BIOL214 E2 People+Structures
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
22 Terms
Helicase
Unwinds the DNA double helix at the replication fork.
Single-Strand Binding Proteins
Stabilize the unwound single strands and prevent them from re-pairing.
Topoisomerase
Relieves the twisting tension created by helicase unwinding the DNA.
Primase
Synthesizes a short RNA primer to provide a 3'-OH group, which is the necessary starting point for DNA polymerase.
DNA Polymerase III
The main replication enzyme. It adds new DNA nucleotides to the 3' end of the growing chain, synthesizing in the 5' to 3' direction. It also has a proofreading function.
DNA Polymerase I
Removes the RNA primers and replaces them with DNA nucleotides.
DNA Ligase
Joins the Okazaki fragments on the lagging strand by forming the final phosphodiester bond.
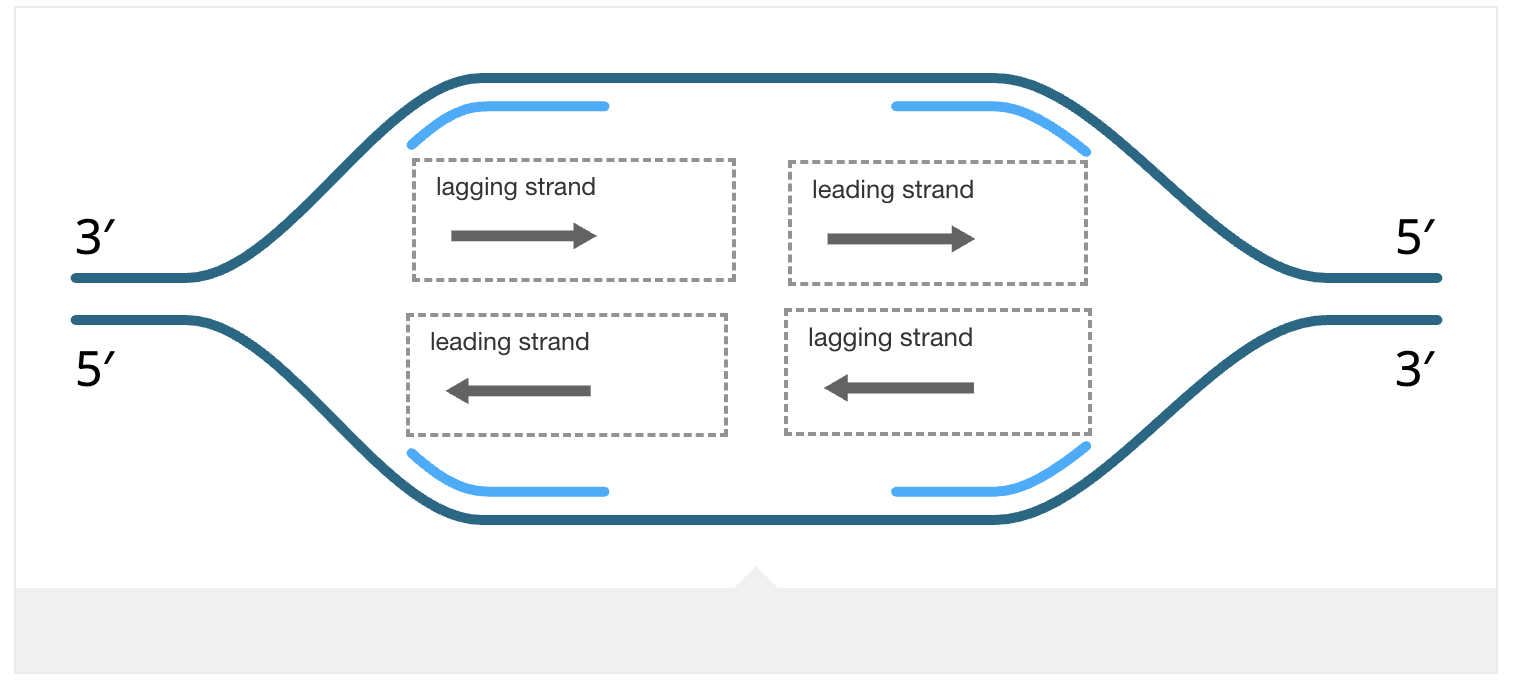
Leading Strand
Synthesized continuously in one long piece, as the polymerase follows the replication fork.
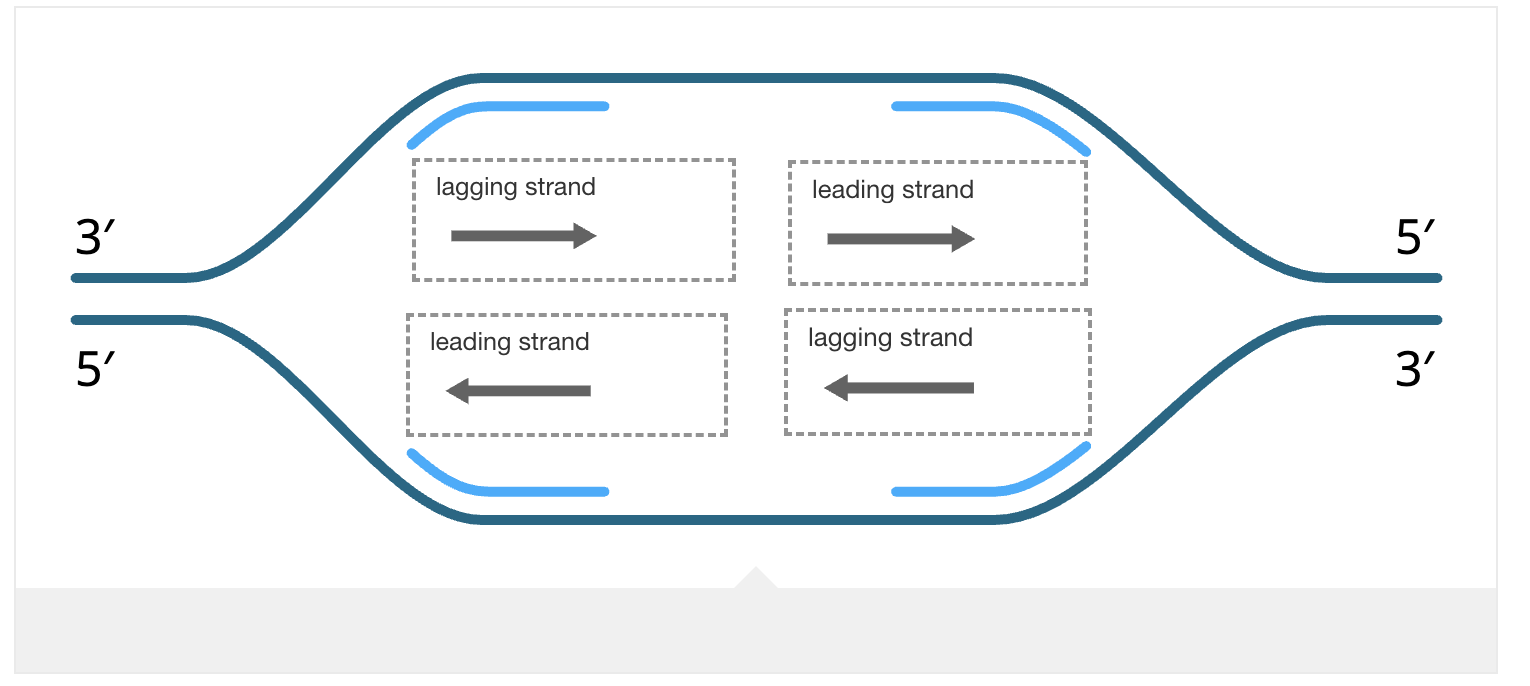
Lagging Strand
Synthesized discontinuously in short pieces called Okazaki fragments, away from the replication fork. Each fragment requires its own RNA primer.
Double Helix
Composed of two strands twisted around each other.
Antiparallel Strands
The two strands run in opposite directions. One strand runs 5' to 3', and its complementary strand runs 3' to 5'. This is necessary to allow the base pairs to form hydrogen bonds.
Complementary Base Pairing
A purine (double-ring base: A or G) always pairs with a pyrimidine (single-ring base: C or T). Specifically, A pairs with T (via two hydrogen bonds) and G pairs with C (via three hydrogen bonds).
Phosphodiester Linkages
Covalent bonds that join adjacent nucleotides, forming the sugar-phosphate backbone of each strand.
Frederick Griffith
(1928) The Transforming Principle Experiment: Worked with two strains of pneumonia bacteria: a virulent "S" (smooth, with capsule) strain and a non-virulent "R" (rough, no capsule) strain. He found that injecting a mouse with a mix of heat-killed S cells and living R cells was lethal, and he recovered living S cells from the dead mouse.
Conclusion: Some "transforming principle" from the dead S cells was able to convert the harmless R cells into the deadly S cells.
Avery, MacLeod, and McCarty
(1944) Experiment: They systematically isolated the major macromolecules (lipids, DNA, proteins, RNA, carbohydrates) from S-cell extract and tested each one's ability to transform R cells into S cells. They found that only the DNA extract could cause the transformation. They confirmed this with a reciprocal experiment where they destroyed one macromolecule at a time; transformation failed only when DNA was destroyed.
Conclusion: DNA is the transforming principle.
Hershey and Chase
(1952) The Definitive Proof Experiment: Used bacteriophages (viruses that infect bacteria). They radioactively labeled the viral protein with Sulfur-35 and the viral DNA with Phosphorus-32 in two separate experiments. After infection, they used a blender to separate the phages from the bacteria and found that only the radioactive phosphorus (DNA) had entered the bacterial cells.
Conclusion: DNA, not protein, is the genetic material that is injected into host cells and directs the production of new viruses.
Chargaff
(1949) Clues to Structure: Analyzed the nucleotide composition of DNA and found that the amount of Adenine (A) always equals Thymine (T), and the amount of Guanine (G) always equals Cytosine (C). This showed DNA was not a simple repeating polymer and provided the rules for base pairing.
Rosalind Franklin
(1953) Clues to Structure: Used X-ray crystallography to produce "Photograph 51," which revealed that DNA was a helix of a consistent diameter with phosphates on the outside.
Semi-Conservative Model (Meselson & Stahl)
(1953) Experiment: Bacteria were grown in a "heavy" nitrogen isotope (N15) and then transferred to a medium with a "light" isotope (N14). After one generation, all the DNA was of an intermediate density. After two generations, there was both intermediate and light DNA.
Conclusion: This pattern proved that DNA replication is semi-conservative, meaning each new DNA molecule consists of one "old" parental strand and one "newly" synthesized strand.
The End Replication Problem (Eukaryotes)
At the very ends of linear eukaryotic chromosomes, the final RNA primer on the lagging strand cannot be replaced with DNA, because there is no 3'-OH group to build off of. This would cause the chromosome to shorten with each replication cycle.
Telomerase
Enzyme that contains its own RNA template and extends the unreplicated 3' end of the template strand. This allows primase and DNA polymerase to complete the lagging strand, preventing the chromosome from shortening.
Five carbon sugar, nitrogenous base(1’ carbon connection), and phosphate group(5’ carbon connection)
Nucleotide Components