CELLBIO Exam 1 (Lecture 1-8)
1/137
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
138 Terms
design a experiment (4 steps)
question/hypothesis
approach/model system
design procedure/execute experiment
make observation/interpret results
experimental approaches (3)
biochemical
microscopy
genetics
biochemical experimental approach
observe structure/behavior of specific molecules and interactions
purify/separate biomolecules
detect proteins with antibodies
identify interactions between biomolecules
monitor biochemical reactions
microscopy experimental approach
visualize shape, location, and behavior of organisms, cells, cell parts, and molecules
observe molecules, cells, tissues, organisms
detect specific molecules, cell parts
live imaging → observe dynamics
genetics experimental approach
evaluate function/structure of cell or cell part when DNA is altered
identify cells/individuals with phenotype of interest and genes
create mutation in region of interest, evaluate the effect on cells
alter expression levels of RNA or protein, evaluate effect on cells
add sequence to gene → adds AAs to protein → fusion protein or tagged version is useful for experiments
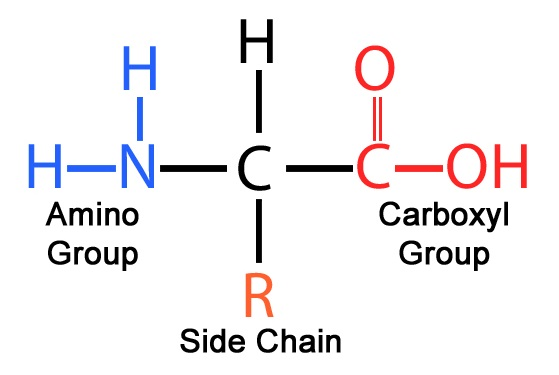
amino acid characteristics determined by
sidechain
charged sidechain → polar, uncharged → nonpolar
negative charge → acidic
positive charge → basic
N-terminus vs C-terminus
N-terminus (Amino terminus): This is the "start" of the protein chain and contains the free amino group (-NH₂). It's called the N-terminus because it has the nitrogen atom from the amino group. It is the first part of the protein synthesized during translation.
C-terminus (Carboxyl terminus): This is the "end" of the protein chain and contains the free carboxyl group (-COOH). It's called the C-terminus because it has the carbon atom from the carboxyl group. It's the last part of the protein synthesized during translation.
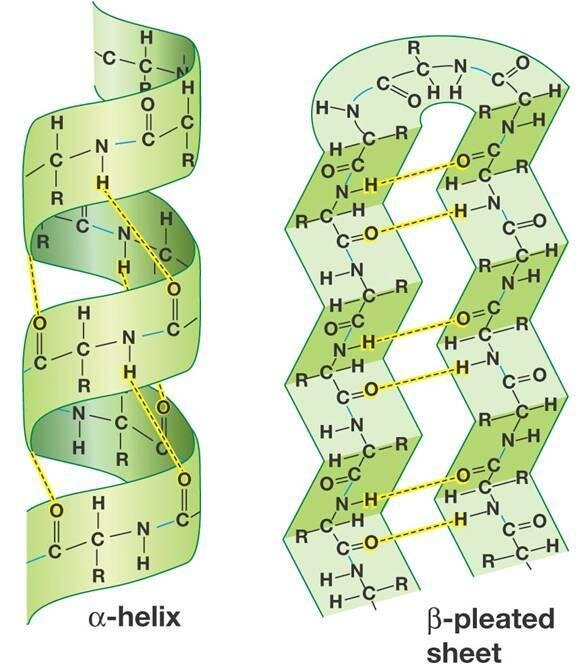
common protein structures
secondary structure
alpha helix - right-handed coiled structure, where the polypeptide backbone twists around an axis, and the side chains (R groups) extend outward from the helix. The structure is stabilized by hydrogen bonds
beta strands - formed by two or more polypeptide chains that align side by side. The backbone of the chains forms a sheet-like structure, where each chain runs in a parallel or antiparallel direction. Hydrogen bonds stabilize the arrangement between the strands.
folded proteins form complexes
monomer - single protein subunit
homodimer
heterodimer
trimer
tetramer
protein structure determines function
function by interaction/binding noncovalently (hydrogen bonds, ionic bonds, hydrophobic interactions, and van der Waals forces) with other molecules like proteins, lipids, nucleic acids
proteins can be covalently modified by
addition of specifc groups to specific AAs
glycosylation
phosphorylation
protein function can be regulated by…
covalent modification or binding to other molecules
model systems
cell or organism commonly used for research as example to understand biology of cell or organism more generally
key characteristics of model systems (5)
easy to maintain/grow
can do experimental manipulation (and observe it)
subject to existing biological data
existing experimental tools/protocols
minimal genetic variation in population
choose model system based on (8)
behavior/features of interest
tools, technique, biological information available
timescale
cost
ethicality
applicability to other systems
simplest that’s sufficient
better than other systems
cell culture (4)
removed from multicellular organism grown in lab to study cell behavior and experiment outside of organism in vitro
manipulate cell behavior without affecting organism
immortalized cell lines (human tumors) → indefinite replication
some cells are difficult to maintain/grow
microscopy
visualize things not visible to human eye
parts of microscope
beam source - light or electrons
sample - must be prepped properly based on microscope type
objective lens - collect signals that go through sample
detector - generate image
light vs. electron microscopy considerations (4)
determined by sample and features of microscope
magnification
detection
resolution
contrast
resolution
shortest distance between 2 points/objects distinguished as separate
resolution = r = 0.61λ/nsinθ
lower r = better resolution
λ = wavelength of beam used
n = property of media between objective and sample (air, water, oil)
θ = property of microscope
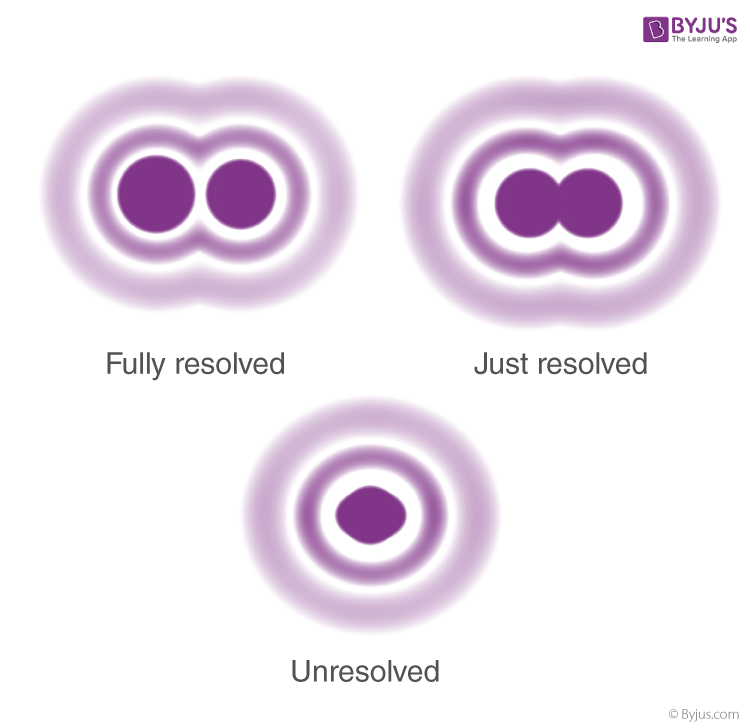
contrast
difference in signal intensity between object and background
increase contrast by
manipulating light/electron beam
manipulating sample by adding stain, fluorescent molecules, or heavy metal
sample preparation steps include (6)
fixation
permeabilization
dehydration/drying
freezing
sectioning
mounting
sample preparation based on (3)
type of sample
type of object visualization desired
type of microscope
Light Microscopy
limit of resolution: 200 nm
alive or dead cells
method of increasing contrast by manipulating sample/light wavelengths are important
types of LM (9 points)
white LM - visible/white light (all λs) → sample → objective lens → detector
brightfield M - manipulate light path to increase contrast → image formed by the contrast between the sample and the surrounding medium
phase contrast - manipulate light to visualize light/dark regions → shifting the phase of light passing through different parts of the sample, making it easier to see internal structures.
Nomarski/differential interference contrast - manipulate light in a different way so image appears 3D → uses polarized light to create 3D-like images of transparent samples. It improves contrast and resolution, making it easier to observe fine details.
fluorescence M - light (specific λ) → sample with fluorescent molecules (excited by specific λ, emit lower energy/longer λ) → object lens (collect lower λ) → detector
different fluorescent molecules have different excitation λs and emission λs, so they can be combined in one experiment
worst: epifluorescence (widefield) - entire sample illuminated at one time, emitted light used to generate image
confocal - one thin section illuminated at a time, images combined to form final image
best: superresolution - only a few molecules illuminated at a time, images combined to form final image, resolution limit <200 nm
approaches to detect protein by fluorescence microscopy
Immunofluorescence using fluorescently labelled Ab that binds to protein of interest
Immunofluorescence using fluorescently labelled Ab that binds to protein tag attached to protein of interest
Direct fluorescence of flourescent protein tag attached to protein of itnrest
Approach 1: Immunofluorescence using fluorescently labelled Ab that binds to protein of interest
Fix sample - limit molecule movement, chemicals react with and crosslink proteins to each other to stabilize sample
Permeabilize sample - with detergent, disrupts lipids in cell membrane to allow Ab to access proteins
Add Ab
Detect with fluorescent microscopy
primary Ab - binds to protein of interest
secondary Ab - has fluorescence, binds to primary Ab
Approach 2: Immunofluorescence using fluorescently labelled Ab that binds to protein tag attached to protein of interest
protein tag - specific AA sequence useful since Ab can bind here and be detected/purified, can work on any protein
protein tag + POI = fusion protein
Generate fusion protein:
DNA encoding protein tag covalently attached to DNA for POI, fusion can occur outside of cells
Introduce fusion DNA into cells via transfection, electroporation, viral infection, microinjection
Fix, permeabilize, add Ab, detect with fluorescent microscopy
Approach 3: Direct fluorescence of fluorescent protein tag attached to protein of interest
fluorescent protein i.e GFP
visualize by fluorescent microscopy without any further sample prep i.e fixing
detect fluorescent protein in live or dead cells
live cells, visualization of dynamics (since no further sample prep like fixing that kills cells)
Electron Microscopy
uses electrons as beam source
electrons very small → resolution possible. down to atomic level
imaging occurs in vacuum → live cell imaging not possible
cells fixed/frozen to preserve structures
heavy metals can be arranged to add contrast
types of EM
transmission EM - samples must be thin/sectioned, electrons pass through the sample to be detected
scanning EM - electrons hit surface of sample produces signals, image detected reveals surface of sample → 3D surface structure
Ab structure
light chain and heavy chain
variable region: tips of Y where antigen can bind
constant regions: everything else
recombination of DNA makes different Abs
B cells
B cells (white blood cell) express one Ab
Ab expressed on surface of B cell recognizes antigen → B cell activated
differentiates into plasma cells that secrete large amounts of Ab that targetpathogen/antigen of that type, mark for degradation
clonal expansion - divides rapidly to form more Ab
immunohistochemistry
use Ab to detect antigens in tissue sample
Ab usually linked to an enzyme or fluorescent dye
enzyme substrate is added or dye is activated for detection by light microscopy
immunogold EM
use Ab to detect antigens
Ab are linked to gold nanoparticle/bead
location of Ab indicated by dense black dot in EM
Western Blot
detect changes in
concentration, modification/substitution/addition to protein, interactions with other proteins
protein isolated
chem/physical based methods to disrupt membrane
detergent solubilize membrane
whole cell lysate (nuclear, cytoplasmic) obtained
lysate loaded onto PAGE for protein separation
with SDS buffer - denature proteins/linearized and uniformly coat in negative charge
lysate can have dye solution - adds color and has glycerol to make it thicker than SDS buffer
protein transfer from gel to membrane (nitrocellulose or unreactive material) that’s more durable via electroelution
Minimize nonspecific Ab binding to membrane by incubating membrane with blocking agent like milk or purified proteins in mild detergent
Incubated in blocking solution and primary antibody for POI
Washes with washing buffer (same as blocking solution/mild detergent remove nonspecifically bound primary Ab) under mild agitation
Incubated with secondary Ab (bound to fluorophore) recognizing primary Ab
Washes with washing buffer
cytoskeleton function (5)
cell shape/’structure
spacial organization
connect cell to external environment
movement of molecules within cells
movement of cells/change cell shape
cytoskeleton filmanent
made of protein subunits that bind to each other noncovalently
actin filaments- 7 nm
intermediate filaments - 10 nm
microtubule - 25 nm
Actin
actin monomer
helical
asymmetrical/polar, 2 ends are different
diverse structures
concentrated near cell edge
Microtubules
tubulin heterodimers
hollow tubes
asymmetric/polar
rigid/straight
one end = minus end that’s attached to microtubule organizing center (MTOC) near nucleus
Intermediate Filaments
tetramers
ropelike structure
symmetric/nonpolar (ends same)
strong
large and heterogeneous group
phospholipid bilayer
hydrophilic/polar head + hydrophobic/nonpolar tails
head = polar group (varies by phospholipid) - phosphate (negative charge) - glycerol
tail = 2 fatty acid chains
form bilayers through hydrophobic effect
cytosolic vs noncytosolic leaflet
a bilayer has 2 leaflets
noncytosolic leaflet - one side extracellular space, one side cytosol
cytosolic leaflet - one side cytosol, one side cytosol, inside = lumen
extracellular environment
outside of cell
water, air
protein, carbohydrate, many other molecules
cells (same, different types)
cell influences EE and EE influences cell
extracellular matrix (ECM) structure (5)
3D molecular network surrounding cells in multicellular organism
composed predominantly of proteins and carbohydrates
synthesized and modified by cells
highly variable components/organization
contributes to structure/function of cells/tissues
ECM Molecules
polysaccharides, proteins with cov. attached hydrates (glycoproteins, proteoglycans)
i.e collagen, elastin, laminin, fibronectin, glycosaminoglycan, hyaluronic acid
ECM function (10)
shape and organization of tissues
mechanical properties of tissues
physical attachment of cells
biochemical signaling
important for
cell migration
cell differentiation
wound healing
development
diseases including cancer
tissue enginering and regenerative medicine
cell junctions
interaction with extracellular environment
link outside to inside
cytoskeleton filaments i.e actin - adaptors - transmembrane proteins through noncytosolic leaflet/PM that bind to ECM
Cell-ECM junctions
transmembrane proteins bind to stuff in ECM
aligned with actin
common junctions: actin linked cell matrix, hemidesmosome
Cell-Cell Junction
contains transmembrane proteins in PM of both cells that bind to each other, can be same or different transmembrane proteins
transmemb prot - adaptor - cytoskeletal filament
intermediate filaments align
common types: desmosome, tight junction, gap junction, adherence junction
cell membrane are diverse, dynamic
diverse: differ in types/amount of lipids and proteins in PM
variation between leaflets, regions, organelles, celltypes
dynamic: lipid/prot move within leaflet: rotation, bending, lateral diffusion
flipping is rare and requires energy
co-translational translocation
prot translation: mRNA read by ribosome, synthesize protein starting at N-terminus
in N-terminal, many proteins made by CTT have specific sequence called N-terminal ER signal sequence or signal peptide
8+ hydrophobic AAs
targets protein for CTT into ER
Signal Recognition Particle made of RNA and protein binds to signal peptide and ribosome → pause in translation
SRP binds to SRP receptor/transmembrane protein complex in ER membrane
SRP binding to SRP receptor all translation to continue + positions ribosome close to translocon/protein complex in membrane with water filled channel
when ribosome not bound to translocon, translocon is blocked by part of protein called plug
when bound, plug moves so protein synthesized moves through channel
SRP/SRP receptor released from complex
ribosome positioned on translocon + protein synthesizing through translocon channel, hydrophobic N-terminal ER signal sequence binds to side of channel near cytosolic side
signal peptidase enzyme cleaves protein after signal sequence → releases peptide into ER lumen
cleaving occurs during or immediately after translation
N-terminal signal sequence released into membrane by lateral opening of translocon, usually gets degraded
After translation, ribosome dissociates from translocon
unidirectional, there’s no going from lumen to cytosol
orientation of protein relative to membrane doesn’t change after translation
protein translocation: proteins that go to GA, lysosomes, endosomes, cell surface all need to pass through the
ER membrane
polyribosome
multiple ribosomes bind to mRNA that then gets cotranslated into ER creating the rough ER
water soluble vs transmembrane proteins
completely cross membrane vs embedded into the membrane
same steps as CTT, ribosome positioned on translocon, SRP receptor leaves
signal sequence opens translocon channel, binds to channel as protein threaded through membrane as loop → released into lumen, signal peptidase cuts peptide after signal sequence which is released and rapidly degraded → protein goes to bind and close translocon
signal sequence bound to translocon channel initiates CTT → transfer halted by stop transfer sequence/additional sequence of hydrophobic AAs further in peptide chain → stop transfer sequence released laterally and drifts into plane of lipid membrane → forms membrane spanning segment that anchors protein into membrane
internal signal sequence
in some transmembrane proteins, ISS used to start CTT which continues until stop transfer sequence is reached → 2 hydrophobic sequences released into bilayer where it stays anchored
complex multipass proteins - many hydrophobic regions span bilayer, additional paris of start and stop sequences that reinitiate translocation along the peptide and stops translocation/polypeptide release
stitched into membrane
single pass transmembrane protein
N-terminal signal sequence coming out of ribosome associated to translocon, gets translocated
hydrophobic transmembrane domain/second transmembrane sequence remains in translocon, doesn’t get into lumen → translation resumes
signal peptidase cuts peptide after NSS
N-terminus in lumen, C-terminus in cytosol
protease protection assays
experimental method to study proteins and processes associated with membrane and membrane bound organelles
can be used to study processes like CTT
determine orientation of membrane proteins in a membrane
protease acts enzymatically on POI → time → protease + degraded POI
“protection” refers to protection of POI or part of POI from protease i.e using a membrane that protects against protease
In vitro PPA
study outside of cellular context
starting sample in tube: POI in membrane bound compartment
experimental sample: +protease, time to starting sample
use SDS-PAGE/WB to detect POI
compare btw starting and experimental sample
control sample for protease activity: +protease +detergent +time
disrupt membrane so protease can degrade POI
if POI not degraded → protease not working
control sample for detergent: +detergent +time
check if detergent has protease activity
membrane disrupted and POI not degraded
ER-Derived Microsomes
study CTT using this in vitro
small membrane bound compartments derived from ER
rough ER with ribosomes on cytosolic side → disrupts → smaller compartments called microsomes
rough and smooth microsomes (±ribosomes)
can be separated by different densities
mini-ERs, support functions like CTT
study CTT in microsomes with PPA
starting sample: rough microsome + components for prot translation: mRNA< amino-acyl tRNAs, GTP, translation factors
+time = protein translation occurs, protein located in microsome following CTT
+protease + time = protease acts
use SDS-PAGE or WB to detect POI
if POI in microsome → not degraded
additional samples as controls or experimental conditions
orientation of internal ER signal sequences (ISS)
N-term - (+)ISS(-) - C-term → N-terminal in cytosol, C-terminal in ER lumen
N-term - (-)ISS(+) - C-term → C-terminal in cytosol, N-terminal in ER lumen
aka positive is inside cytosol
ISS vs TMD
internal signal sequence - anchor and targeting sequence for CTT, hydrophobic AAs
transmembrane domain - anchor in CTT, hydrophobic AAs
protein processing in ER (3)
3 major processes
folding
disulfide bond formation
glycosulation
folding (protein processing ER)
can occur cotranslationally as soon as it exits ribosome
molecular chaperones are proteins that help others fold properly
bind to unfolded protein, incorrectly folded protein, and unassembled component of protein complex
chaperone binding/unbinding often coupled with ATP hydrolysis
in ER: BIP, calreticulin, calnexin
disulfide bonds (protein processing ER)
covalent bonds between two sulfurs on 2 cysteine AA sidechains
formed due to oxidizing environment
two -SH sticking out, remove Hs, S-S
protein disulfide isomerase - protein in ER that helps formation of correct disulfide bonds
form between 2 cysteines on same protein or 2 different proteins
glycosylation (protein processing ER)
most proteins synthesized at ER have covalently attached carbohydrate
carbohydrate attached as single unit to asparagine AA in protein
asp/N - X - S or T
linkage catalyzed by enzyme in ER membrane
N-linked glycosylation helps proteins fold properly
N-linked glycosylation helps proteins fold properly
carbohydrate attached to Asn → two glucoses of three removed by enzymes → monoglucose carb bound by chaperone calreticulin/calnexin to help protein fold → protein separated from chaperone by removal of final glucose by glucosidase
if protein misfolded, it will bind to glucosyl transferase (adds back glucose), so then chaperones can rebind and refold
proper folded protein - no glucoses on it
ubiquitination
covalently attaching ubiquitin - small protein of 76 AAs to other proteins
attachment is between C-terminal of ubiquitin and sidechain of lysine AA or N-terminal of target protein
catalyzed by ubiquitin ligase (also known as E3 enzymes, E1 and E2 also involved)
removed by deubiquitinating enzymes
single ubiquitin attachment = monoubiquitination
polyubiquitination
since ubiquitin has lysine residues, one ubiquitin can be attached to another ubiquitin to form a chain
use K6, 11, 27, 29, 33, 48, 63 or N-terminus (M1)
linear or branched
added by ubiquitin ligase, removed by deubiquitinating enzymes
ubiquitination alters target protein structure and function
creates binding sites for other proteins with specific ubiquitin binding domains
linkage types determines outcome of ubiquitin change
has many roles in
regulation of transcription
DNA repair
nuclear transport
protein degradation
cell death, signaling, division
ER-Associated Degradation (ERAD)
process of degradation of misfolded proteins in ER
removes nonfunctional proteins from ER
proteins in lumen and ER membrane that aren’t able to fold correctly are substrates for degradation
ERAD process (4 steps)
recognition of misfolded protein
proteins translated by CTT, start unfolded in lumen and fold
molecular chaperones help fold + recognize if protein unable to fold within a reasonable timeframe
retrotranslocation
Transport of recognized misfolded protein across lipid bilayer into cytosol through retrotranslocon.
polyubiquitination
Unfolded protein is polyubiquitinated
proteasomal degradation
Polyub chain targets protein for degradation by proteasome
Proteasome is large protein complex present in cytosol, protein enters middle of proteasome that has multiple proteases that completely degrade the protein
Ubiquitin proteins are removed before degradation so they can be reused/recycled again
guanine nucleotide-binding protein (g-proteins)
bind guanine nucleotides
bind GDP or GTP (diphosphate vs triphosphate)
GTPases that catalyze hydrolysis of GTP→GDP
enzymatic activity/GTPase activity
GTPase activity
G-proteins have enzymatic activity: GTP → GDP
G-proteins must release GDP molecule → empty G-prot → rebind GTP if conc. high enough
in cells, GTP concentration is typically way higher than GDP conc
GTP bound form = active conformation
GDP bound form + empty form = inactive conformation
GTPase Activating Proteins (GAP)
increase GTPase activity
stimulate hydrolysis of GTP → GDP by G-prot
favor inactive state
guanine nucleotide exchange factors (GEFs)
stimulate GDP release by G-prot → empty G-prot
favor active state
G-Proteins act like molecular switch
either on or off
can be switched quickly between two states
key for temporal and spatial regulation of cellular processes
vesicle transport
exchange of molecules occurs through vesicles: small membrane compartments, between endomembrane system: ER, GA, nuclear envelope, endosomes, lysosomes, PM, biomolecules
vesicles are formed at one membrane and fuse at a different membrane
aka membrane trafficking, vesicle trafficking
topology is maintained during transport
orientation of molecules relative to cytosolic/noncytosolic side of membrane is maintained
luminal in ER → as it moves, it will stay inside lumen/noncytosolic
transmembrane in ER → as it moves, N/C terminus will stay oriented i.e N-terminus noncyt/C-terminus cyt
vesicles formed at particular cellular membranes have specific target membranes (3)
secretory
endocytic
retrieval
secretory pathway
two types: ER → PM, ER → lysosome
ER → PM
ER → GA (early compartments → late comparments)
exocytosis: late GA → PM → release molecules into extracellular space
secretory vesicle = specialized vesicle that delivers specialized cargo molecules to extracellular space
ER → lysosome
ER → GA
GA → early or late endosomes
early endosome → late endosome
late endosome → lysosome
endocytic pathway
molecules located in extracellular space or at PM to move into cell towards lysosomes
PM → lysosome
PM → early endosome → late endosome → lysosome
retrieval pathways
in order to function, need molecules located in specific place
molecules can be transported to other compartments in a process that is part of their function or nonspecific process
used for molecules that need to be returned from one location to a different location in order to function properly
GA → ER
late GA → early GA
early or late endosomes → late GA
early endosome → PM directly or early endosome → recycling endosome → PM
secretory vesicle → late GA
vesicle budding
vesicle budding from donor membrane - process of curving/bending membrane into cytosol to form small separate membrane bound compartment
can be initiated with transmembrane cargo proteins with specific signal sequence
adaptor protein complex binds to signal sequence in cargo protein, link cargo protein with coat protein complexes
self assembly of coat protein complex leads to membrane curvature/bending and cargo clustering within forming vesicle bud
coat recruitment GTPase
regulate association of coat proteins, specifically with donor membrane
coat recruitment GTPases located in cytosol in inactive or GDP bound state
nucleotide exchange and activation of coat recruitment GTPase is stimulated by GEF in membrane
coat recruitment GTPases swap GDP for GTP → conformational change causes hydrophobic part of protein to become exposed → associates with lipid bilayer of donor membrane
GTP bound coat recruitment GTPase binds to coat protein complex bringing it close to the membrane
vesicle scission
pinching off of vesicle occurs in small region still connecting vesicle to donor called neck
vesicle targeting
association of vesicle with a specific membrane and the movement of vesicle close to target membrane
transmembrane proteins in vesicle membrane and target membrane mediate targeting
vesicle tethering - first association of vesicle with target membrane is through long range association
vesicle docking brings vesicle and target membrane close together
vesicle tethering
involves Rabs - GTPases that can be in GDP bound/inactive or GTP bound /active state, have a covalently attached lipid group, GDP dissociation inhibitors - shield hydrophobic lipid group and keep Rab soluble in cytosol when Rab bound to GDP
Rab GEF promotes nucleotide exchange GDP → GTP, regulates association of Rab with a specific membrane
In GTP bound state - Rab unbinds GDI and exposed lipid embeds in a membrane i.e vesicle, target, or vesicle & target membrane
Rab GTP in membrane can bind to 1+ Rab effector proteins - proteins that bind Rab only in GTP state
some Rab effector proteins are membrane bound proteins that act as tethering proteins to link vesicle and target membranes together
vesicle docking
close association of vesicle and target membrane
accomplished by transmembrane proteins called SNAREs in both vesicle and target membrane
associated with vesicle = v-SNAREs
consist of single protein with one helical domain
associated with target membrane = t-SNAREs
consist of 3 different proteins each with one helical domain
4 helical domains of SNAREs interact to form four-helix bundle that acts like zipper to bring membranes very close together
association of v-SNAREs and t-SNAREs is very specific
vesicle fusion
after vesicle targeting brings vesicle very close to target membrane, vesicle fusion can occur
association of SNAREs involved in membrane fusion too
formation and zipping up of four helix bundle excludes water from space between vesicle and target membranes
very close association of two membranes allows for fusion of cytosolic leaflets of membranes
fusion of noncytosolic leaflets occurs to complete fusion
following fusion, lipids from vesicle are part of donor membrane, lumen of vesicle merged with lumen of target membrane
vesicle process + vesicle uncoating + vesicle movement
vesicle budding
vesicle scission
vesicle…
UNCOATING: coat proteins come off of vesicle, happens at different times/ways for different coat proteins
MOVEMENT: vesicle gets moved by motor protein along cytoskeletal filament (microtubule)
vesicle targeting
tethering
docking
vesicle fusion
vesicle coats
COPI, clathrin
outer layer = coat proteins
coat proteins, coat recruitment GTPases
inner layer = adaptor proteins
bind to transmembrane cargo proteins
COPI and clathrin look different because coat proteins have different structures
resetting system for vesicle transport
in order to recycle components, need to be reset and get back to right place
coat recruitment GTPases hydrolyze GTP, release membrane, go bind to another donor membrane
coat proteins disassemble and can be recruited somewhere else
Rabs hydrolyze GTP and unbind effectors
v-SNAREs and t-SNAREs separated (need energy) by a protein
TMDs on each and 4 helix bundle
why is there specificity of coat protein based on vesicle formed at a membrane
this is about vesicle formation: adaptor → coat recruitment GTPase → GEF
location of GEFs for coat recruitment GTPases is the key
adaptor proteins are binding to at membrane
NOT coat protein
why would a vesicle fuse with different target membranes
vesicle targeting: tethering/docking
based on Rabs (and Rab GEFs) and SNAREs (v-SNAREs)
NOT coat protein
golgi apparatus
multiple flattened compartments (cisternae) arranged in stacks
vesicles form/fuse with cisternae
cis/trans side
cis golgi network - cis golgi - medial golgi - trans golgi - trans golgi network
proteins made by CTT in ER are transported ER → cis golgi → trans golgi
protein gets processed moving through golgi
protein gets processed in golgi
sequential processing as protein moves cis → trans
protein processing carried out by enzymes localized to specific GA compartments
N-linked glycosylation
addition/modification of O-linked glycosylation: covalent attachment of carbohydrate to oxygen atom of Ser/Threo sidechain
sulfation + sulfate
phophorylation + phosphate
lipidation + lipid
proteolysis - cut protein in specific places
golgi vesicle transport destinations (4)
vesicles at cis-golgi fuse with ER
intra-golgi - can fuse with adjacent compartment of GA
trans-golgi fuse with endosomes
trans-golgi fuse with PM
cis golgi → ER
COP1 coat protein complex
cargo protein require specific signal sequence for transport