Chapter 13 - The Molecular Basis of Inheritance
1/28
Earn XP
Description and Tags
The Molecular Basis of Inheritance
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
29 Terms
what were chromosomes believed to be made up of up until the 1940s?
proteins
Transformation
alteration of genotype and phenotype due to the assimilation of external DNA
Bacteriophage
virus which infects bacteria
Virus
DNA (sometimes RNA) surrounded by a protein coat that is a parasite and needs a host in order to survive
what enters the cell when a bacteriophage injects into the cell
DNA (not proteins)
what did the data collected by Rosalind Franklin in Maurice Wilkins lab show?
that DNA was a helical molecule
DNA is composed of two strands
position of phosphate groups
width and spacing of the bases
what did James Watson and Francis Crick contribute to the findings of the structure of DNA
observed DNA’s basic components
determined that the two strands of DNA are antiparallel
determined base pairing and hydrogen bonding in base pairing
applied Chagraff’s rules
observed Franklin’s data on the width of the helix and spacing of bases
applied Franklin’s conclusions about the position of phosphate sugars
what accounts for the large amount of diversity in organisms
sequence of bases in a DNA molecules
DNA packaging
plays an important role in both organization and regulation of expression of genetic material
what is chromatin made up of
made up of DNA and proteins
heterochromatin
highly condensed and mostly inaccessible to transcription and translation proteins
euchromatin
more dispersed and contains accessible regions which are actively transcribed
semi-conservative model of replication
each new molecule of DNA contains 1 parental srand and 1 newly formed strand
3 theories of replication and explain each
conservative mode - the two parental strands reassociae after acting as templates for new strands which restores the parental double helix
semiconservative model - the two strands of the parental molecule separate and each functions as a template for synthesis of a new, complementary strand
dispersive model - each strand of both daughter molecules contains a mixture of old and newly synthesized DNA
what would determine if conservative or dispersive replication was correct
if conservative replication was correct, DNA of two densities would have ben found after the first replication
if dispersive replication was correct, DNA of one density would be found after two replications
origins of replication
sites containing specific sequences where DNA replication begins
specific proteins bind these sequences and open a replication bubble
origin of replication in prokaryotes vs eukaryotes
prokaryotes - 1 origin of replication
eukaryotes - 0 up to 1000 origins of replication
more linear so it has multiple origins of replication
replication fork
at each end of the replication bubble where DNA unwinds
DNA Helicase
enzyme that unwinds DNA strands at the replication fork so that the parent strands can act as replication templates
single-strand binding proteins
help to keep the single strands of DNA from rebinding to one another
topoisomerase
enzyme that helps relieve strain ahead of the replication fork by repeatedly binding, swiveling, breaking, and then rebinding
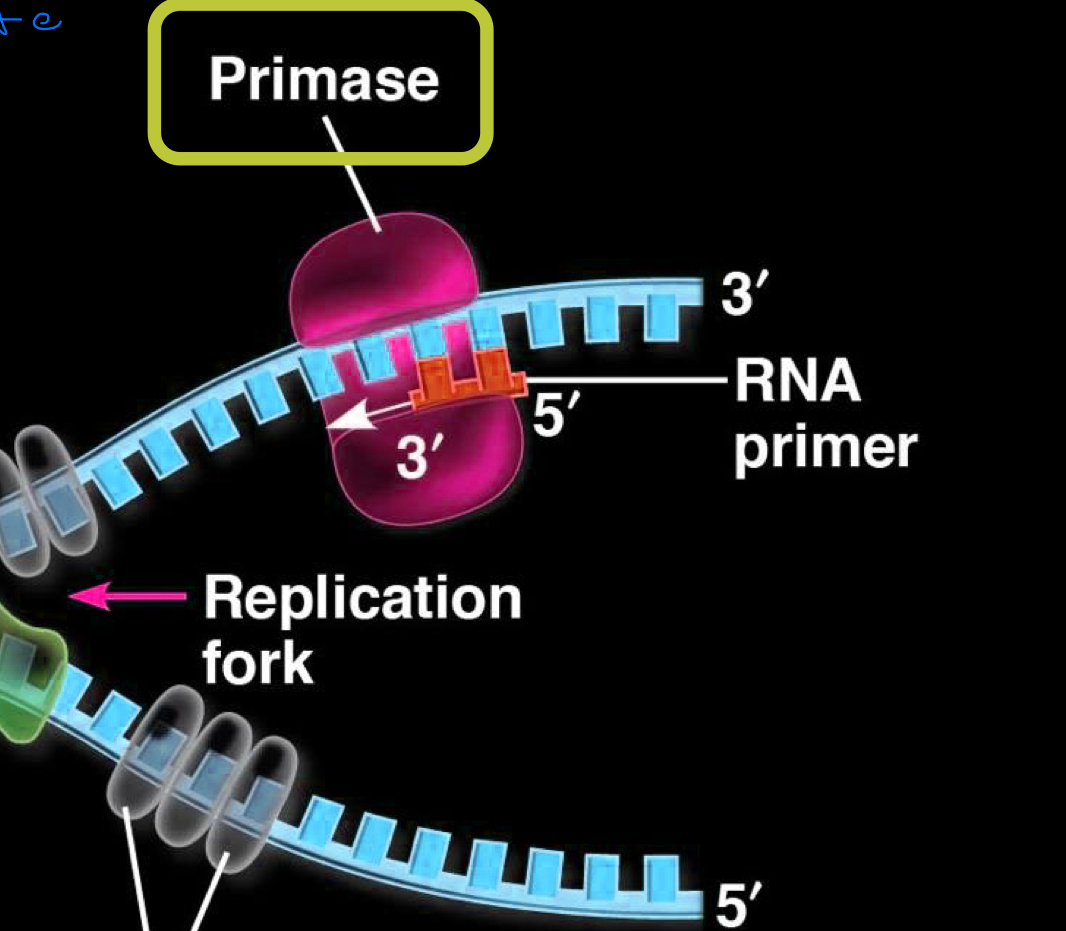
primase
enzyme which creates a short RNA primer (5-10 nucleotides) which complements the template strand of DNA
new DNA strand is then synthesized by DNA polymerase starting from the end of the RNA primer
DNA polymerase
synthesizes new strands of DNA
builds from 5’ to 3’ direction starting from 3’ end towards 5’ end
several different DNA polymerases exist
eukaryotes have 11+ different types of DNA polymerases
dNTP
added by DNA polymerase onto DNA nucleotides to mark that its a deoxyribose and not a ribose
adding dNTP releases a pyrophosphate which is hydrolyzed to 2 molecules of inorganic phosphate
antiparallel
DNA strands run antiparallel to one another (like a two way street)
leading strand
is synthesized continuously towards the replication fork
is built as a sliding clamp protein and a single DNA polymerases exist eukaryotes move towards the replication fork
lagging strand
cannot be synthesized continously since nucleotides are added away from the replication fork
produces short fragments called Okazaki fragments
100-200 dNTP (DNA nucleotides) long in eukaryotes
okazaki fragments
short fragments synthesized in the lagging strand of DNA replication
100-200 dNTP (DNA nucleotides) long in eukaryotes
lagging strand synthesis steps
Primase binds and a short RNA primer is synthesized to create a useable 5’ end
a DNA polymerase adds dNTP to the 5’ primer end to create the first Okazaki fragment
DNA polymerase 3 and the sliding clamp protein disassociate when they reach the next RNA primer
Okazaki fragment 2 is synthesized closer to the replication fork
DNA polymerase 1 removes the RNA primer and replaces it with DNA nucleotides for fragment 1
DNA ligase creates a bond between the 5’ and 3’ end of neighbouring fragments which joins them together