BIO 120.11 | Module 1: Cell Locomotion
1/90
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
91 Terms
Most common type of pilus
Conjugation pilus
Type of pili for twitching motility in Bacteria & Archaea
Type IV pilus
Protein structure shorter but higher in number than pili
Surface attachment
Fimbriae
Resembles tiny grappling hook
found in SM1 group (unculturable Archaea)
Attachment (networked biofilm)
Hamus / hami
Thin (2-10 nm), long protein filamentous structure
All Gram-negative
Many Gram-positive and Archaea
Pilus / pili
Functions of pili
cantd
Conjugation (genetic exchange)
Adhesion
Nanowires (Conducting electrons for metabolism)
Twitching motility (Type IV) - Bacteria & Archaea
DNA-binding (Type IV)
Type of pili for DNA-binding
Type IV
T/F: Pili is shorter but higher in number than fimbriae
FALSE
Fimbriae is shorter but higher in number than pili
Function of fimbriae
Surface attachment
Function of hamus
Attachment (networked biofilm)
Basis of packaging prokaryotic chromosome
Supercoiling
5 internal prokaryotic cell structures
ccrip
Cytoplasm
Chromosome
Ribosome
Inclusion bodies
Plasmid
Aqueous mixture of
Macromolecules (lipids, nucleic acids)
Small organic molecules (macromolecule precursors)
Various inorganic ions
Ribosome
Cytoplasm
Large complexes of protein and RNA
Site of protein synthesis
Quantity is growth-dependent
Thousands/cell depending on growth phase/rate
Ribosomes
Describe number of ribosomes per growth phase
Lag phase = relatively low
Log phase = thousands
Stationary = start to decline
Decline/death = decline (lowest)
Size of ribosome in bacteria
70S
50S
5S rRNA, 23S rRNA, 31 proteins
30S
16S rRNA, 21 proteins
Not additive bc it’s a sedimentation rate (Svedberg)
Main genetic element that aggregates in prokaryotic cytoplasm to form nucleoid
Chromosome
Typical configuration of prokaryotic chromosome
One copy
Haploid (1 set)
Closed circular dsDNA
Small & compact genome
500 - 10,000 genes
0.5 - 10M bp
Basis of packaging: supercoiling
Genes are sometimes clustered (operons)
T/F: Prokaryotic chromosomes are haploid
TRUE
Positively charged proteins that tightly pack DNA to form nucleosomes in Archaea
Histones
T/F: Plasmid replicates separately from chromosome
TRUE
T/F: Archaea have histones, bacteria have histone-like proteins
TRUE
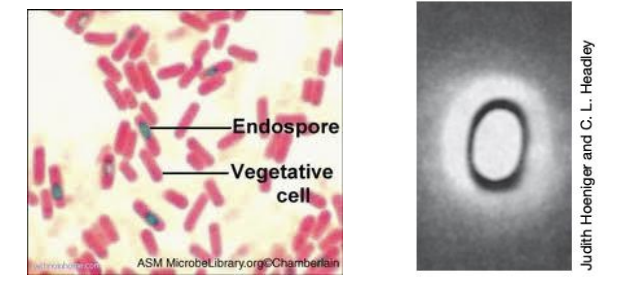
Highly differentiated, dormant, light refractive, non-reproductive survival structure
Endospore
One or more linear or circular dsDNA
Smaller than chromosome
Copies: 1, few, or >100
Replicates separately from chromosome
Plasmid
Endospore structure
Layer of spore-specific proteins
Spore coat
Describe genome of bacteria
500 - 10,000 genes
0.5 - 10 M base pairs
T/F: 70S ribosome and 16S rRNA are found in both Bacteria and Archaea and is the basis of tree of life.
TRUE
16S rRNA gene sequence of bacteria is the reason why it is considered a separate domain and no longer under Kingdom Monera
Example of special properties conferred by plasmid
Unique metabolism
Antibiotic resistance
Toxin resistance
Virulence factors
Bacteriocin
Conjugation
Endospore structure
Made of peptidoglycan
Cortex
Reason why Cyanobacteria was reclassified from being a blue-green algae to a bacteria
It has 16S rRNA sequence
Endospore structure
Develops from vegetative cell CM
Core wall
Where can u find sulfur granules
Periplasm, but it appears to reside in cytoplasm
Harsh conditions tolerated by endospores
drenc
Drying
Radiation
Extreme heat
Nutrient depletion
Chemicals
Typically contains genes that are not essential but often confer special properties on a cell
Plasmid
When does sulfur granules form
When elemental sulfur accumulates from sulfide oxidation (H2S → S0)
Endospore-forming genera
Gram (+) Bacillus (aerobic), Clostridium (anaerobic)
Describe endospore structure
Exosporium: outer proteinaceous layer
Spore coat: layer of spore-specific proteins
Core wall: derived from vegetative CM
Cortex: made of peptidoglycan
DNA: contained in the core
Endospore structure
Outer proteinaceous layer
Exosporium
What happens to sulfur granules when sulfide becomes limiting
Oxidized
Vegetative cells vs. Endospores
Vegetative cell | Endospore | |
Microscopic appearance | Nonrefractile | Refractile |
Calcium | Low | High |
Dipicolinic acid | Absent | Present |
Enzyme activity | High | Low |
Respiration rate | High | Low/absent |
Macromolecular synthesis | Present | Absent |
Heat resistance | Low | High |
Radiation resistance | Low | High |
Chemical resistance | Low | High |
Lysozyme | Sensitive | Resistant |
Water content | High (80-90%) | Low (10-25%) |
Small acid-soluble spore proteins | Absent | Present |
T/F: The number of plasmids a bacterial cell has would have implications on the amount of energy it spends, i.e., more plasmids = more energy exhausted
TRUE
Increased no. of plasmids = increased energy expenditure
In archaea, histones are positively charged proteins that tightly pack DNA to form _
nucleosomes
Forms from accumulation of elemental sulfur due to sulfide oxidation
Oxidized when sulfide becomes limiting
Periplasm (appears to reside in cytoplasm)
Sulfur granules
Forms when there’s excess of carbon
Polymer- and storage-form of glucose
Carbon and energy reservoir
Glycogen
4 functions of inclusion bodies
Highly specialized functions
Energy reserves
Reduction of osmotic stress
Space-saving storage units
Endospore structure
Contained in the core
DNA
Forms when there’s excess of lipids, carbon
Carbon / energy source
Poly-B-hydroxybutyric acid (PHB), Poly-B-hydroxyalkanoate (PHA)
3 types of endospores
Terminal: at the very end or pole of cell
Subterminal: near the end but not the end; in between the center and end of cell
Central: in center
Poly-B-hydroxybutyric acid (PHB), Poly-B-hydroxyalkanoate
Glycogen
Carbon storage polymers
Describe life cycle of endospore-forming bacteria
Vegetative cell
Harsh conditions (ercdn) can trigger cell to enter sporulation and become a
Sporulating cell with developing endospore inside,
such that eventually mature endospore will be released
Once conditions have become favorable enough for growth again,
Mature endospore can revert back to its vegetative cell state and enter germination
TLDR: Germination (vegetative), Sporulation (endospore)
Inclusion bodies for inorganic materials
Polyphosphate granules
Sulfur granules
Carbonate minerals
Enclosed in single layer membrane (instead of unit), composed of proteins
Inclusion bodies
Forms when phosphate is in excess
Phosphate source for nucleic acid, phospholipid, ATP synthesis
Polyphosphate granules
Confers buoyancy
Allows cells to position themselves in region of water column best suited to their metabolism
Conical-shaped, made of 2 diff proteins
Gas vesicle
e.g., Benstonite of Gloeomargarita
Contains barium, strontium, magnesium
Ballast
Way to sequester carbonate (source of CO2) to support autotrophic growth
Carbonate minerals inclusion bodies
4 types of inclusion bodies
Carbon storage polymers
Polyphosphate, sulfur, carbonate minerals
Gas vesicles
Magnetosomes
Biomineralized particles of magnetites, greigites
Allow cells to efficiently locate the microaerophilic zones they thrive in
Allow cells to orient themselves in magnetic field
Magnetosome
Conical-shaped structure composed of 2 different proteins
Gas vesicle
Contains barium, strontium, magnesium
Ballast
Way to sequester carbonate (CO2) source to support autotrophic growth
Benstonite of Gloeomargarita
Movement of cells by some kind of self-propulsion
Motility
Importance of motility
Allows bacteria to find/exploit new resources/habitat
Allows bacteria to escape unfavorable conditions (e.g., toxic chemicals, predators)
2 types of motility
Swimming
Surface motility
Individual movement in liquid powered by rotating flagella / archaella
Swimming
T/F: Flagella rotate at constant speed
FALSE
Flagella does not rotate at constant speed; depends on strength of PMF
Which is faster: bacteria or cheetah
Bacteria = 60 cell lengths/sec; 1000 rev/sec
Cheetah = 25 body lengths/sec
Discontinuous movement
CCW: runs, CW: tumbles, randomly reorients, CCW: runs again
name also species
Peritrichously flagellated cells, e.g., E. coli
Continuous movement
Faster than peritrichous
Reversible / unidirectional
Polarly flagellated cells
CCW: runs
CW: runs in reverse direction
name also species
Reversible polarly flagellated, e.g., Pseudomonas
CW: runs
Stops, randomly reorients
CW: runs again
name also species
Unidirectional, e.g., Rhodobacter
Typical colony morphology of gliding bacteria
Rod-shaped or filamentous
Involves crawling over surfaces
Requires attachment to surface
Results in distinctive colony morphology
Cells can move out and away from center of colony
Slower than swimming (<10mm/sec)
Surface motility
4 types of surface motility
Swarming
Twitching
Gliding
Sliding
Surface movement powered by extension and retraction of type IV pili
Found in Bacteria and Archaea
name also species
Twitching; Pseudomonas aeruginosa
Smooth continuous motion along long axis of cell
Without aid of external propulsive structures (e.g., pili, attachment organelles)
Not found in Archaea; mostly in rod-shaped or filamentous Bacteria
name also species
Gliding; Oscillatoria
Multicellular surface movement over semisolid medium
Mediated by flagella, archaella, type IV pili
name also species
Swarming; Proteus mirabilis, P. aeruginosa
T/F: Cells tumble frequently in the dark
TRUE (if they’re scotophobotactic)
Passive surface translocation powered by growth / cell division
Facilitated by surfactants (rhamnolipids) and other compounds (exopolysaccharides)
name also species
Bacillus subtilis
Ability to sense and respond to stimuli in their environment
Stimuli: physical or chemical
Response: move towards (attractant), move away (repellent)
Taxis
Ecological significance of taxis
Directed movement enhances cell’s access to resources
Allow cells to avoid harmful substances that could damage or kill them
7 types of taxis
pocsham
Phototaxis
Osmotaxis
Chemotaxis
Scotophobotaxis
Hydrotaxis
Aerotaxis
Magnetotaxis
Response to chemical stimuli
Sense environmental stimuli and transmit signals to flagella/archaella, causing it to alter its rotation
name also species
Chemotaxis; Gliding bacteria
Movement in response to gradient of light intensity
Allows phototrophic organisms to position themselves most efficiently to receive light for photosynthesis
name also species
Phototaxis; Filamentous Cyanobacteria
Identify movement
No attractant =
Attractant present =
No attractant = random movement
Attractant present = directed movement (to it)
Response to absence of light (fear of the dark)
Cells tumble frequently in the dark
Mechanism that prevents phototrophic organisms from swimming away from lighted zone into darkness
name also species
Scotophobotaxis; Chlorochromatium aggregatum
Directed movement with respect to gradients of O2
Aerotaxis
Directed movement with respect to gradients of available water
Hydrotaxis
Directed movement with respect to ionic strength gradient
Osmotaxis
Causes bacterial cells to point up or down so that they can swim either towards or away from O2 at surface
Allows bacteria to align themselves with earth’s magnetic field lines
Exhibits aerotaxis
Magnetotaxis
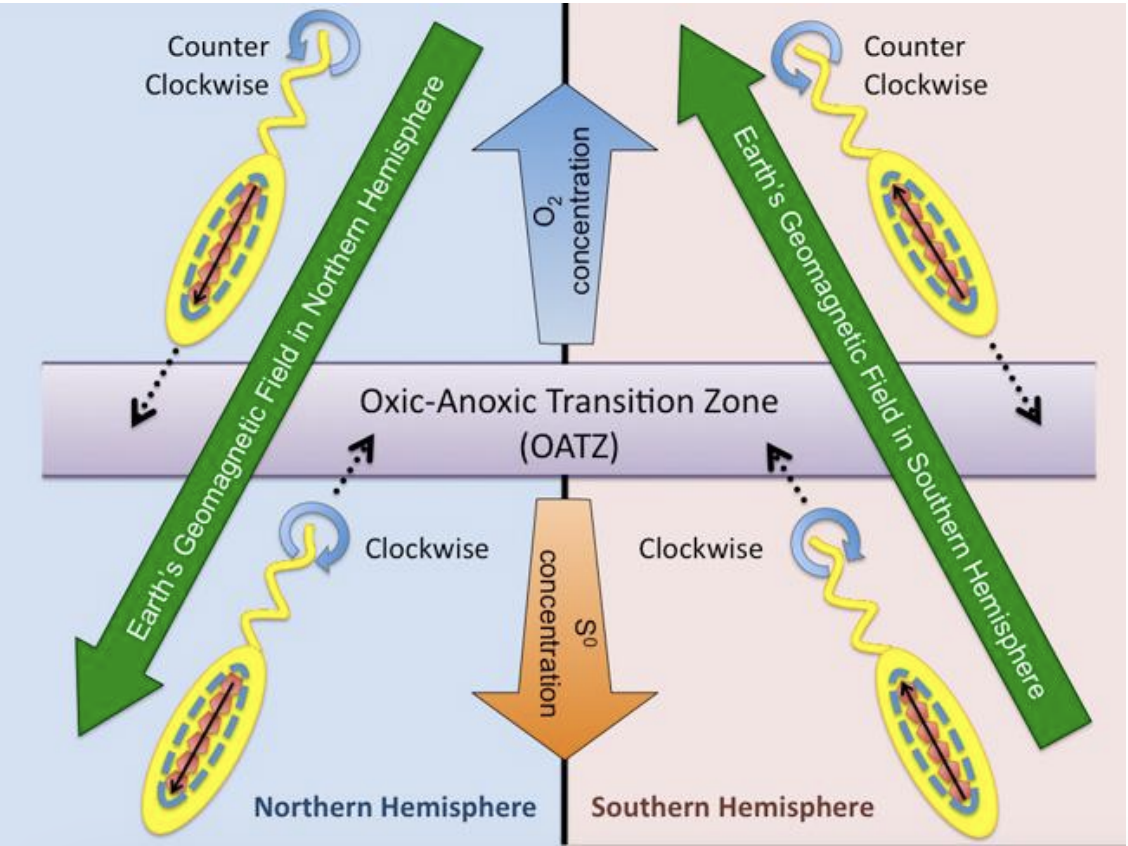
T/F: Magnetotactic bacteria do not actually exhibit directed motility towards magnetic fields but exhibit aerotaxis
TRUE
Towards oxic-anoxic transition zone (OATZ)
Movement towards attractant
Biased random walk (random walk = more tumbles)
2 types of bacteria coming together
Consortium