CH 6 (Part 1) - Viruses, Viroids, & Prions
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
34 Terms
What domains are susceptile to viruses?
Cell types in ALL taxonomic domains are susceptible to virus infection.
Discovery of viruses
(Ivanovsky & Beijerinck) tobacco mosaic virus; “non-filterable particles”
tobacco plant infection by virus - found virus in filtrate (tiny - 20 nm)

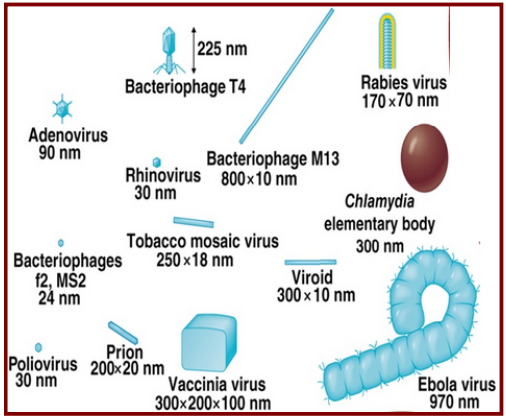
Virus size range
20 to 900 nm
(0.02 to 0.90 um)
Viruses
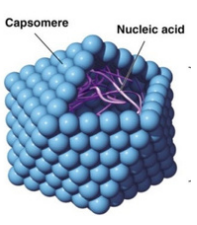
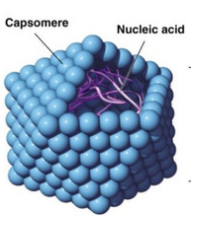
acellular; they are obligate (they must do it) intracellular parasites. ALL viruses consist of nucleic acid (DNA/RNA) in a protein shell ⟶ a capsid; comprised of repeating protein subunits, capsomeres.
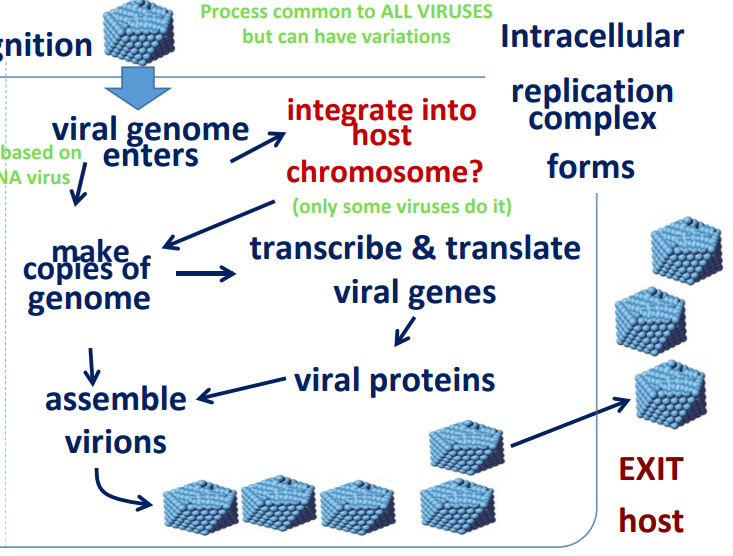
Process common to ALL VIRUSES but can have variations
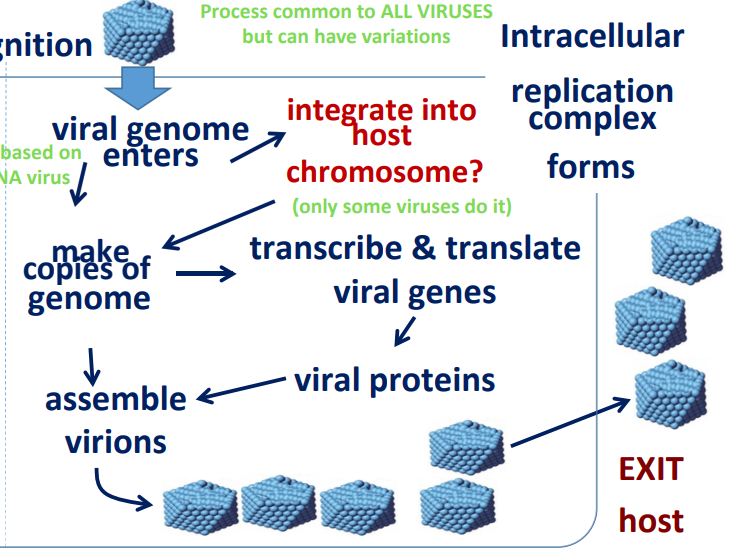
What is needed from the host?
DNA/RNA polymerase (can vary based on DNA/RNA virus)
Ribosomes
tRNA
Nucleotides (*varies)
Other: viruses lack a metabolism; rely on host
host range
Each species of virus infects a particular group (range) of host species
Broad: rabies virus
Narrow: HIV only infects humans
trophism
Animal viruses ⟶ tissue specificity
how many types of cells in a host species can be infected?
Broad: Ebola
Narrow: HIV (infects T-helper cells)
based on: glycoproteins or capsid proteins on its surface recognize those of the host cells.
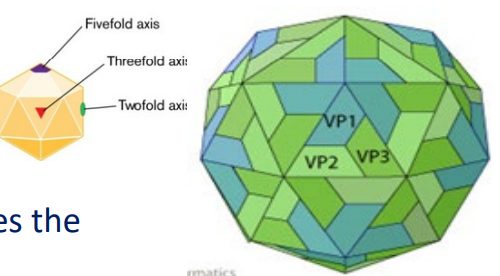
Symmetrical viruses
icosahedral or filamentous capsid
geometric pattern

Simplicity of capsid structure
minimizes the number of genes
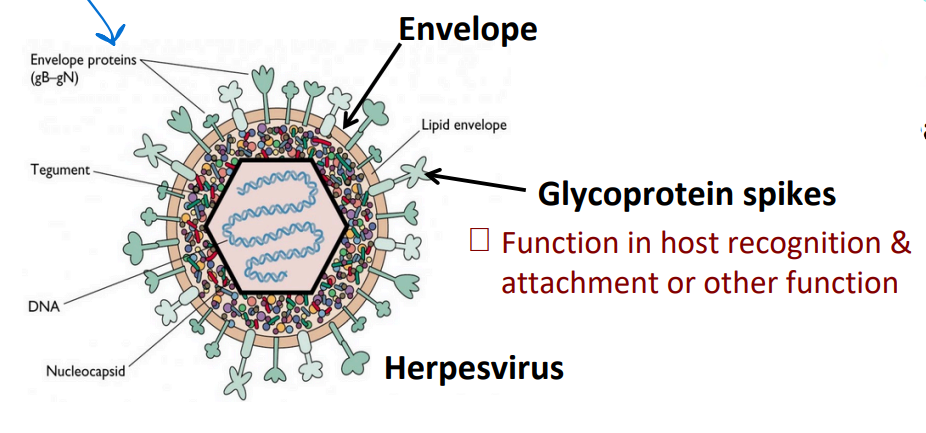
Enveloped viruses
an envelope surrounds the capsid; derived from the host membrane
“Naked” viruses LACK envelopes
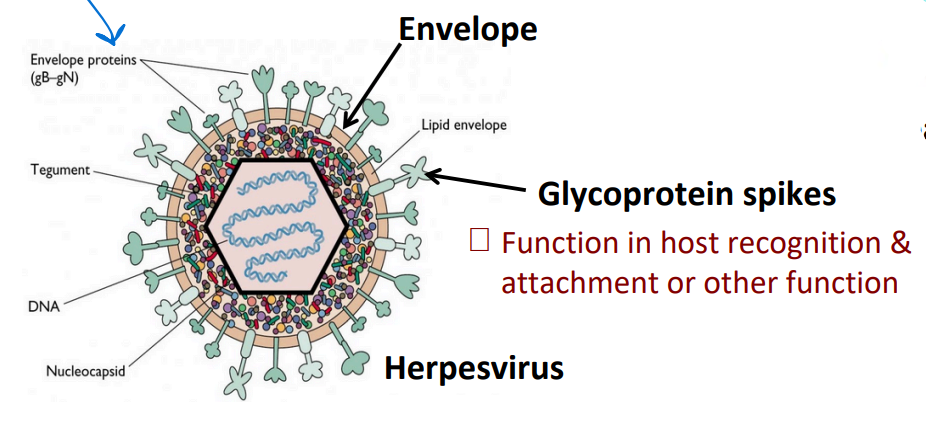
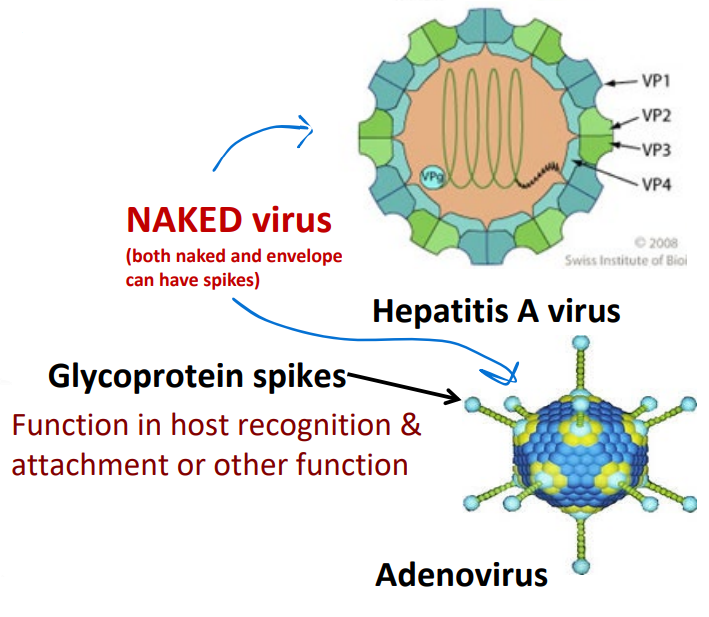
Naked viruses
lack an envelope
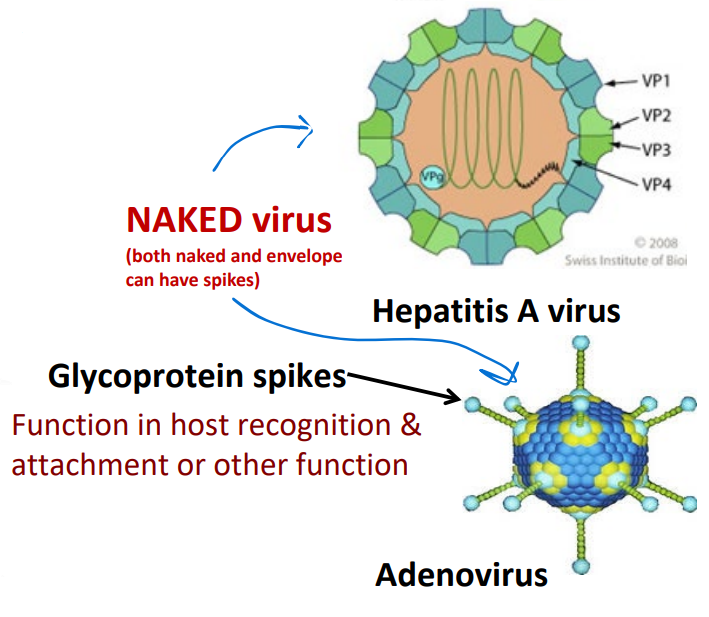
Glycoprotein spikes
Function in host recognition & attachment or other function
BOTH naked and enveloped can have spikes
Filamentous viruses
The capsid ⟶ long tube of protein,with genome coiled inside; vary in length
Ex: Ebola, M13, TMV
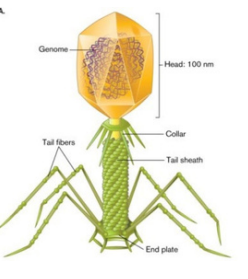
Tailed viruses
addition of a genome delivery device tothe icosahedral head.
Ex: T4 bacteriophages: helical “neck” & tail fibers
complex viruses - many structures
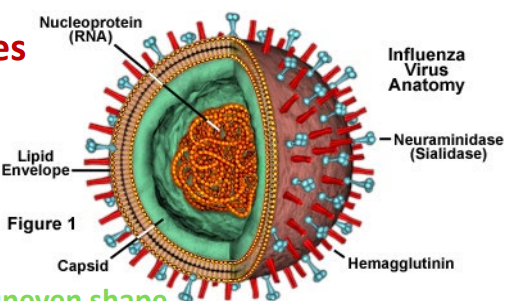
Asymmetrical viruses
lack capsid symmetry —→ uneven shape
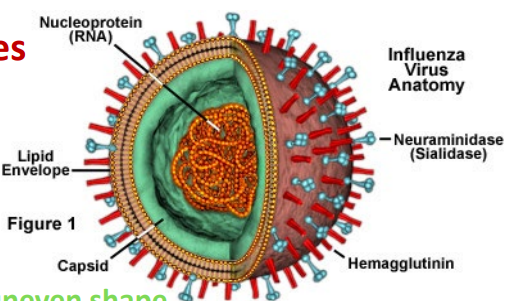
influenza viruses (as well as others)
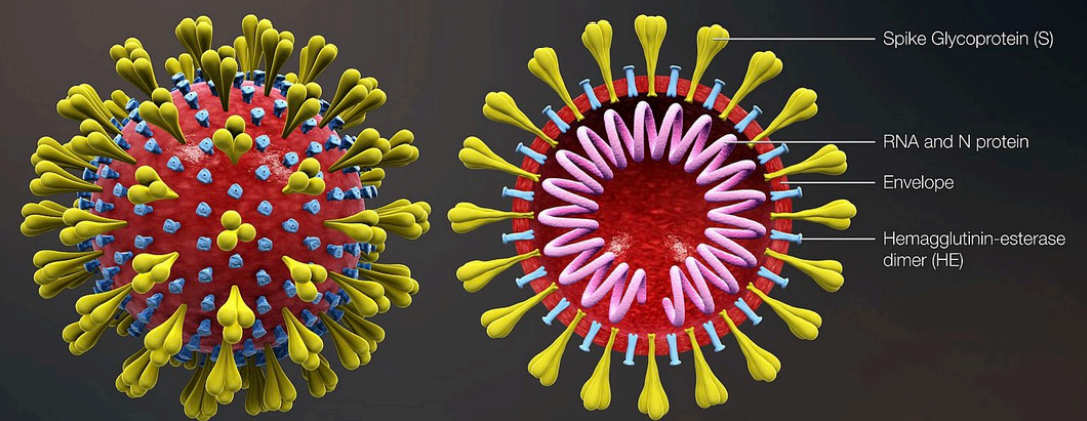
Coronavirus
enveloped viruses
nucleocaspid proteins - protects genome (stuck to genome)
Viral Genomes
Small viruses: ≤ 10 genes
Large viruses: > 100 genes
average virus has 11,000 nucleotides
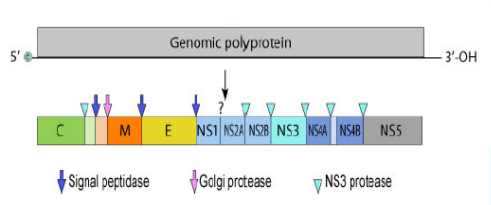
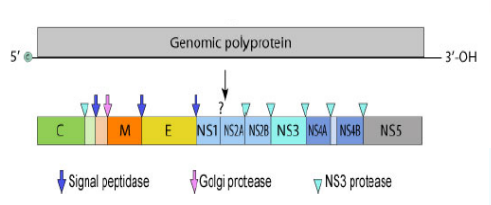
Zika Virus Genome
Non-segmented, single-stranded (+) RNA genome
10,794 ntbases long
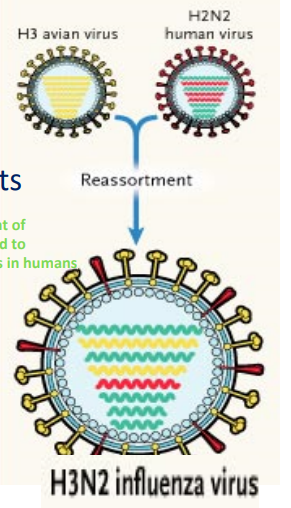
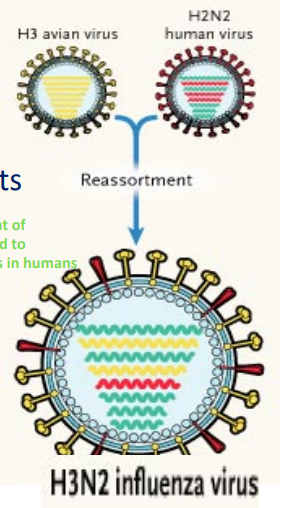
Influenza Virus Genome
Segmented, single-stranded (-) RNA genome; 8 segments
11 proteins encoded;13,500 ntbases total
mutation and reassortment of segmented genomes ⟶ lead to variants that are infectious in humans
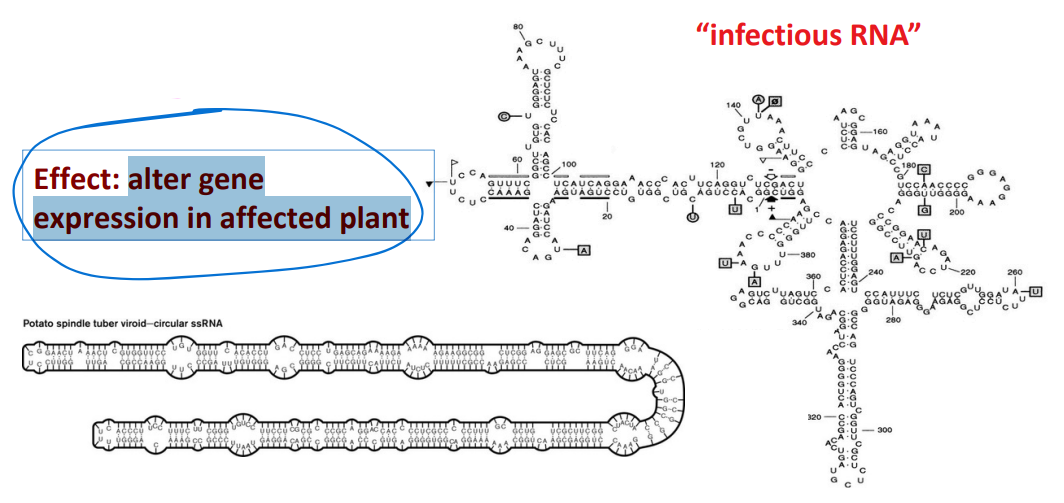
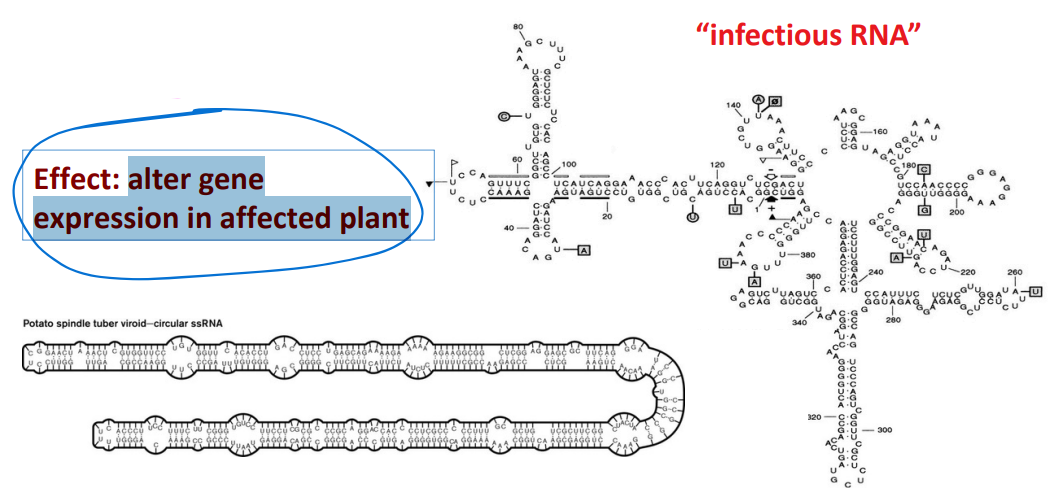
Viroids
are RNA molecules without a capsid surrounding the RNA; infect plants. They are not viruses.
Are replicated by host RNA polymerase
The RNA does not encode for proteins; 300-400 nucleotides long
Some have catalytic ability.
EFFECT: alter gene expression in affected plant

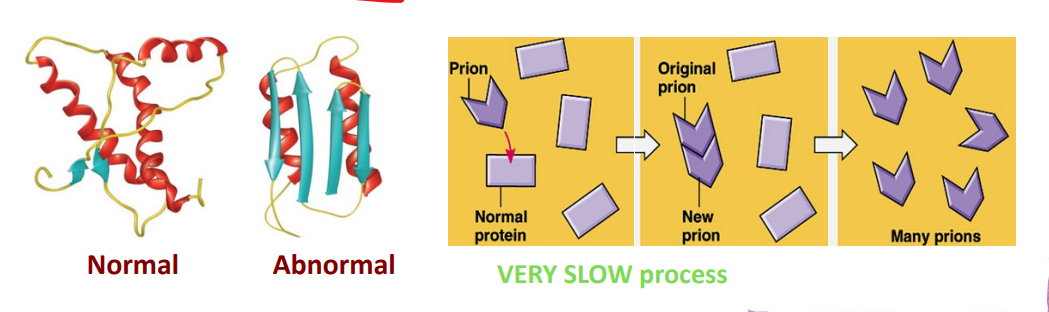
Prions
proteins that infect animals; they have NO nucleic acid component.
Cause degenerative brain diseases; transmitted in food, prepared from infected animals
misfolded form of a normal brain cell protein
converts a normal protein into the prion version, creating a chain reaction; produces harmful aggregates in the cell. (VERY slow process)
Are highly resistant to physical/chemical agents
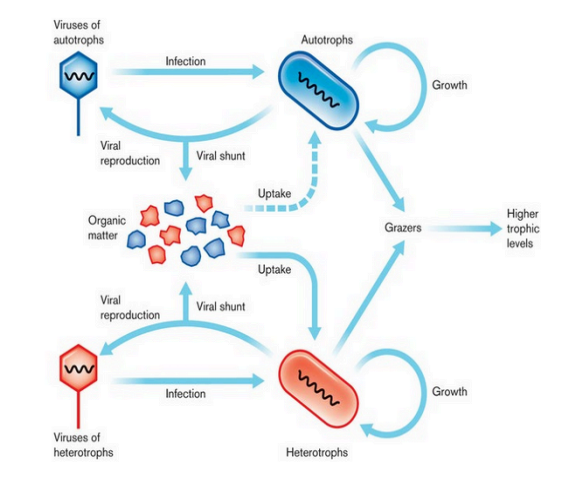
Important roles in ecosystems (Viral Ecology)
limit host population densities; recycle nutrients; increase host diversity; gene transfer
Marine Ecosystems: cycling of nutrients
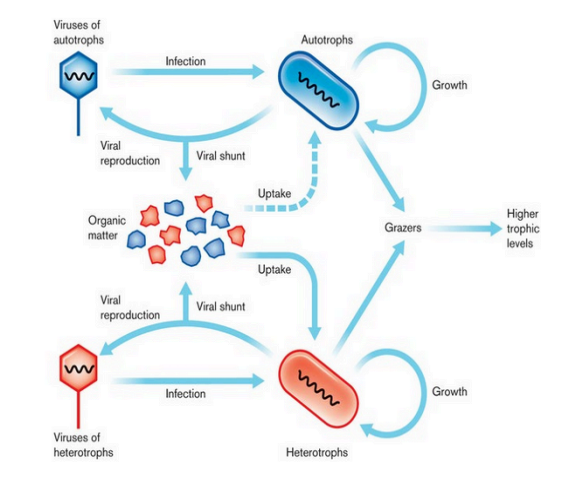
viral shunt (Viral Ecology)
viral infection of hosts convert them to detritus, rich in organic & inorganic molecules.
marine ecosystems
Weird viruses
Very large viruses
can have some metabolism - like a normal cell (but not always
virus that can infect other viruses
International Committee on Taxonomy of Viruses; based on:
Genome
Capsid symmetry
Envelope
Host range
Virion size
1971: Baltimore classification
genome (RNA or DNA) & route used to express messenger RNA (mRNA)
+ RNA = mRNA = “sense” RNA —→ codes for protein (final step)
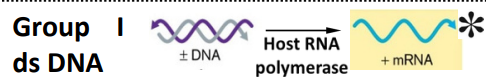
Group I: ds DNA
± DNA —→ host RNA polymerase —→ + mRNA
use host DNA polymerase to make copies of genome
Group II: + ss DNA
+ DNA —→ host DNA polymerase —→ ± DNA —→
host RNA polymerase —→ + mRNA
use host DNA polymerase to make copies of genome

RDRP
RNA-Dependent RNA Polymerase
VIRAL enzyme, not found in cells
Group III: ds RNA
± RNA —→ viral RDRP —→ + mRNA
use RDRP to make copies of genome from

Group IV: + ss RNA
+ ssRNA —→ viral RDRP —→ - RNA —→ viral RDRP —→
+ mRNA
more efficient to have >1 copy of RNA so to make a bunch, you need to make a template ( - RNA )

Group V: - ss RNA
- RNA —→ viral RDRP —→ + mRNA
use RDRP to make copies of genom

Group VI: Retroviruses
+ RNA —→ RT —→ - DNA —→ RT —→ ± DNA —→ host RNA polymerase —→ + mRNA
need double strand DNA to integrate into host chromosome
NOT lysogenic b/c they integrate into host chromosome and start replicating actively
