BIO 311 - Genetics Exam 3
1/157
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
158 Terms
mutation
any permanent change in the nucleotide sequence of a strand of DNA —> increases diversity
*can be in coding and noncoding regions
benefits of mutations
allows development of new genes
new species
adaptation to new environments
point mutations
mutations that only are only caused by one base pairing
silent point mutation
change in DNA result in NO CHANGE in amino acids
missense point mutation
change in DNA results in change in ONE amino acid
neutral mutation
missense mutation that does not affect phenotype
nonsense point mutation
change in DNA results in an early “stop” codon causing a shortened polypeptide/protein lacking proper function
Frameshift mutation
a mutation that changes the reading frame of the DNA sequence —> IDELS
insertion frameshift mutation
an extra nucleotide is added
deletion frameshift mutation
a nucleotide is missing
somatic mutation
mutations that affect non-sex cells (*only in sexually reproducing multicellular organisms)
generally not heritable unless organism reproduces through mitosis (asexually)
germline mutation
mutations that affect gametes —> heritable
(*only in sexually reproducing multicellular organisms)
germline cells
spermatogonia and oogonia
primary spermatocytes and oocytes
secondary spermatocytes and oocytes
spermatids
ova and sperm
effects of somatic mutations
*very common —> ~1 mutation per day
cell phenotype is unchanged
non-functioning
targeted for apoptosis
somatic mutations and cancer
somatic mutations tend to lead to cancer under the right conditions
in some cases, like hemimegaloencephaly, this enlargement is non-cancerous
but can still cause changes in behavior or nutrient intake to brain
chromosome mutations
large scale mutations that affect the structure of an entire chromosome
gene mutations
small-scale mutations affecting individual genes
duplication (chromosomal mutations)
when a region of a chromosome is copied twice over into a chromosome
inversion (chromosomal mutations)
when a region of a chromosome is flipped upside down and reinserted into the chromosome
deletion
when a portion from chromosome is missing
insertion
when a portion of one chromosome is inserted into another chromosome
translocation
when two pieces of two different chromosome switch places
base substitution
another term for point mutation; can be classified based on their effect on the protein (silent, missense, nonsense) or mechanism
transition (base substitution mechanism)
substituting a purine (A-G) for another purine or pyrimidine (C-T) for another pyrimidine (*more common)
*because A-G and C-T are a similar structure, mistakes are more common
transversion (base substitution mechanism)
substituting a purine for a pyrimidine and vise verse (*less common)
reading frames
6 total, 3 for each strand of DNA
can start at any of the 3 sports in the first codon
the correct reading frame is the one that results in the longest uninterrupted reading frame
closed reading frames
reading frames that are not used for gene expression — usually marked by interspersed stop codons
open reading frames (orfs)
the correct reading frame that has the promoter region in front of it
expanded nucleotide repeats
mutations that result from duplicated repeat regions, often within genes (typically in non-coding regions but can cause mutations in coding regions)
ex: CAG (micro-satellite sequences) repeated mistakenly
can affect phenotype if found within a gene or even outside the gene —> can also be caused by toxic RNA transcript
genetic diseases caused by expanding nucleotide repeats
spinal and bulbar muscular atrophy (CAG)
fragile X syndrome (CGG)
Huntington disease (CAG)
Fragile-X syndrome
results from tandem repeats (CGG) in the X-chromosome
more common in males (heritable)
symptoms are mostly cognitive (impairment, delayed development, learning disability)
CGG repeats create CpG islands that are more prone to DNA methylation —> silencing of the gene —> lose level of development
strand slippage
misalignment of the sequences; occurs during DNA synthesis
slips back and recopies DNA again
forward mutation
mutation that affect a wild-type (normal) phenotype is called
reverse mutation
reversal of a forward mutation back to wild type (normal)
loss of function
mutations result from missing or nonfunctioning protein product
GW2 gene (growth width) - loss of function
normal function: stops growth of the plant
loss of function: allow plant to continue growing
gain of function
results in a function that is not normally present - due to entirely new protein or protein produced in inappropriate tissue or time during development
Craniosynostosis - gain of function
results from improper timing of fusion of cranial plates in infants — due to improper timing of protein production
gains function earlier than expected
can cause significant cognitive repair
mutation rates
refers to the frequency in which wild-type alleles are permanently changed
rate reference can be within the cells of an individual or within populations or within taxa
smaller genome = higher mutation rate (increased reproductive rate)
large genome = lower mutation rate (DNA repair mechanisms and more accurate DNA replication)
factors of mutation rate
frequency at which changes in DNA take place
probability of DNA repair
detectability (need to be able to see/measure it)
*mutations are more common in non-coding regions and in wobble position
adaptive mutation
in stressful environments, bacteria may accumulate mutations at a faster rate (induced) which may help them survive
ex: antibiotic resistance
spontaneous mutation
mutations that arise under normal conditions, spontaneously without external influence or mutagens
ex: error in DNA repair, transcription, polymerization, replication
induced mutation
mutations that arise from outside environmental factors
ex: toxins or UV damage
tautomers
different versions of a nucleotide in which hydrogen atoms shift position, causing them to form bonds with incorrect base pair
ex: instead of G-C —> G-T
primary cause of spontaneous mutations
wobble base-pairing
incorrect nucleotide base-pairing with only two congruent pairings instead of three
ex: T-G and C-A
incorporated error
base substitution causes a mispaired base to be incorporated; can be fixed
replicated error
a mistake made during DNA replication where incorrect nucleotides are incorporated into the new DNA strand, which can lead to permanent mutations if not corrected.
strand slippage in insertions and deletions
occurs when the DNA polymerase slips during replication, leading to insertions or deletions in the new DNA strand; forms a hump in the new strand
misalignment in crossing over in insertions and deletions
misalignments in crossing over can lead to unequal exchange of DNA segments, resulting in insertions or deletions in the chromosomes; typically chromosomal mutations
results in a shortened chromosome and an elongated chromosome
depurination in spontaneous mutations
chemical change in DNA that causes a loss of a purine (A & G) base from a nucleotide
usually replaced with an A from ATP (incorporated error), which can cause improper replication in both strands (replicated error)
steric hindrance
causes the loss of purines in DNA; loss of pyrimidines is much less common
deamination
a chemical reaction that leads to a loss of an amino group (NH2) from the nitrogenous base, which causes inappropriate base-pairing
ex: cytosine —> uracil & 5-methylcytosine —> thymine
mutagens
chemicals in the environment that may damage DNA or alter its structure
ex: processed foods, cigarettes, UV rays, mustard gas
base analogs
chemicals with structures similar, or analogous, to natural nitrogenous bases
these chemicals are identical to nitrogenous bases in the perspective of DNA polymerase so it incorporates them into a growing DNA strand during replication
ex: 5-bromouracil is a thymine analog that normally pairs with adenine but can also incorrectly pair with guanine
alkylating agents
chemically modify nucleotide bases by adding alkyl groups, causing incorrect base pairing
ex: methyl groups (CH3) or ethyl groups (CH3-CH2)
ex: guanine + ethyl group = G-T pairing
oxidative radicals
reactive forms of oxygen that can cause chemical changes in DNA
ex: hydrogen peroxide is a reactive oxygen species
guanine + hydrogen peroxide = G-A
interculating agents
insert themselves in between adjacent bases, which causes indels
ex: acridine orange
radiation
is a form of energy release
particle radiation
subatomic particles such as neutrino or protons are released from a material
acoustic radiation
energy is released as gravitational waves
electromagnetic radiation
release of energy in the form of photons which act as a particle and a wave
the shorter the wavelength the higher the energy and higher frequency
the longer the wavelength the lower the energy and lower frequency
ionizing radiation
when radiation is high enough, it can dislodge electrons from atoms, which causes the formation of a free radical or ion
this can cause DNA breaks and damage
pyrimidine dimers
non-ionizing radiation mutation caused by UV radiation, which can halt DNA replication by forming a kink in the DNA
Ames test
used to test for carcinogens (causes cancer) in a substance
first bacteria is modified to prevent them from producing histidine (amino acid necessary for growth), then the substance is put in
if mutations occur, we expect reverse mutation, so that the bacteria regain the ability to produce histidine —> carcinogen
*some chemicals are not carcinogenic until they are metabolized, so rat liver extract is added to the test
transposable elements (transposons)
mobile DNA that can move around in the genome and can cause mutations by inserting into a gene or promoting chromosomal rearrangements (deletions, duplications, inversions)
*highly variable in sequence but often have flanking repeats (caused by staggered cuts)
*similar to viruses —> make staggered cuts and insert themselves into target DNA
often contain code for a transposase enzyme — this cuts DNA open to allow the transposon to insert itself
terminal inverted repeat
sequences 9-40 bp long that are inverted complements of each other that are found at the ends of many transposable elements and play a crucial role in their mobility
*DIFFERENT than flanking direct repeat
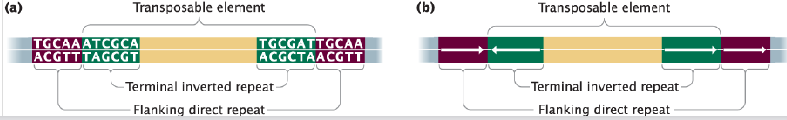
DNA transposons
a class of transposable elements that move within the genome through a "cut-and-paste" mechanism, allowing them to insert into new locations (target DNA) and potentially alter gene function.
retrotransposon
use reverse transcription
uses transcription to make an RNA intermediate
uses reverse transcription to make a DNA intermediate
which is then able to insert itself into target DNA
defenses against transposons
methylating DNA regions where transposons are common —> silence and prevent reverse transcription
use piwi-interacting RNA (piRNA) —> combines with argonaut protien to identify transposons and recruit methylating machinery
process of transposition
staggered cuts in target DNA (by transposase)
transposon is joined to single-stranded ends
DNA is replicated to fill in the gaps
replicative transposition
a copy is mad so original transposon may remain in original location (class 2)
nonreplicative transposition
transposon is cut out of original site and locates elsewhere
transposable elements in deletion
pairing by looping and crossing over between two transposable elements oriented in the same direction leads to deletion
transposable elements in inversion
pairing by bending and crossing over between two transposable elements oriented in opposite directions
mismatch repair
mismatch recognition (accessory proteins)
strand discrimination (correct sequence strand)
mismatched DNA cut out & replaced by DNA polymerase, sealed by ligase
in prokaryotes, special sequences of GATC nucleotides are methylated on OLD strand
direct repair (DR)
corrects chemically altered nucleotides by restoring their normal structure —> easy fixes
photolyase in some bacteria can fix thymine dimers caused by UV radiation —> cuts covalent bonds
does not replace altered nucleotides, restores them
base-excision repair (BER)
repair mechanism where the altered based is cut out and replaces
endonuclease cuts phosphodiester bond
DNA glycosylase cuts out base
other enzymes remove deoxyribose sugar
DNA polymerase beta adds new nucleotide
*base analogs can be repaired via BER
Nucleotide-excision repair (NER)
entire nucleotides are cleaved out and replaced
any irregularities n the 3D shape of DNA can lead to nucleotide
excision repair —> intercalating agents (chemical mutagens)
Enzymes scan DNA for “bumps”
Additional enzymes (helicase) separate the strands and SSBPs stabilize them
sugar-phosphate backbone is cleaved on both sides of damage
gap is filled by DNA polymerase and sealed by DNA ligase
ex: can be used to fixe thymine dimers (along with direct repair)
double strand break repair (DSBR)
a critical cellular process that repairs breaks in both strands of the DNA helix, restoring genomic integrity. It typically involves mechanisms such as homologous recombination and non-homologous end joining.
can be extremely dangerous — lead to DNA replication stalling and chromosomal rearrangements
Homologous directed repair in DSBR
a process of DNA repair that uses non-damaged sister chromatid as a template strand to fix the damaged DNA
nucleotides are removed from broken ends of DNA strands
homologous chromosome strand invades
DNA polymerase replicates homologous strand to complete broken strand
ligase closes broken strands
ex: BRCA 1 and BRCA 2 are frequently mutated breast cancer cells that are involved in HDR
nonhomologous end joining in DSBR
DNA repair mechanism where the broken strands are simply reattached
uses Ku protein binding
more error prone; could have loss of DNA
occurs in G1 phase of cell cycle
p53 protein
a tumor suppressor protein that prevents cell cycle from going unregulated
cancer
the unregulated growth and division of cells, usually due to a disruption in cell cycle
tumors
large masses of cancerous tissue growth
most common form of cancer
breast cancer
most deadly form of cancer
lung cancer; due to a lack of early, cheap screening tests and environmental factors
how is the type of cancer determined
cancer type is determined by the initial site of development
benign tumor
a tumor that remains in the same location
malignant tumor
a tumor whose cells have moved to another part of the body and formed new tumors in those sites
metastasis
the spread of cancer cells from the original site to other parts of the body
cancer as a genetic disease
many mutagens cause cancer - thus many mutagens are carcinogens
some cancers are associated with chromosomal abnormalities
some cancers run in families
Note: *other cancers that are not genetically passed on are typically caused by environmental factors
retinoblastoma in adults
usually only effects one eye as the cancer is only caused by environmental mutagens
adults need two mutated alleles in order to develop cancer, which is why the effects only occur in one eye
retinoblastoma in children
usually effects both eyes as the cancer is often caused by inherited genetic mutations, leading to a higher risk in family members
children already inherit one mutated allele and only need one more in order to develop cancer
Knudson’s 2 hit hypothesis
A model suggesting that two genetic hits or mutations are necessary for the development of certain cancers, such as retinoblastoma. This hypothesis explains how inherited mutations combined with environmental factors can lead to cancer.
Knudson’s multistep hypothesis
proposed thar cancer requires multiple mutations to develop
most cancers require more then mutations — often at different loci
*HOWEVER, most tumors develop from spontaneous mutations or mutagens
Clonal evolution
when tumors develop, the cells have mutations that promote proliferation allowing the tumor to grow rapidly
bypass checkpoints in cell cycle unnoticed
higher number of cells
can lead to cells that grow at a faster rate, making them more likely to mutate
aneuploidy
cells that have an uneven amount of chromosomes; these chromosomal mutations can contribute to clonal evolution
duplicating copies of genes that promote cancer
deleting copies of genes that prevent cancer
environmental factors in cancer
include various elements such as chemicals, radiation, and lifestyle choices that can increase the risk of mutations leading to cancer
individuals that migrate from one country to another take on cancer rates of that country
age can also effect cancer rates due to increased exposure to toxins and the aging of DNA repair mechanisms
allele affinity and cancer
some alleles that are linked to lung cancer are associated with higher rates of addiction due to their influence of dopamine receptors, which can increase likelihood of addiction
oncogenes
behave like dominant alleles - 1 mutated version is enough to promote cancer
molecules that promote division