BME 429 - Receptor and Ligand Interactions
1/47
Earn XP
Description and Tags
North Carolina State University - BME 429 (Cellular Engineering) - Dr. Ashley Brown
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
48 Terms
Cell
The basic biological unit of living organisms. Tissue and development changes occur at the cellular level
What do membrane-bound organelles do for animal cells?
Enhance compartmentalization and regulation of cell functions by separating various biochemical processes.
What are some reasons why we might want to control cell signaling?
Stem cell differentiation
Immune engineering
Targeted cell therapies (ie, cancer)
Preventing/treating fibrosis
Cell manufacturing
Paracrine signaling
Cells communicate over relatively short distancesthrough the release of signaling molecules that affect nearby target cells.
Synaptic signaling
A type of paracrine signaling, where nerve cells transmit signals over a synapse.
Autocrine signaling
A cell signals to itself.
Endocrine Signaling
Long-distance cell signaling. Signals are produced by specialized cells and released into the bloodstream, which carries them to target cells in distant parts of the body.
Cell-Cell Contact
Cell signaling through gap junctions or cell membrane protein interactions.
Ligand
A molecule used for cell signalingthat binds to a receptor on a target cell, initiating a response.
Binds to just 1 or a few target receptors
Are often proteins, but can also be ions and phospholipids
Receptor
A receiving molecule/protein on the target cell that binds to a specific ligand, triggering a cellular response.
Recognizes just 1 or a few specific ligands
How does a ligand bind to receptor?
Binding of a ligand to a receptor changes its shape or activity, allowing it to transmit a signal / directly produce a change inside the cell
Doesn’t use covalent bonds nor chemical reactions
Upstream
Used to describe molecules and events that come earlier in the relay chain
Downstream
Used to describe molecules that come later relative to a particular molecule of interest in the relay chain
Phosphorylation
Addition of a phosphate group to one or more sites on the protein
Phosphate groups are typically linked to one of the 3 AAs with hydroxyl (-OH) groups in side chains (tyrosine, threonine, serine)
Not permanent
Kinase
An enzyme that catalyzes the transfer of the phosphate group; usually represented by a “K”
Phosphatase
Enzymes that remove a phosphate group from their targets
Intracellular receptors
Receptors found inside the cell, such as in the cytoplasm or nucleus
Most of their ligands are small and hydrophobic (must be able to cross plasma membrane to reach these receptors)
Are unique because they can cause changes in transcription of genes very directly by binding to DNA and altering transcription themselves
Cell surface receptors
Membrane-anchored receptors found inside the plasma membrane
Many different kinds of molecules can act as their ligands, since the ligand doesn’t need to pass through plasma membrane
3 domains/protein regions of a cell-surface receptor
Extracellular ligand-binding domain
Hydrophobic domain extending through membrane
Intracellular domain, which often transmits a signal
3 common types of cell-surface receptors
Ligand-gated ion channels
G protein coupled receptors
Enzyme-linked receptors
Receptor tyrosine kinases (subclass)
Ligand-gated ion channels
Ion channels that can open in response to the binding of a ligand
Contains a membrane-spanning region with a hydrophilic channel through middle
Allows ions to cross membrane without having to touch hydrophobic core of phospholipid bilayer
Binding of ligand causes protein structure changes to allow for ion passage or channel closure
Common in neurons (neurotransmitters as ligands)
G protein-coupled receptors (GPCRs)
Large family of cell surface receptors that share a common structure and method of signaling
7 transmembrane domains
Signals through G protein
All types of G proteins bind the nucleotide guanosine triphosphate (GTP), which they can break down (hydrolyze) to form GDP
G protein active when attached to GTP
G protein inactive when attached to GDP
Enzyme-linked receptors
Cell surface receptors with intracellular domains that are associated with an enzyme
In some cases, the intracellular domain of receptor is an enzyme that can catalyze a reaction
Other enzyme-linked receptors have an intracellular domain that interacts with an enzyme
Receptor tyrosine kinases (RTKs)
An important class of enzyme-linked receptors found in humans and many other species
Transfers phosphate groups specifically to AA tyrosine
In many cases, the phosphorylated receptors serve as docking platform for other proteins that contain special types of binding domains
Used in growth factor binding
Activity must be kept in balance: overactive RTKs are associated with some types of cancer
Intracellular ligands
Steroid hormones (estradiol [female sex hormone], testosterone [male sex hormone], vitamin D)
Nitric oxide
Extracellular ligands
Water-soluble: polar or charged
Mostly peptide (protein) ligands
Growth factors
Hormones (insulin)
Neurotransmitters
Agonist
A drug that causes the receptor to respond in the same way as the naturally occurring substance
Antagonist
A drug that binds to the receptor, but doesn’t produce a response and prevents the receptor binding to the ligand, having an inhibitory effect on the naturally occurring substance
R + L <=> C
Receptor + Ligand <=> Receptor/Ligand Complex
Molecular recognition
Molecules “recognize” each other when they come close enough to “feel” the presence of each other, either through physically bumping into one another or through interactions of the fields (electrostatic) between each other
Uses non-covalent interactions, so can be reversible
Specificity
A description of how favorable the binding of the ligand/receptor for the receptor/ligand is compared with its possible binding to other types of receptors/ligands that may also be present
Affinity
Refers to how strong the binding is (as judged by Kassociation or Kdisassociation or ∆G°).
Association or dissociation constants (KD) typically referred to as the “affinity” or “binding” constants
ka
Association rate constant for formation of RL or C, “on rate”, kforward
kd
Dissociation rate constant for complex RL or C, “off rate”, kreverse
Stoichiometry
Refers to how many molecules of ligand can bind to a single receptor
Cooperativity
AKA synergism, refers to situations where binding of 1 or more molecules to the receptor enhances/weakens the binding of additional molecules to the same receptor
Cooperative binding effects (both positive and negative) are also known as allosteric effects (eg, cofactors, calcium)
Reversible vs. Irreversible binding
All non-covalent binding processes are reversible, meaning ligand can both bind to and dissociate from the receptor
Equilibrium reached when the time following mixing is long compared to the t1/2 (half-life) binding and dissociation
However, sometimes non-covalent binding is so tight that the association is effectively irreversible and doesn’t reach equilibrium within the relevant time frame
Kinetics
Term used to describe both the rates at which processes occur and the field associated with the study of rates
Binding and dissociation processes will be characterized not only by the equilibrium constants but also by how fast association/dissociation occurs
1:1 Stoichiometry for Receptor/Ligand
The simplest case in which 1 ligand binds to 1 receptor
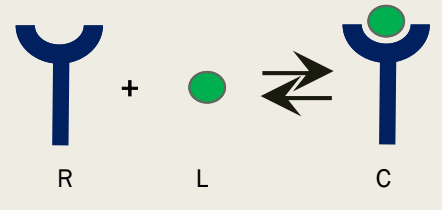
What are binding and other equilibrium constants related to?
Rates of interchange between states involved in the equilibrium process
Rate
A description of how frequently something happens
Zero order reaction rate (unimolecular rate)
Independent of concentration
Per time (e.g. sec-1 (Hz), min-1)
k = [conc/time] (M/s)
Eg, decomposition of ammonia (happens at high temperatures and is independent of concentration because it has to do more with adsorption to the surface area of platinum
First order reaction rate
Dependent upon the concentration of a single species
Δ[concentration]/time (e.g. mM product produced/min) or Δquantity/Δtime
(e.g. micromoles produce produces/sec)
k = time-1 (sec-1)
Ex:
Production of an enzyme product complex from an enzyme-substrate complex
Dissociation of a 1:1 receptor-ligand complex to form free ligand and free receptor (while 2 separate species are proceeded, the rate at which they are produced will be dependent upon a single concentration: that of the complex)
Second order reaction rate
Dependent upon the concentration of 2 species
Δ[concentration]/time for either products; rate constant depends on both
inputs
k = concentration-1 * time-1. For example (M-1*sec-1)
Ex: Binding of a ligand to a receptor to form a 1:1 ligand complex (the rate is dependent on both concentrations; L and R can associate only if they bump into each other, and the probability that they will is determined by their concentrations)
Ka
Equilibrium association rate = [RL] / [R][L] ← these are concentrations at equilibrium (product over reactant)
Ka = 1/Kd
Kd
Equilibrium dissociation rate = [R][L] / [RL] ← these are concentrations at equilibrium
(also ‘product over reactant’, but the ‘product’ this time is the unbound ligand/receptor)
Also = free ligand concentration where the total population of free and complexed receptors will be equal (half maximum binding)
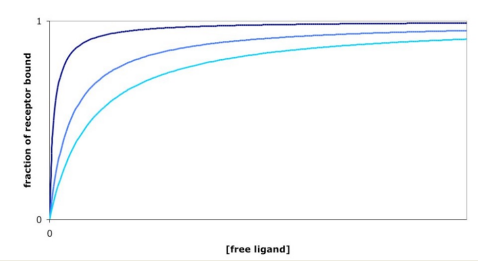
Which is the weakest binder?
Light blue
Formula for influence of temperature on equilibrium
ΔG° = RTln(Kd) = -RTln(Ka)