Enzymes as Drug Targets
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
34 Terms
The deficiency or excess of a metabolite can be controlled by inhibiting an enzyme in the pathway. What kind of enzyme would you block in metabolic disorders?
A → B → C
(Enzyme I) (Enzyme II)
The enzyme of the slowest or the rate-limiting step.
True or False: It is possible to inhibit 100% of enzyme synthesis.
False!
Which of the following is not an advantage of using enzymes as drug targets?
a. may resemble substrates, which is helpful as a start for drug design
b. need multiple enzymes to lead the design of drugs for other similar substrates
c. more likely to have 3D structure determined than the receptor (sometimes receptor is not determined until CTs)
d. more easily purified in proteins since they are soluble in cytosol
b. need multiple enzymes to lead the design of drugs for other similar substrates
only need 1 enzyme can lead to other drug designs
Match the Type of Enzyme Inhibitor to its definition:
A covalent bond forms between enzyme and substrate (large binding energy) that inhibits enzyme for longer than the substrate’s lifetime.
a. Reversible
b. Covalent Irreversible
c. Covalent Reversible
b. Covalent Irreversible
Match the Type of Enzyme Inhibitor to its definition:
A non-covalent bond forms between enzyme and substrate (small binding energy) that temporarily inhibits enzyme.
a. Reversible
b. Covalent Irreversible
c. Covalent Reversible
c. Covalent Reversible
Match the Type of Enzyme Inhibitor to its definition:
A typically non-covalent bond, inhibition of enzyme activity that is reversible.
a. Reversible
b. Covalent Irreversible
c. Covalent Reversible
a. Reversible
What type of enzyme inhibitor is Enzyme Inhibitor (slows down or blocks enzyme catalysis) associated with?
a. Reversible
b. Covalent Irreversible
c. Covalent Reversible
a. Reversible
specifically in the alteration of enzyme catalysis
Example: the blocking of the active site by salicylic acid residue of aspirin in COX-2
What type of enzyme inhibitor is Enzyme inactivator (irreversible inhibitor) associated with?
a. Reversible
b. Covalent Irreversible
c. Covalent Reversible
b. Covalent Irreversible
irreversible modification of enzyme active site
Example: the acetylation of SEr530 of COX-2 by aspirin
In what cases do you inhibit an enzyme? (Select all that apply)
a. uncontrolled cell growth
b. when you want to change the enzyme in the pathway
c. infestation by a foreign organism
d. to decrease the amount of enzymes in a pathway
e. deficiency or excess of a metabolite
a. uncontrolled cell growth
c. infestation by a foreign organism
e. deficiency or excess of a metabolite
Which of the following Ideal Characteristics of Enzymes is FALSE? Explain why.
a. Enzymes can be used for many different diseases.
b. Is essential for growth in foreign organisms but is not present or essential in humans.
c. Similar enzymes that catalyze similar reactions but differ in substrate preference (i.e. a substrate of a virus vs in the human body) are suitable targets.
d. Enzymes that bind to 1 ligand only (really rare)
a. Enzymes can be used for many different diseases.
FALSE —> you want enzymes specific for 1 disease (drug specificity)
Catalytic Efficiency results from tight or low binding of enzyme and substrate during transition state. How can you determine the tightness of enzyme-substrate binding?
very tight binding
can measure using transition state energy
High Transition State Energy: ____ bound
Low Transition State Energy: ____ bound
loosely; tightly
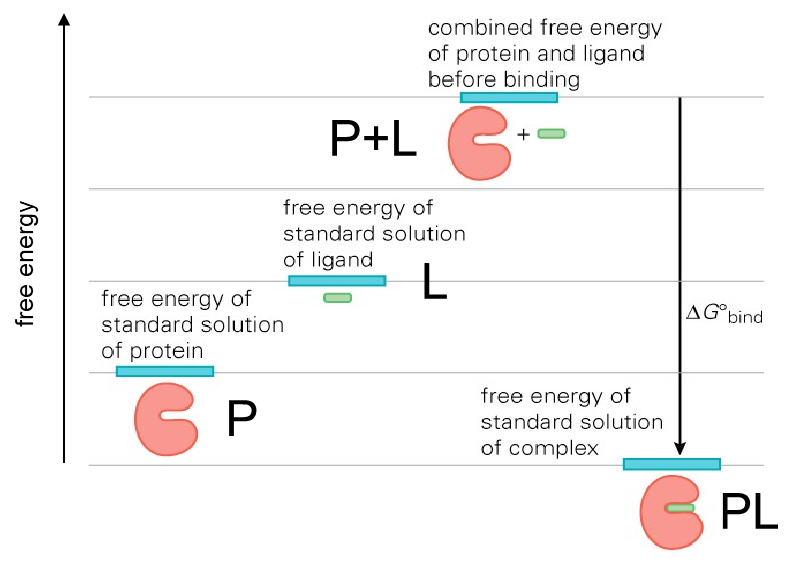
Why does the energy of the PL (protein ligand) complex need to be low (lower than the sum of each component P + L)?
Low complex energy —> tight binding
Needs to be low enough to provide high enzyme occupancy
Why have enzymes not evolved to bind substrates very tightly?
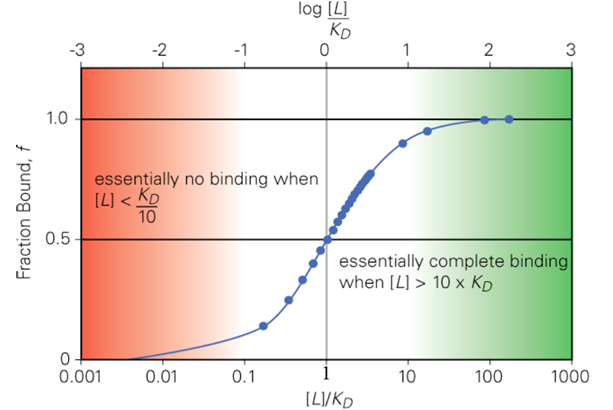
When substrates have an affinity that results in > 90% enzyme occupancy, it does not increase the efficacy of the substrate turnover.
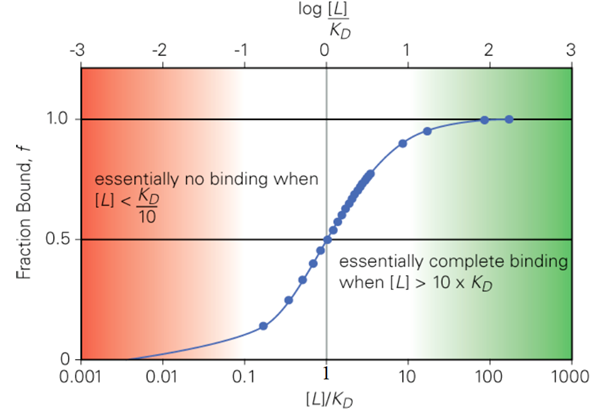
When [L] = KD, what is f as the value of [L]/KD?
f = 0.50
[L]/KD = 1
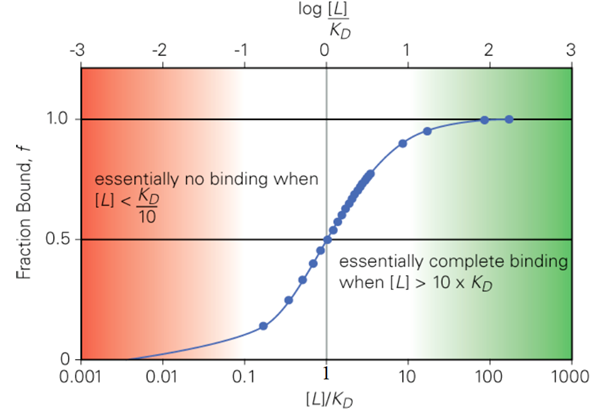
When L = KD, how much of the protein is bound?
a. 25%
b. 50%
c. 75%
d. 100%
b. 50%

When L = 10 KD, how much of the protein is saturated?
a. 9%
b. 99%
c. 91%
d. 19%
c. 91%
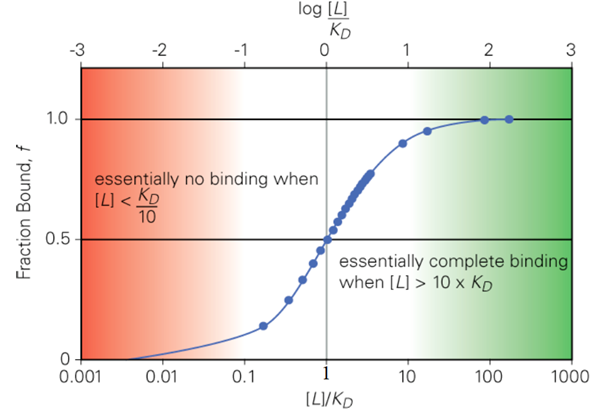
When L = 0.1 KD, how much of the protein is bound?
a. 9%
b. 99%
c. 91%
d. 19%
a. 9%
How do you calculate the degree of inhibition of an enzyme?
f = [L]/[L]+KD
f = fraction bound
[L] = ligand concentration
KD = dissociation constant
True or False: Substrates are NOT a good starting point for inhibitor design, but it’s the fastest way to obtain a competitive inhibitor molecule.
True! Substrates are weak ligands —> weak binding of E-S complex
energy that is needed to effect enzyme & substrate activation is subtracted from the total energy of interactions btwn substrate and enzymes
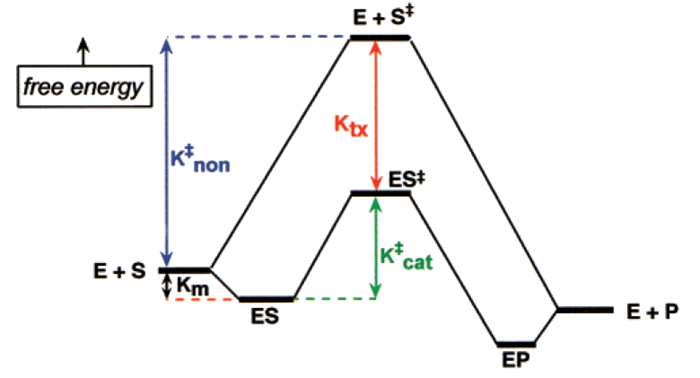
The energy with which enzyme binds TS and the resulting equilibrium constant is estimated as:
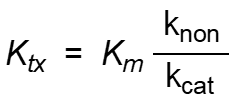
Define all the factors in the equation.
Ktx = dissociation constant of enzyme - TX complex
Km = dissociation constant of enzyme-substrate complex
knon = rate constant of non-catalyzed reaction
kcat = rate constant of catalyzed reaction
Which of the following is not able to be mimicked from the synthesized (mimic) transition states?
a. Conformations
b. Geometry of the reaction site —> sp2, sp3
c. Electronic state (± charge from bond breaking or making)
d. Energy of the transition state
e. Longer separation between interacting parts in bond-cleavage reactions
d. Energy of the transition state
Match the definition to its term:
An interaction between a protein and substrate/drug in which multiple substrates can be bonded to the active site of the protein/enzyme. These are catalytic interactions.
a. Allosteric Interactions
b. Orthosteric Interactions
b. Orthosteric Interactions
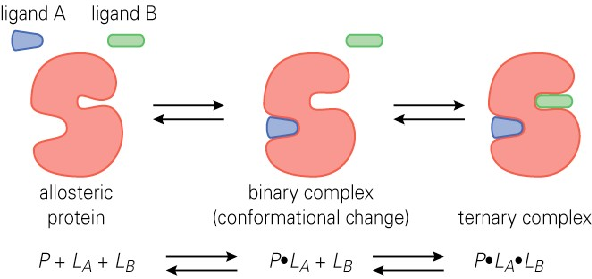
Match the definition to its term:
An interaction that involves the enzymes/proteins having multiple binding sites. The binding modulates the structure of the catalytic site, and binding of substrate at the catalytic site is dependent on the occupancy of the allosteric site.
a. Allosteric Interactions
b. Orthosteric Interactions
a. Allosteric Interactions