA
What are ncRNAs originating from nucleosome free regions?
o promoter-associated RNAs
o Transcription from promoter of protein-coding gene results in mRNA and ncRNA
o Short ncRNA produced in the same direction as RNA (into the coding sequence)
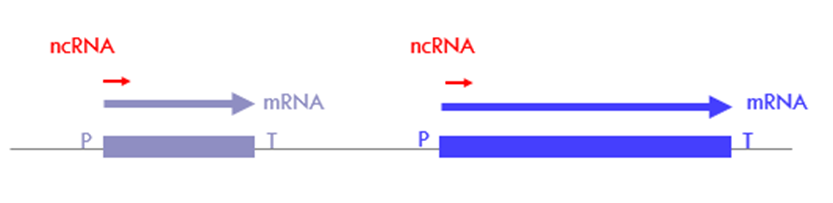
What are the promoter associated ncRNA classes?
paRNA/TSS-ncRNAs (promoter-associated RNA/ transcription start site ncRNA)
PROMPTs (promoter upstream transcripts)
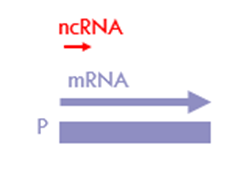
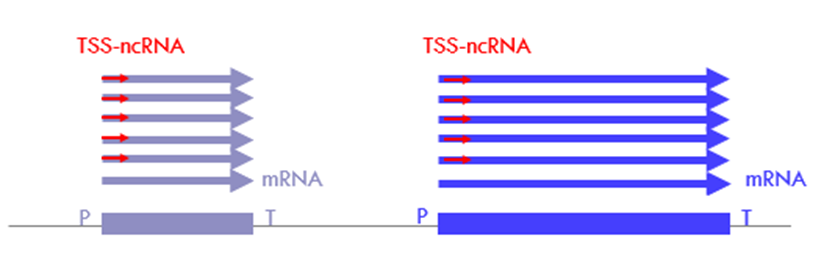
What are TSS-ncRNAs?
o transcribed in the same direction as the coding/sense sequence
o may make up to 80-90% of sense transcription
o 20– 60 nt long
o non-functional
o products of abortive transcription – important check-point in the regulation of gene expression
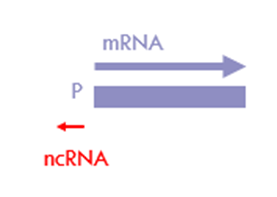
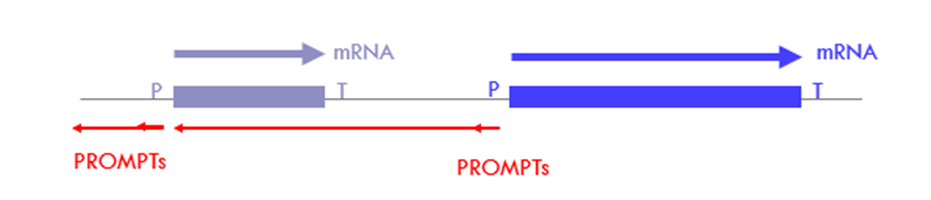
What are PROMTs?
o transcribed in reverse orientation to protein coding/sense sequence
o 0.5 – 1.5 kb long
o non-functional
o products of bidirectional promoters
biproducts of transcription from protein coding genes
How does transcription termination regulate TSS-ncRNAs?
Important for the regulation of promoter proximal pausing
Important for inducible genes
Every single polymerase starting from the promoer has the capacity to reach the end and produce mRNA
If this is necessary, promoter-proximal pausing and abortive transcription will be overridden and polymerase will produce mRNA
If it is not necessary, transcriptional termination will kick in to reduce transcription synthesis of full length mRNA
Most polymerases will start and only produce short ncRNAs – very quickly terminated
What does transcription termination play a role in?
Gene activation and silencing
How does termination regulate PROMTs?
Don’t have polyadenylation signals which trigger transcription termination of protein-coding genes in intergenic regions
If ncRNAs are not terminated transcription may continue into regions located downstream
In this situation, we have 2 polymerases transcribing towards each other (one for PROMTs, one for mRNA)
Polymerase transcribing mRNA will be blocked by polymerase transcribing PROMTs
Uncontrolled pervasive transcription results in transcription interference and is lethal
Also creates a lot of tortion on DNA which may lead to DNA breaks and genomic instability
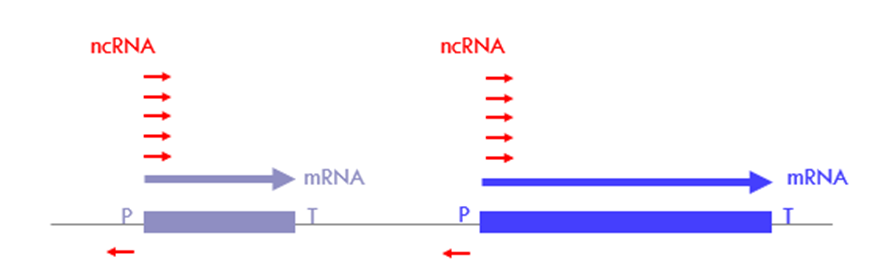
What are ncRNAs originating from nucleosome free regions (NFR)?
• Transcription from promoter of protein-coding gene results in mRNA and ncRNA
• Eukaryotic promoters are very often bidirectional and transcription is initiated in both directions (Sense or antisense direction)
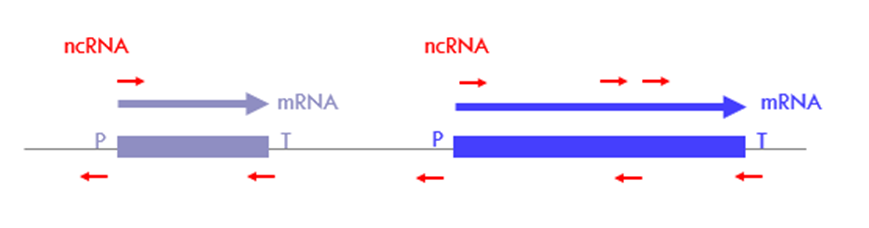
Where can RNA Pol II initiate transcription?
at any NFR, not only from promoter
Can happen at any place in introns or exons if the repositioning of nucleosomes is affected, eg in pathological conditions
Observe transcription interference and deregulation of gene expression
How is transcription interference avoided?
To avoid transcription interference and support proper transcription and expression of genes
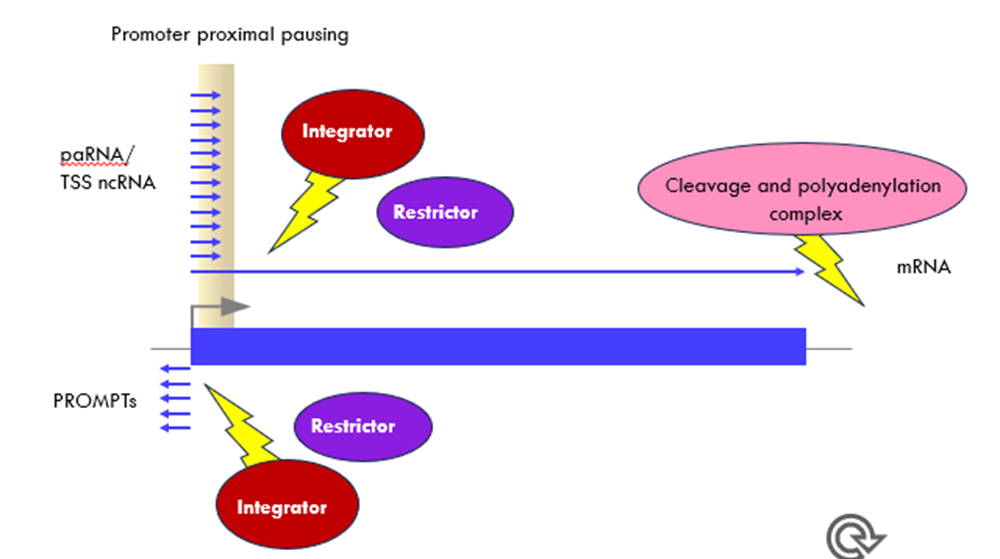
2 complexes act on the promoters which are involved in transcription termination of ncRNA
Integrator and restrictor (different from cleavage and polyadenylation complexes at 3’ end)
paRNA – promoter-associated RNA
TSS ncRNA – transcription start site ncRNA
PROMPTs - promoter upstream transcripts
Take home message?
Non–productive transcriptional events at the protein-coding genes are immediately restricted to avoid transcriptional interference and deregulation of gene expression.
What is an example of ncRNAs processed from pre-mRNA intermediates?
snoRNAs
Where are human snoRNAs found?
Embedded into introns
What does synthesis of human snoRNAs depend on?
Synthesis of human snoRNAs depends on expression of their host
Very few human snoRNAs are independently expressed
Many in the introns of protein-coding genes of long-coding RNAs
What does synthesis of snoRNAs in lower eukarotes depend on?
They are mainlu independent
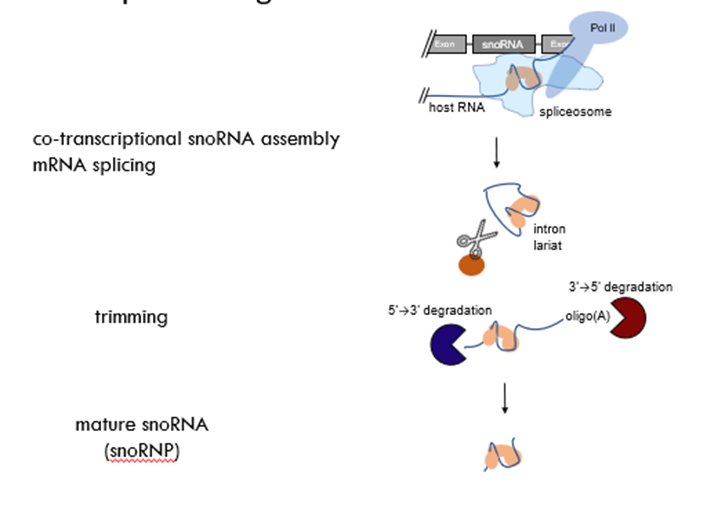
How is snoRNA processed?
• When pre-mRNA is transcribed and spliced, if the intron contains snoRNA sequence, the sequence will be bound by specific snoRNA proteins
• When the intron is released and targeted for degradation, 5’-3’ and 3’-5’ exonucleases cant go through the structure because it is bound by snoRNA proteins
• It is therefore released to the nucleoblast
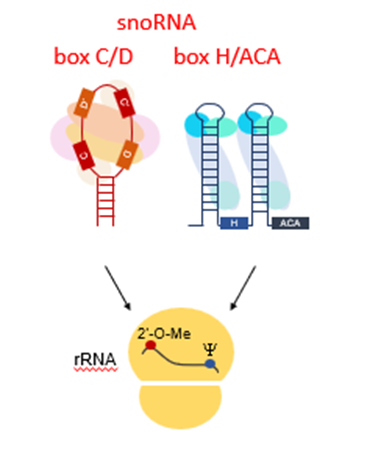
What are the 2 major classes of snoRNA?
boxC/D and boxH/ACA
What do boxC/D snoRNAs do?
RNA methylation
What do boxH/ACA snoRNAs do?
RNA pseudouridinylation
What is RNA methylation and pseudouridinylation required for?
required and essential for ribosome function and stability
What do snoRNAs recognise?
RNA targets via short complementary sequences
What are orphan snoRNAs?
snoRNAs with no known targets
How does the number of snoRNA units increase?
Increases with organism’s complexity
How many snoRNAs do we know in humans?
550 snoRNAs have been found
What is Prader-Willi syndrome?
Neurodevelopmental disease
autism-like syndrome
the most common genetic disease causing morbid obesity
fully manifests in early childhood
no cure
molecular processes leading to the syndromes are not known
What causes the PWS locus?
Large deletion in chromosome 15
Usually contains a lot of ncRNAs and protein coding genes
Minimum deletion to cause PWS contains a cluster of orphan snoRNAs
Some of these form human specific long ncRNAs called sno-lncRNAs
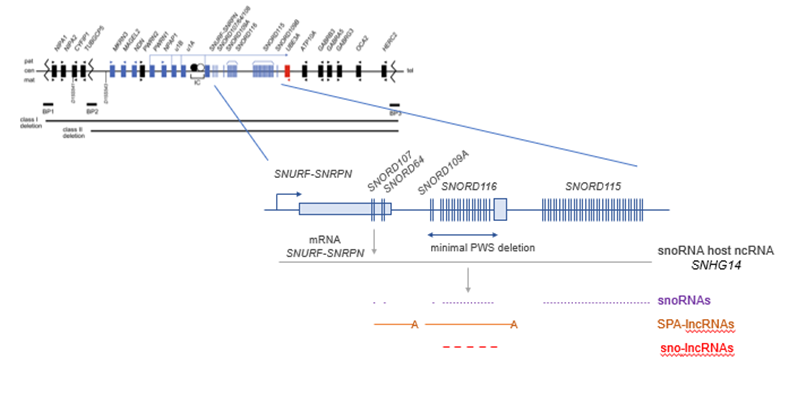
How do sno-lncRNAs form?
2 snoRNAs are in the same intron
Exonucleases cannot go through snoRNA molecules
End up with a long ncRNA with snoRNAs at either end instead of a cap and polyadenylate
Makes it very stable
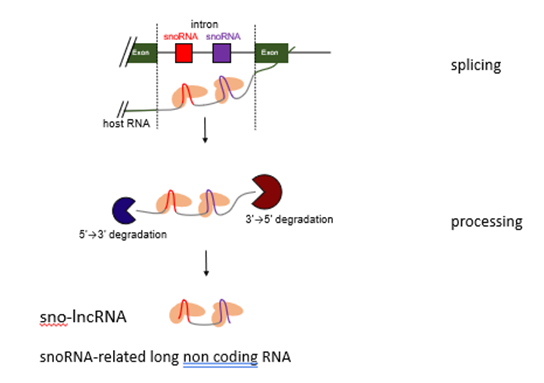
What do sno-lncRNAs do?
sno-lncRNAs are one of the most abundant RNA species in human stem cells
the intervening sequence may sequester splicing and transcription factors
sno-lncRNAs regulate transcription of genes involved in neurodevelopment
primate specific – not present in other species
Take home messages?
• snoRNAs functional and essential
• the balance between snoRNP assembly and degradation regulates their synthesis
• in higher organisms functions of many snoRNAs are still unknown
What are independently expressed ncRNAs?
• ncRNAs expressed independently of protein-coding genes
• transcribed from their own promoters
What are examples of Independently expressed ncRNAs?
o small nuclear RNAs (snRNAs)
o enhancer RNAs (eRNAs)
o long intergenic RNA (lincRNAs)
What are snRNAs?
• present in all eukaryotic organisms (U1, U2, U4, U5, U6)
• form riboprotein complexes
• required for splicing - essential
• short (~150 nt) and fold into “structural RNAs”
• non-polyadenylated, terminated by Integrator
• transcribed by RNA pol II apart from U6 which is transcribed by RNA pol III
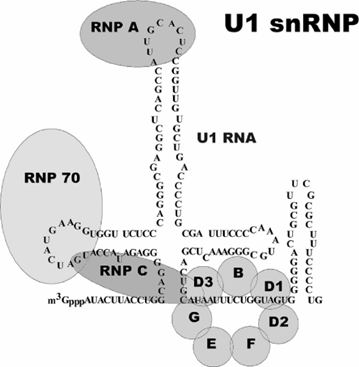
What are enhancers?
• enhancers are cis-regulatory elements in the genome that cooperate with promoters to control target gene transcription
• active enhancers are highly transcribed
• are bound and mediated by mRNA specific transcription factors
What are enhancer RNAs terminated by?
terminated by either cleavage and polyadenylation complex or Restrictor or Integrator complexes
What are the 2 major types of eRNAs?
polyadenylated and non-polyadenylated
What are non-polyadenylated eRNAs?
bidirectionally transcribed eRNA of less than 2 kb (~350nt) terminated by Restrictor or Integrator complexes
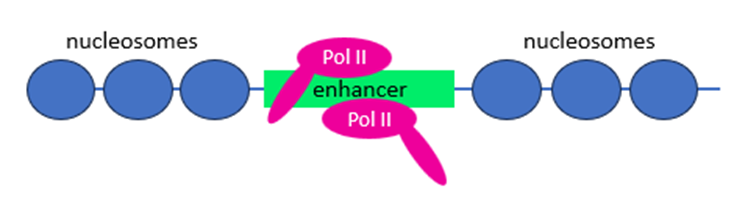
What are polyadenylated eRNAs?
unidirectionally transcribed eRNA longer than 4 kb, transcribed from highly active enhancers, terminated by cleavage and polyadenylation, Restrictor or Integrator complexes

What are some suggested functions of eRNAs?
functional RNAs or by-products of transcription?
o Transcription keeps enhancers accessible for interaction with chromosomes
o OR
o eRNAs play roles in regulation of gene expression
What are the putative eRNA functions?
o Molecular glue keeping enhancer and promoter together
o Sequestering functions – bring transcription factors in proximity of promoters to enhance initiation processes
o Phase separated environment to facilitate gene expression
What diseases are eRNA mutations associated with?
Cancers
eg breast cancer
Altered eRNAs transcribed from enhancers of genes like ESR1 (estrogen receptor 1) and MYC are linked to cancer progression and endocrine therapy resistance.
Cardiovascular diseases
eg athersclerosis
eRNAs transcribed from enhancers of inflammatory genes (e.g., IL6, VCAM1) are implicated in vascular inflammation and plaque formation
Neurodegenerative diseases
Alzheimer’s Disease
Dysregulated eRNAs involved in enhancer-promoter interactions of genes such as APP (amyloid precursor protein) may contribute to amyloid plaque formation.
Metabolic diseases
eg Diabetes
Dysregulated eRNAs associated with enhancers of genes like PPARG (key regulator of adipogenesis) and INS (insulin) may impair glucose homeostasis.
Take home messages?
• active enhancers are transcribed
• both transcription of the enhancer region and produced eRNA may be required for regulation of the target gene
What are long intergenic ncRNAs (lincRNAs)?
• Independent transcription units with defined start site, exons and introns, and termination site
• Longer than 200nt
• 13k transcripts arising from 9.5k gens
• Many functional and tissue-specific
• May evolve into functional mRNAs
lincRNAs vs mRNAs?
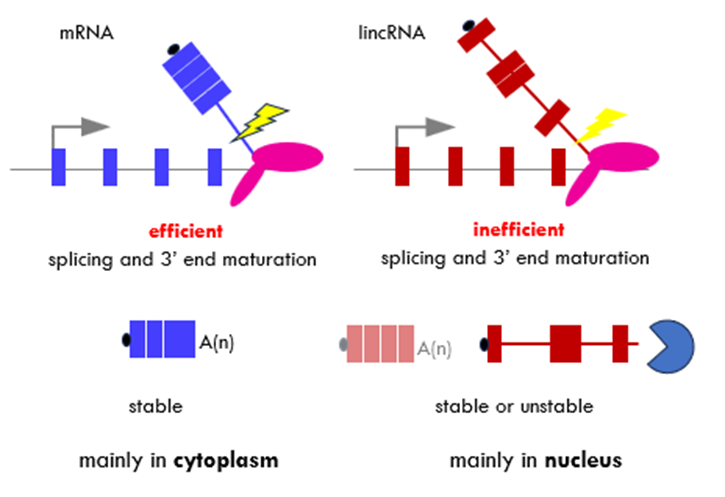
synthesis employs the same processing machineries – capped, cotranscriptionally spliced and terminated in 3’ processes processed by cleavage and polyadenylation machinery
processing of mRNA is much more efficient so is exported to the cytoplasm
only some lincRNAs will be spliced and maturated so they are mainly in the nucleus
many will be associated with the site where they are synthesised
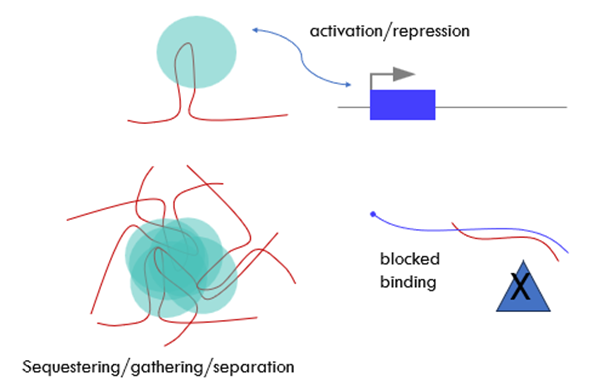
What are lincRNAs functions?
lincRNAs functions are mostly associated with their high-order structure and/or ability to bind proteins (or DNA and RNA)
Regulation of transcription
Roles in nuclear organization by binding different parts of the genome
Signalling
Decoy/sequestering – making them accessible or inaccesiblle
Guiding
Scaffolding
What are examples of lincRNAs?
MALAT1
regulates alternative splicing and modules gene expression
Firre
interacts with chromatin and scaffolds epigenetic regulators
HOTAIR
acts as a scaffold for histone modifying complexes
As-Uchl1
regulates Uchl1 expression post transcriptionally and enhances its translation in response to stress
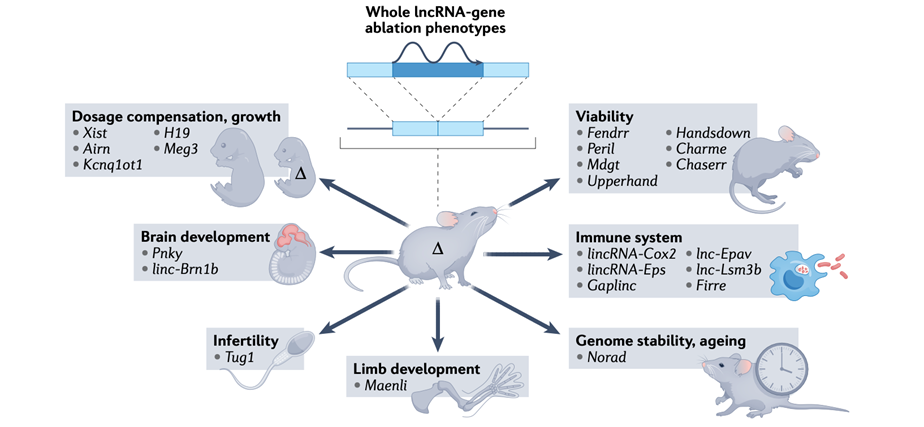
What do lincRNAs affect when removed from mice?
Brain development
Infertility
Limb development
Genome stability, ageing
Immune system
Viability
Growth
Take home messages?
• lincRNAs are “non-coding copies” of mRNAs: are capped, spliced and polyadenylated
• lincRNAs are usually localized in the nucleus
• many lincRNAs are functional