Molecular Genetics 1.2
1/23
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
24 Terms
What phase of the cell cycle does replication take place?
Synthesis phase (S phase)
How many potential origins of replication are there?
30-50,000 but only a subset are activated per S-phase
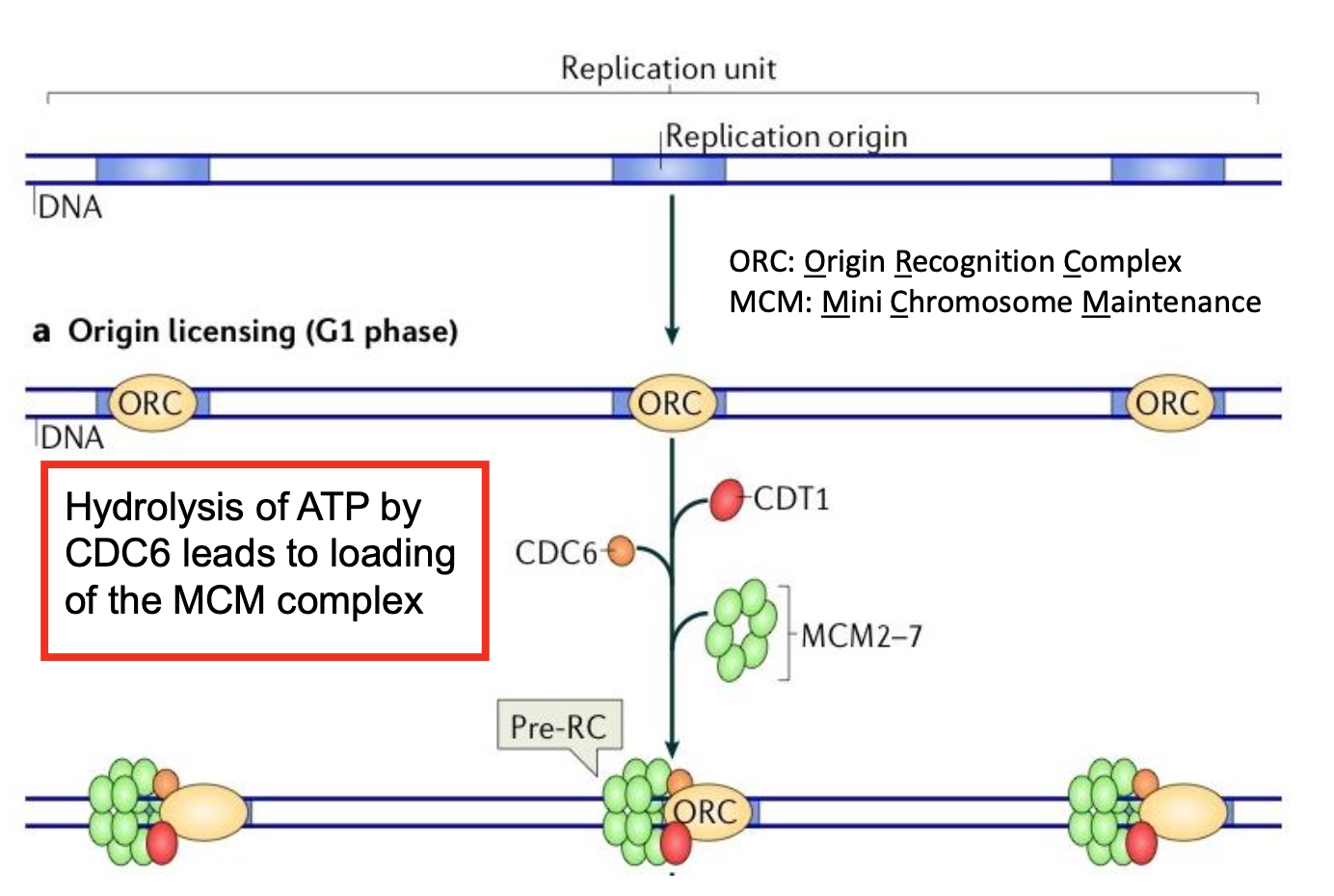
What happens in Pre-RC assembly in G1 phase?
Origin Recognition Complex -ORC- (6-submit ATPase complex) binds DNA
Cdc6 & Cdt1 recruit MCM2-7 double hexamer
Hydrolysis of ATP by cdc6 leads to loading of the mini chromosome matrix - MCM - complex
DNA sequence:
No strict consensus sequences
Enrichment of OGREs (Origin G-rich Repeated Elements)
Formation of G4 DNA - can act as recruit platforms for origin-binding proteins but as a barrier to fork progression if required.
G4 needs to be unwound -
Helices such as PIF1 and BLM involved in unwinding G4s during replication
DNA shape is DYNAMIC - always changing
Origins are fired during S-Phase entry, what does this cause?
Origins fire once, then are inactivated for the rest of S-Phase
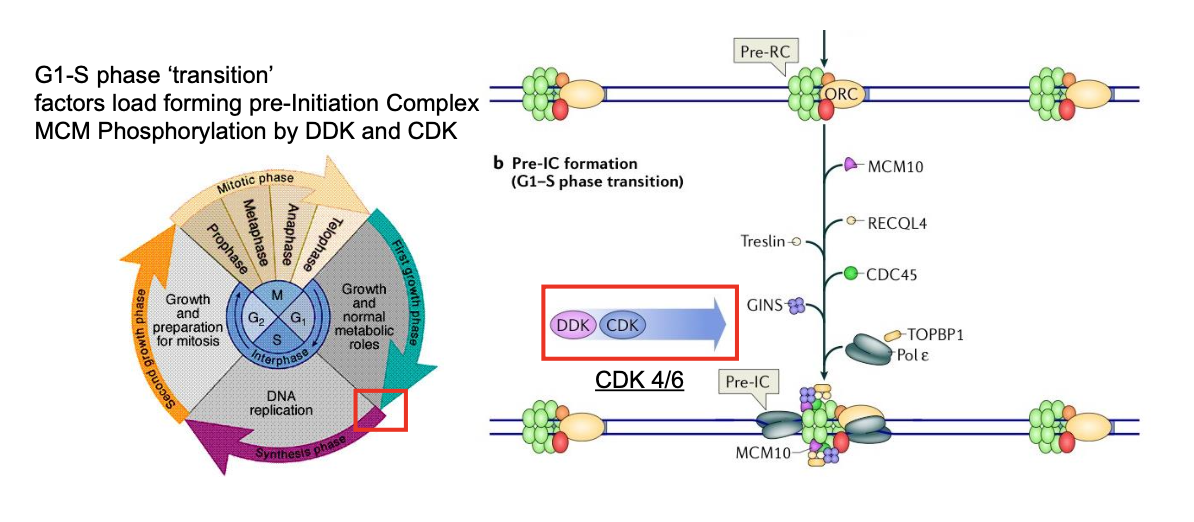
Activation of Kinases:
Cyclin Dependent Kinase 2/Cyclin E
Dbf4-dependent Cdc7 kinase (DDK)
phosphorylate initiation factors
minichromosome maintenance protein complex (MCM)
CMG complex is formed and encircles the leading strand template
ATP hydrolysis drives unwinding
Initiation factors (TopBP1, Treslin, RecQL4) stabilise CMG assembly
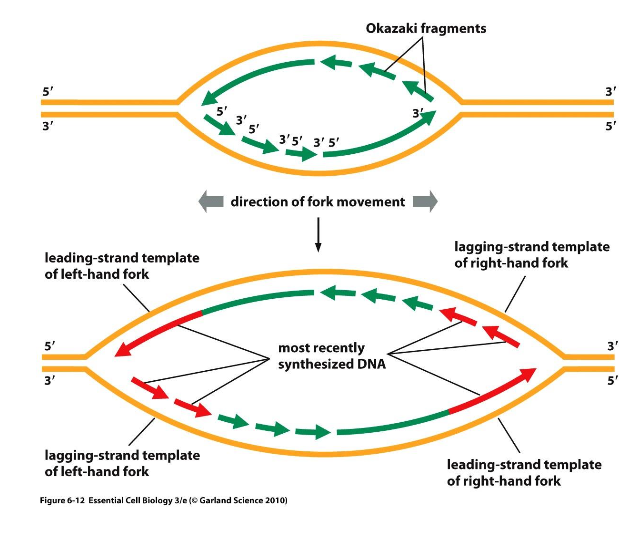
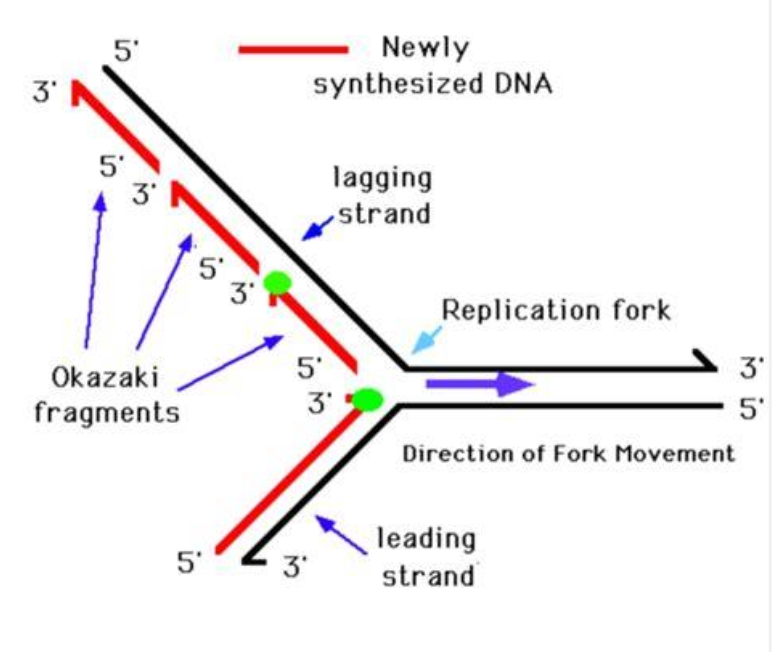
What are replication forks and bubbles?
Replication forks form at the point of synthesis initiation - a Replication Origin
Two DNA strands are generated via co-ordination of Leading Strand and Lagging
Strand synthesis
Leading strand DNA synthesis is continuous in a 5ʹ – 3ʹ direction producing
as a single, continuous molecule, until the
replication fork terminates.
Lagging strand DNA synthesis is discontinuous in a 3ʹ – 5ʹ
direction producing as a series of “short” molecules Okazaki fragments, until the replication fork terminates
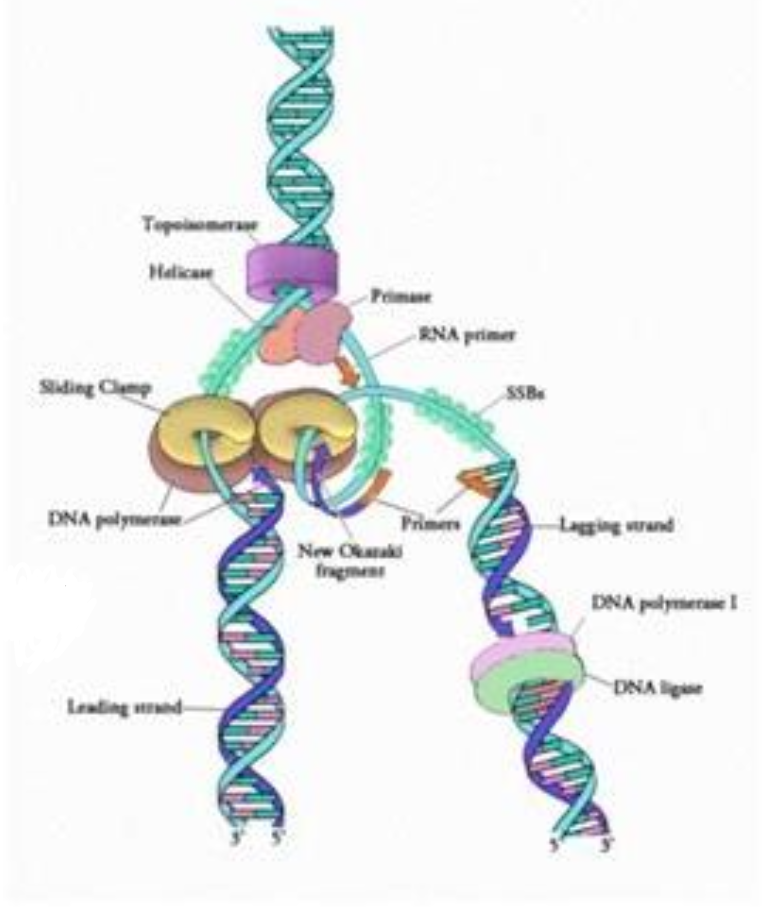
How is the replication fork built and name key molecules involved?
Unwinding: CMG separates dsDNA into ssDNA
Replication Protein A binds ssDNA stabilising via prevention of formation secondary structures
Polymerases:
Pol α-primase: RNA-DNA primer
Pol ε: continuous leading strand synthesis
Pol δ: discontinuous lagging strand (Okazaki fragments)
Processivity factors:
Proliferating cell nuclear antigen clamp keeps polymerase bound
Replication Factor C loads PCNA
Topoisomerases I/II relieve supercoiling ahead of fork
How is the lagging strand completed?
Lagging strand continuity is critical for genome integrity Okazaki fragments ~150 bp
Primer removal
RNase H1 digests RNA except last ribonucleotide
Flap Endonuclease 1 (FEN1) removes remaining flap structure
Gap filling
Pol δ synthesizes DNA
Nick sealing
DNA Ligase I joins fragments
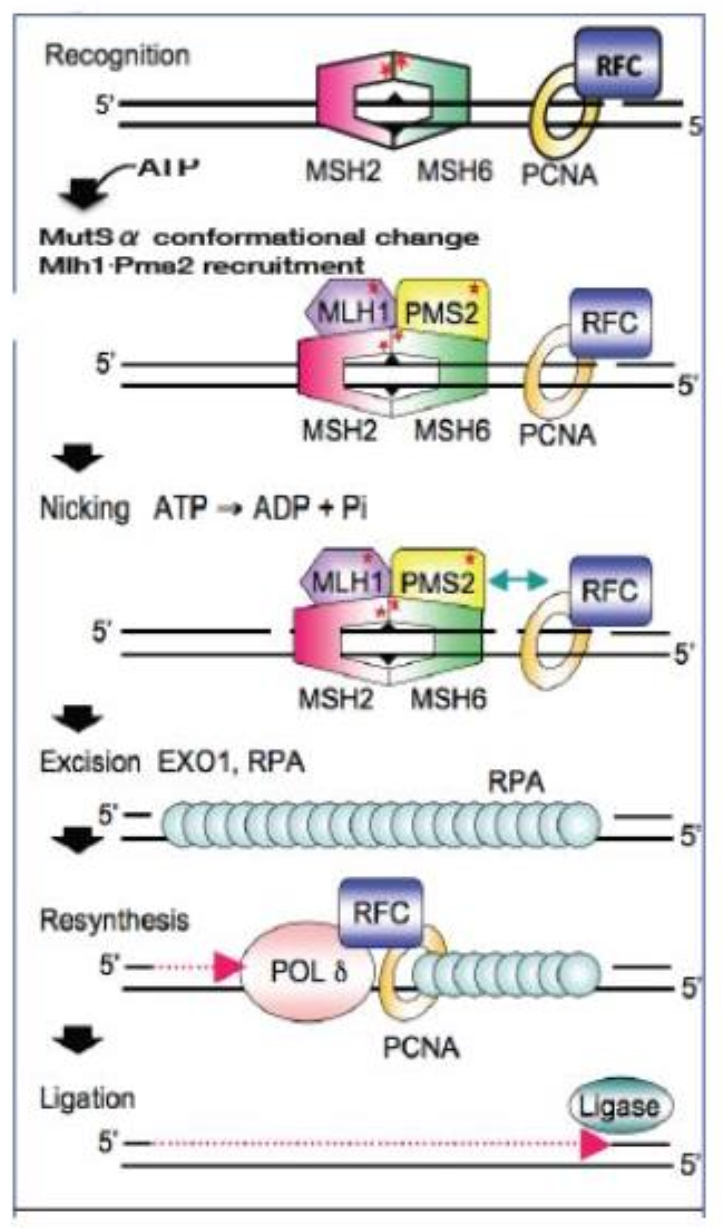
Tell me about proof reading and error fixing in DNA transcription
Polymerase proofreading:
Pol δ & Pol ε have 3’→5’ exonuclease
domains
Mismatch Repair (MMR):
MutSα (MSH2/MSH6) detects mismatches and INDELs
MutLα (MLH1/PMS2) directs excision and resynthesis
Error rates:
Pol error: ~10⁻⁵ / bp
With proofreading: ~10⁻⁷ / bp
With MMR: ~10⁻¹⁰ / bp
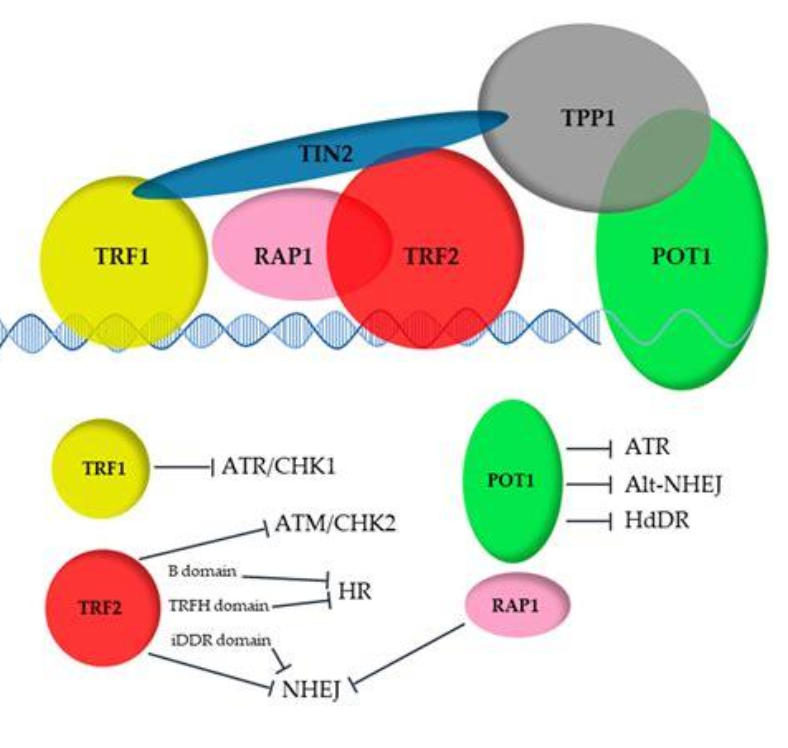
Replicating telomeres
Lagging strand cannot fully replicate ends of DNA so Telomerase (TERT + TERC RNA template) extends 3’ overhang
Shelterin complex (TRF1, TRF2, POT1, TPP1, TIN2,
RAP1):
Protects ends from DNA damage response
Prevents inappropriate repair/recombination
Telomere shortening results in cellular senescence, aging
Telomerase reactivation in observed cancer immortality
Alternative Lengthening of Telomere (ALT) pathway in telomerase deficiency
True or false? Transcription & RNA polymerases require primer molecules to begin polymerisation?
FALSE! They do not require primers
In which direction do RNA polymerases synthesise transcripts?
Synthesised in a 5’ to 3’ direction, therefore creating an RNA equivalent of the gene DNA sequence. Transcript sequences are always written in a 5’ to 3’ direction, from left to right.

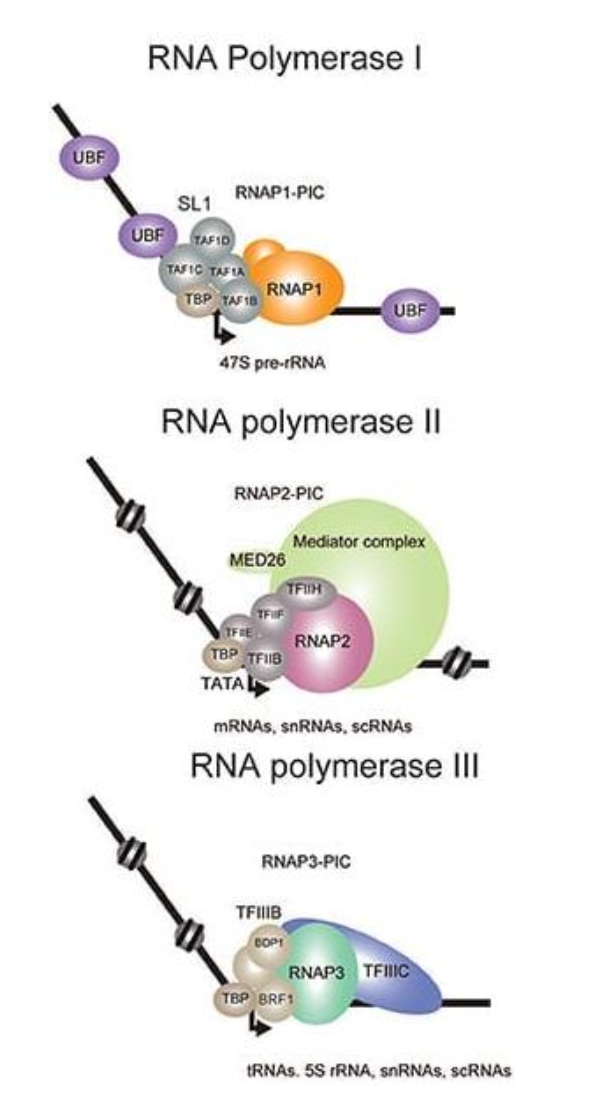
What are the key molecules in transcription?
RNA Pol I: rRNA
RNA Pol II: mRNA, lncRNA, snRNA
RNA Pol III: tRNA, rRNA and small RNAs
RNA Pol 2
Catalyses the formation of phosphodiester bonds in the sugar phosphate backbone of RNA
DNA-directed synthesis of mRNA during transcription
550kDA complex of 12 subunits
RNA polymerase cannot enter any part of transcription alone
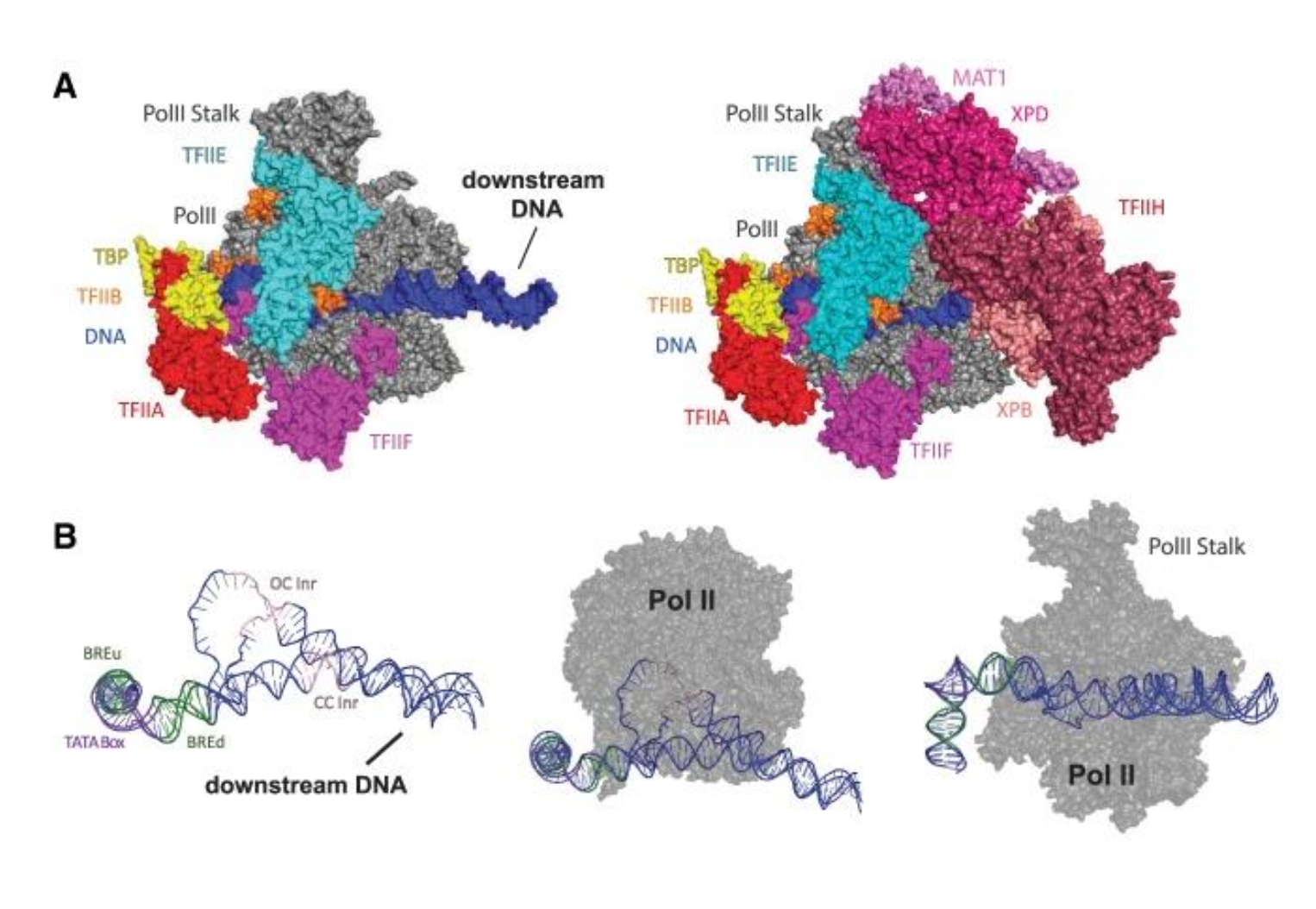
What is the difference between gene and gene specific transcription factors?
General transcription factors (GTFs)
Usually specific for each class of RNA polymerase
May directly or indirectly interact with the RNA polymerase, at a promoter, to initiate formation of a Pre-Initiation Complex (PIC) and facilitate the transcription process
Gene-specific transcription factors Respond to cell signals and determine which genes are transcribed. Can act to activate or repress transcription
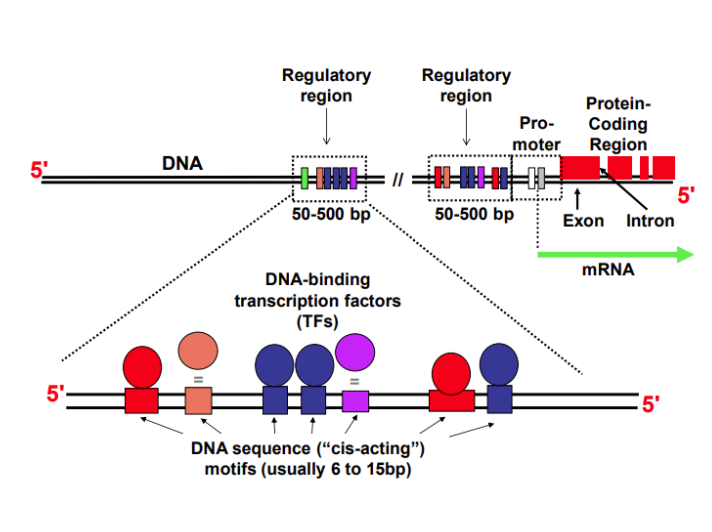
What are promotor elements and give examples?
Sequence of DNA to which proteins bind to initiate transcription downstream
TATA box (-25 to -30) - TATA(A/T)A(A/T)
Binds TATA Binding Protein
Positions RNA Pol II at transcription start site
Initiation focused at a single TSS
Initiator - PyPyAN(T/A)PyPy (A = TSS)
Overlaps with Transcription Start Site
Recognised by TAF/TAF2
Supports multiple dispersed start sites
Downstream Promotor Elements (+28 - +32)
Functions in TATA-less promotors
Recognised by TAF6/TAF9
What are the two routes for nucleoside generation in humans?
De novo synthesis pathway
Nucleoside salvage pathway
what is the de novo pathway for nucleoside synthesis?
Generation of new dNTPs from a multistep series of enzymatic reactions. The final key enzyme in this process is HRR.
What is the nucleoside salvage pathway?
Nucleoside bases which have already been used are covered back to their cognate nucleoside monophosphate (NMP).
Important as:
More energy efficient - ATP saving
Some organs and tissues have no capacity to undergo de novo synthesis
DNA replication occurs in vivo and in vitro, true or false?
True
What does in vivo replication look like?
Replication of genomes
Occurs during S phase
Doubling DNA in nucleus so that when it splits in two, each daughter cell has the same amount of DNA
Faithful replication of chromosomal DNA
Tightly controlled repair and maintenance of genomic DNA
What does in vitro replication look like?
Important for research and medical applications
PCR
Can look deeper into DNA/RNA sequencing, cloning and genomics
What are the 3 key principles of DNA replication?
semi-conservative
bi-directional
semi-discontinuous
successful replication is…?
and what does failure of these lead to?
Accurate
Complete
Regulated
Failure leads to:
Most likely cell cycle will pause and cell will undergo apoptosis - mediated cell death. But if it does not die then this can lead to:
Mutagenesis
Cancer/disease
Genome instability