BIOL 3301 Chapter 12 Gene Transcription and RNA Modification
1/97
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
98 Terms
gene
at molecular level, a ? is a. segment of DNA used to make a functional product, like RNA or a polypeptide
transcription
first step in gene expression
note: not every gene in your genome is turned on/expressed in all cells all the time
your liver cells aren’t the same as kidney cells bc they have different proteins encoded in your genome.
transcription
act or process of making a copy
in genetics, copying of a DNA sequence into an RNA sequence
DNA is structure is NOT altered from this process
it can continue to store information
protein coding/structural genes
code the amino acid sequence of a polypeptide
transcription of protein coding gene produces mRNA
mRNA base sequence determines the amino acid sequence of a polypeptide during translation
1 or more polypeptides constitute a protein
synthesis of functional proteins determines an organism’s traits
mRNA base sequence
determines the amino acid sequence of a polypeptide during translation
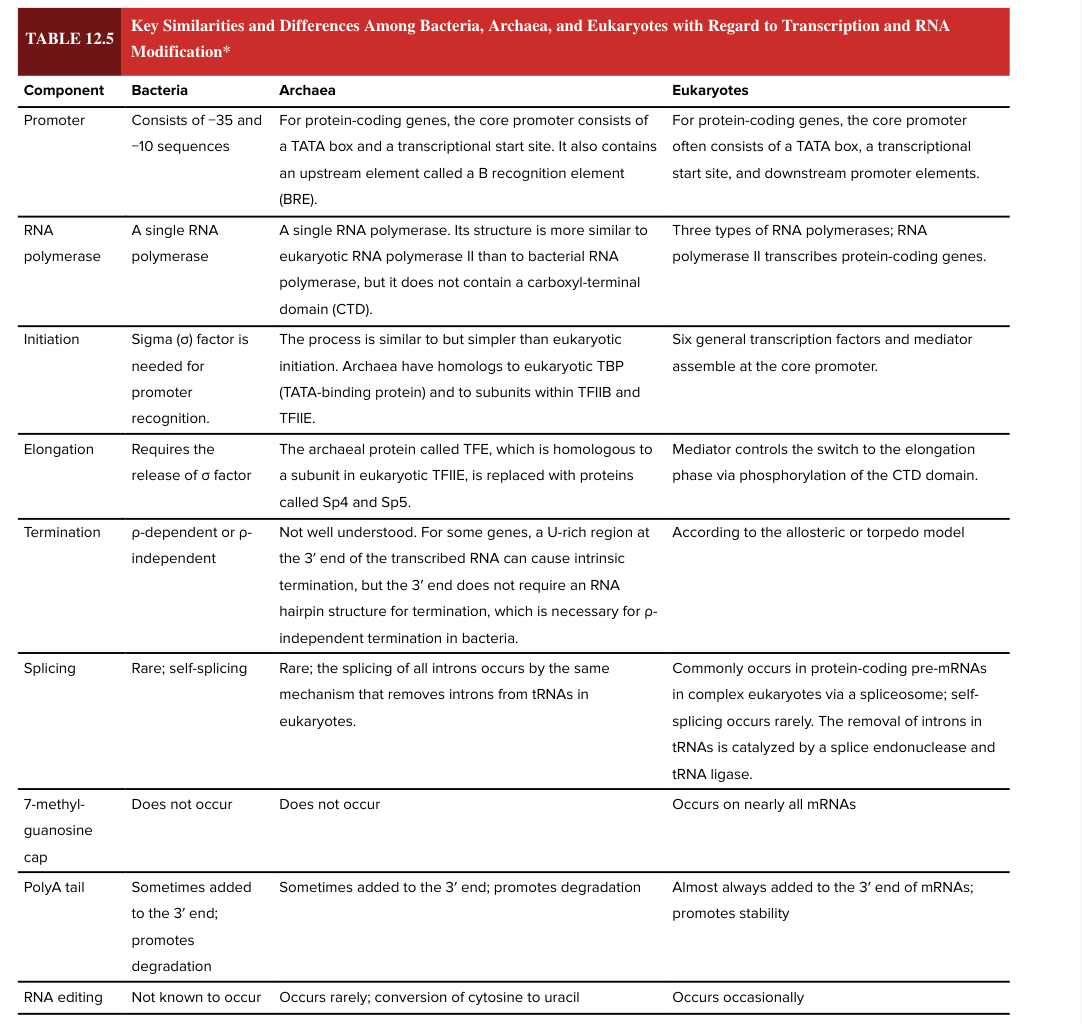
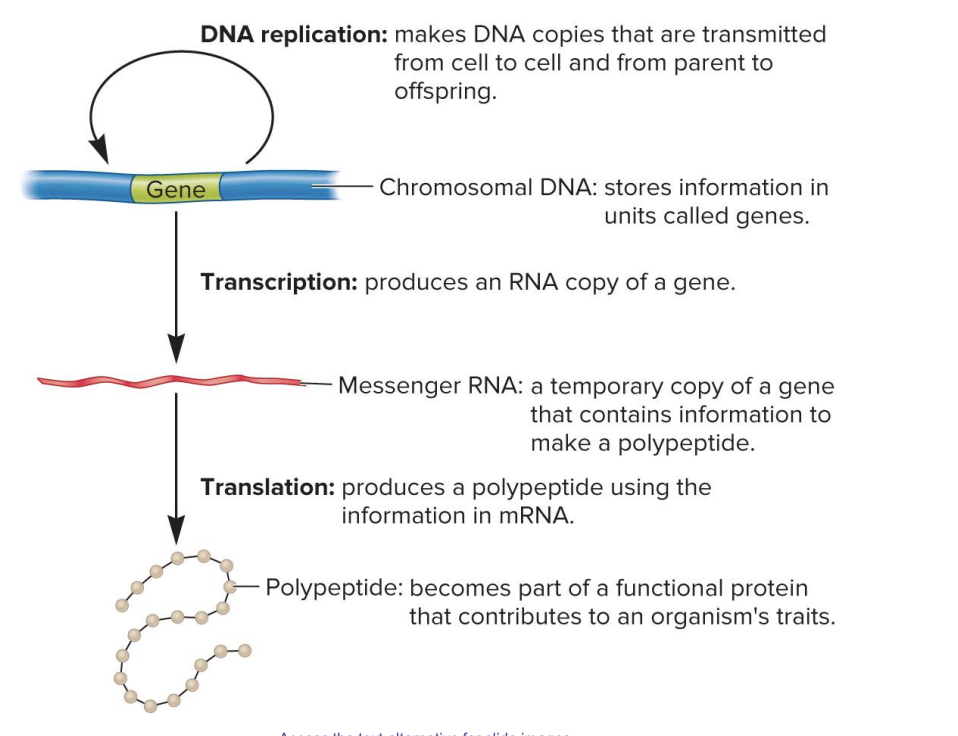
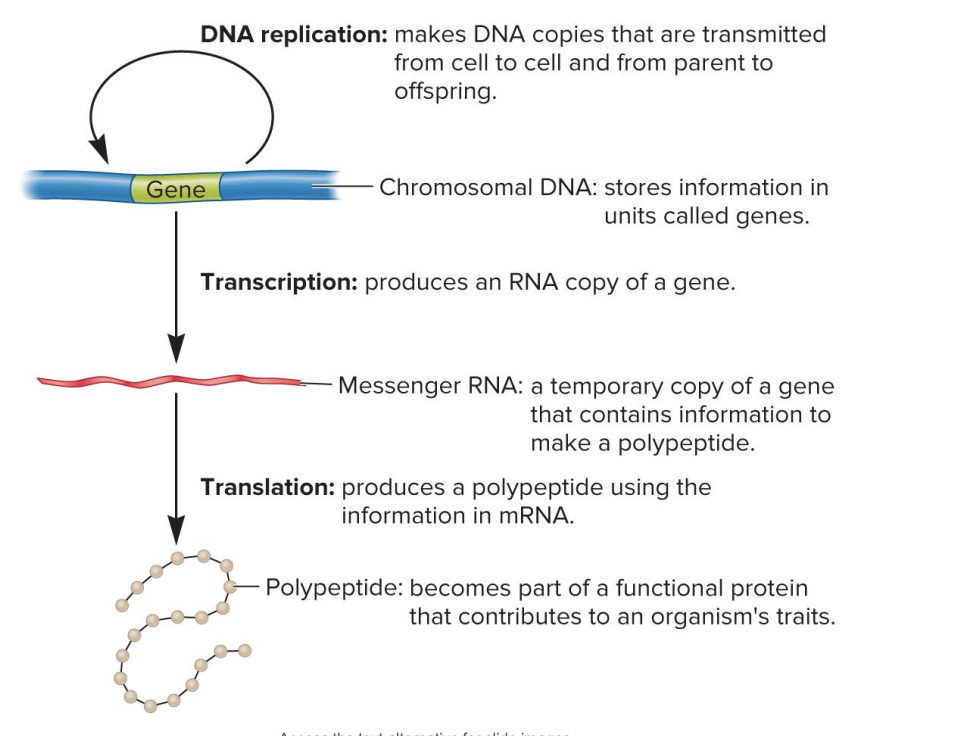
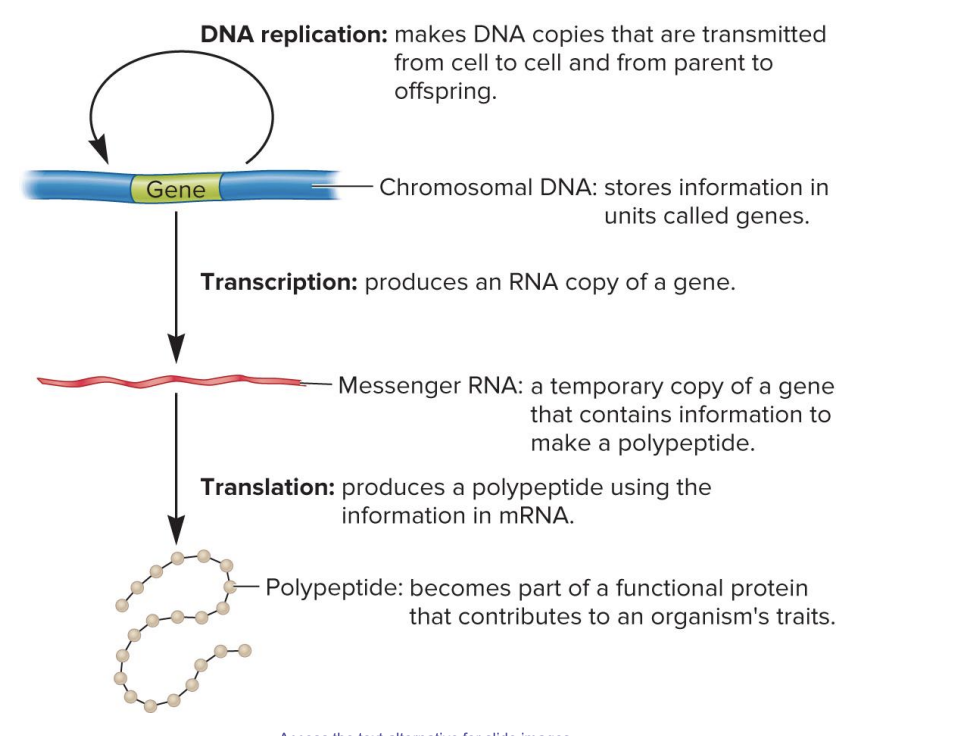
DNA replication
makes DNA copies that are transmitted from cell to cell and from parent to offspring
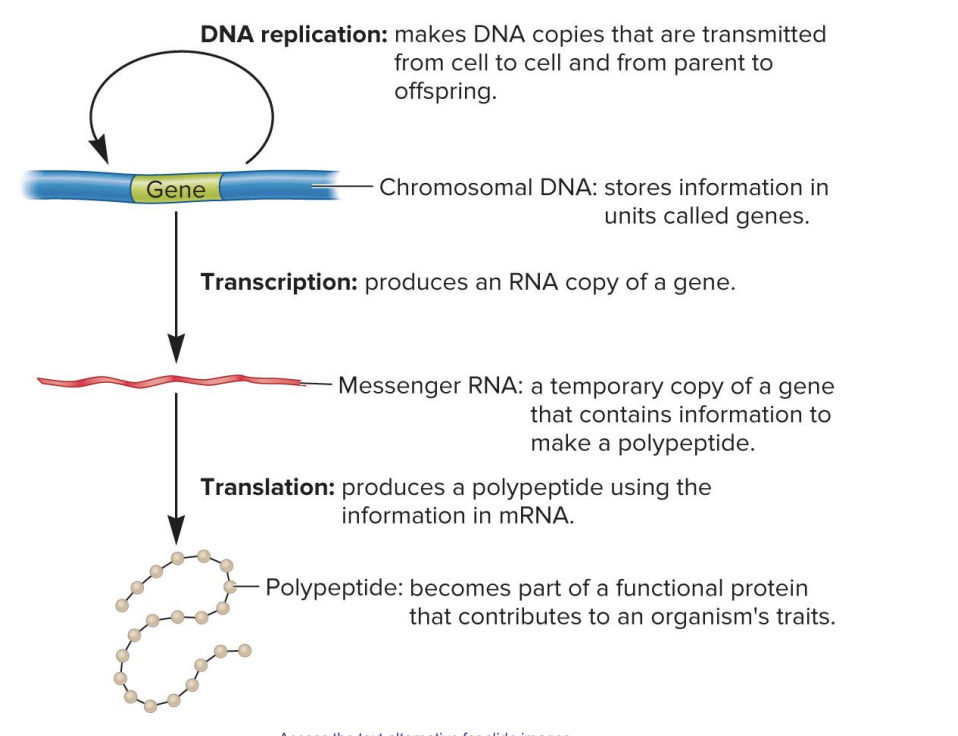
chromosomal DNA
stores information in units called genes
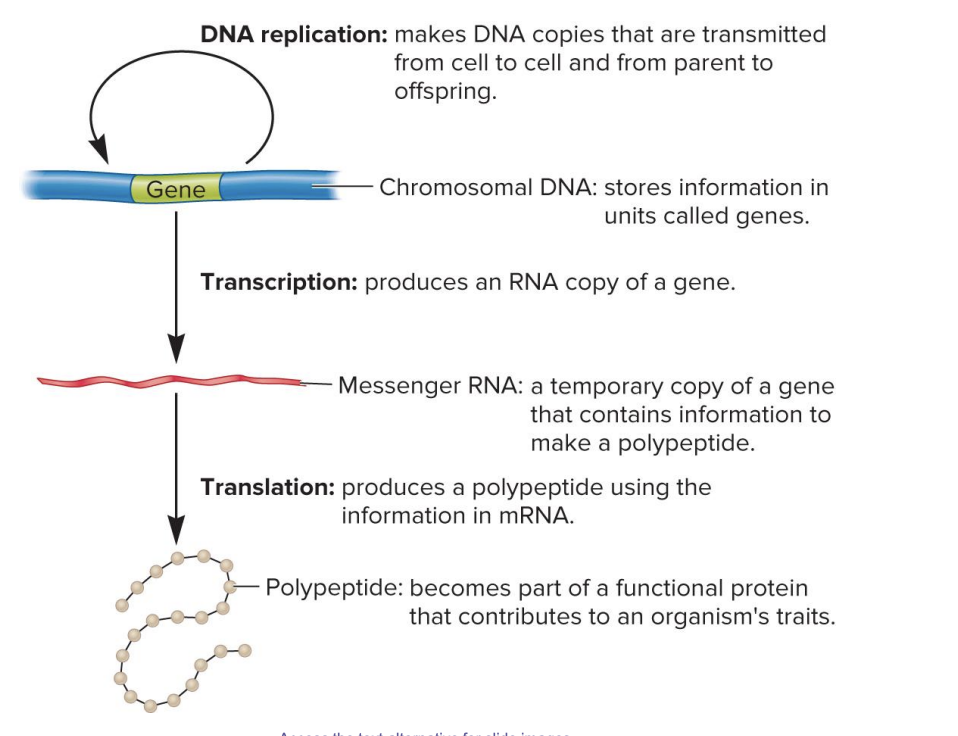
transcription
produces an RNA copy of gene
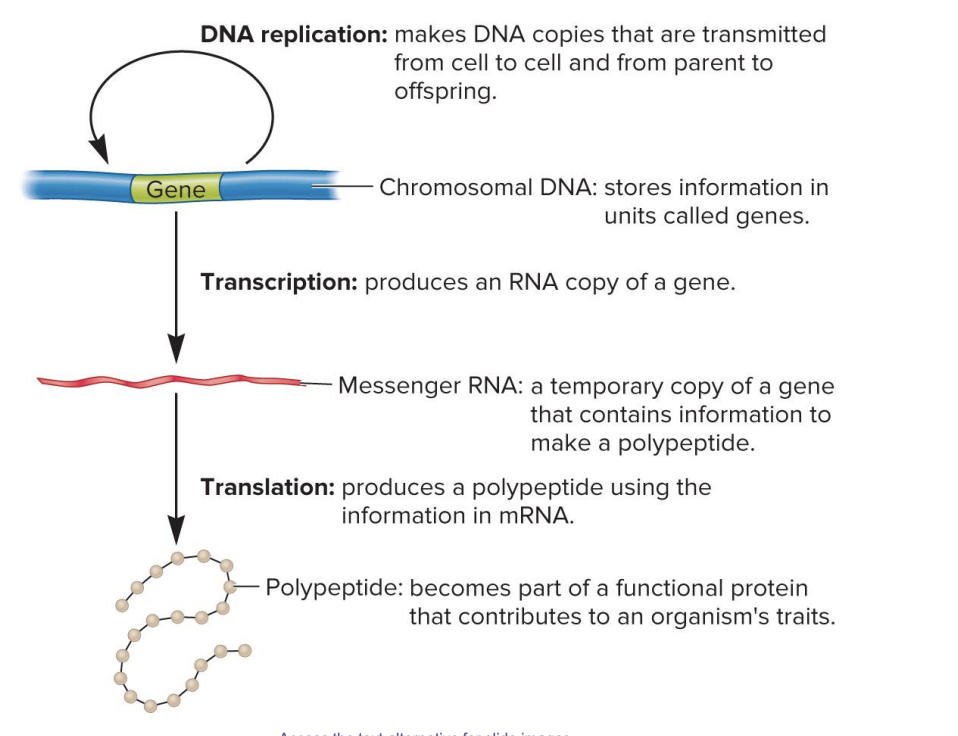
messenger RNA
a temporary copy of a gene that contains information to make a polypeptide
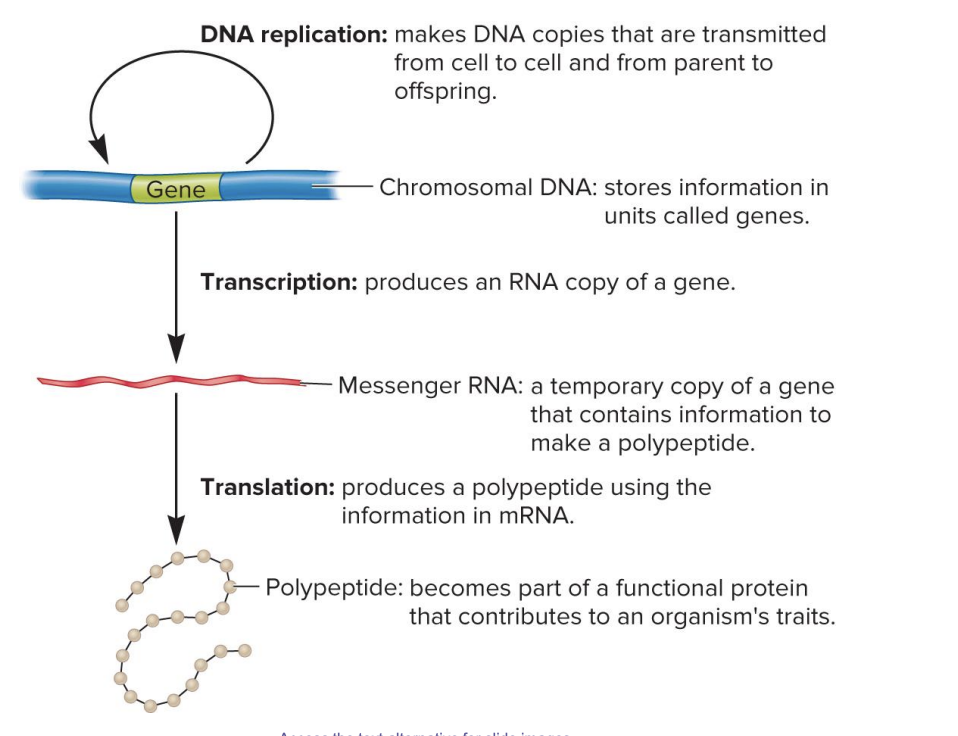
translation
produces a polypeptide using the information in mRNA
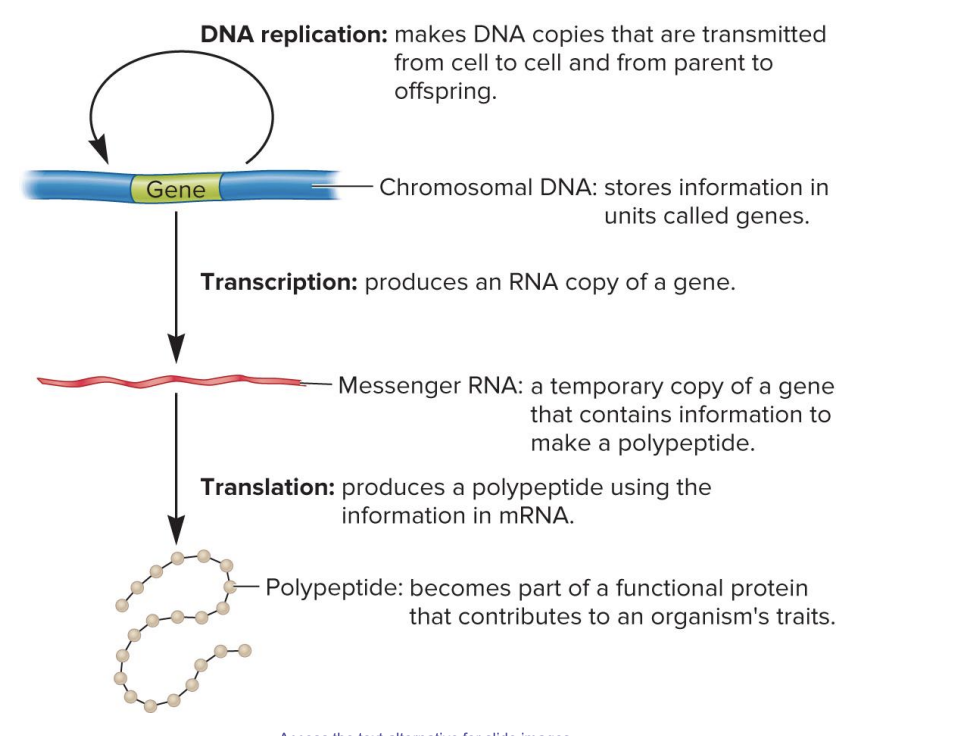
polypeptide
becomes part of a functional protein that contributes to an organism’s traits
central dogma of genetics
DNA replication
transmitted from cell to cell and from parent to offspring
chromosomal DNA
stores info in genes
transcription
produce RNA copy of gene
messenger RNA
temporary copy of gene w/ info to make polypeptide
translation
produce polypeptide using mRNA info
polypeptide
becomes part of functional protein that contributes to an organism’s traits
central dogma of genetics
DNA → RNA (transcription) → Polypeptide (translation) → Protein (functional role)
The DNA stores the information.
The information is copied into mRNA through transcription.
The mRNA is used to make a polypeptide through translation.
The polypeptide folds into a protein that performs a function, contributing to the organism's traits.
transcription
DNA base sequences define the beginning and end of a gene and regulate the level of RNA synthesis.
The DNA sequence:
Specifies where transcription begins (promoter).
Specifies where transcription ends (terminator).
Determines what should be made (e.g., mRNA, tRNA, etc.).
Regulates how much RNA is made
enhancers: increase RNA production
silencers: decrease RNA production
Sequences in DNA signal to proteins that:
Recognize and read the sequences.
Follow the instructions from the gene information to produce proteins or other functional molecules (like tRNA).
Proteins (RNA polymerase and transcription factors) must recognize and act on DNA for transcription to occur.
transcription factors: proteins that recognize specific DNA sequences and control transcription rate
gene expression
overall process by which the information within a gene is used to produce a functional product which can, in concert with environmental factors, determine a trait
(A gene's information is first transcribed into RNA, which is then translated into a protein. This protein, functioning within a specific environment, ultimately contributes to the expression of a particular trait)
transcription
in a DNA molecule, the DNA sequence itself says where the beginning and end are, how to start and stop, and what should be made from that sequence and also how much
those sequences are signals to the proteins that can “recognize” those sequences and actually do the work of “reading” the information in a gene and following the instructions in that information to make more proteins, or other functional molecules (like tRNAs)
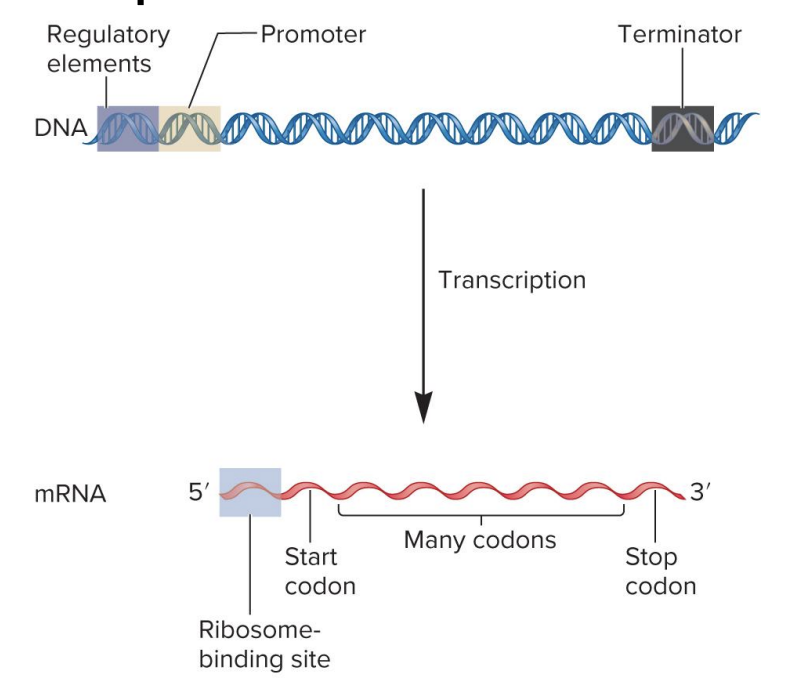
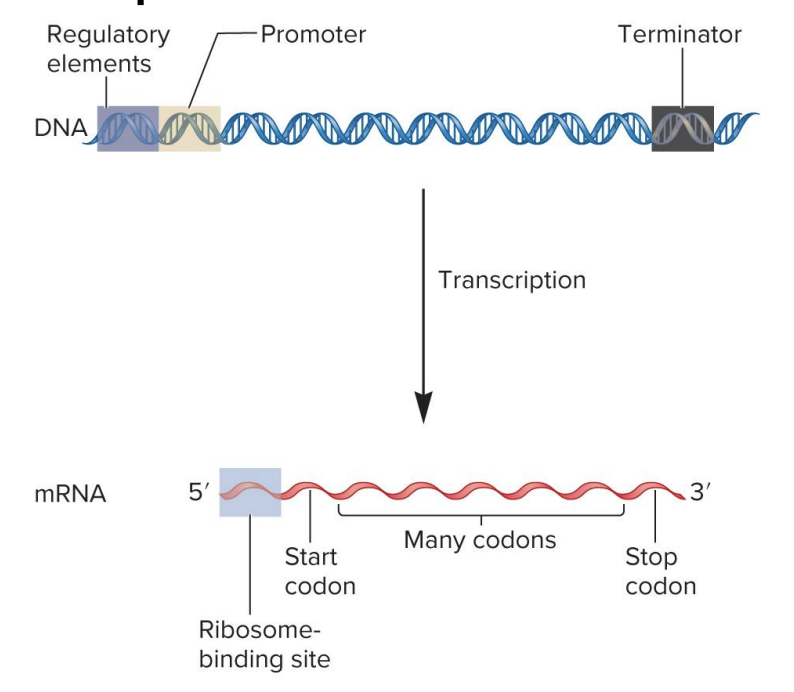
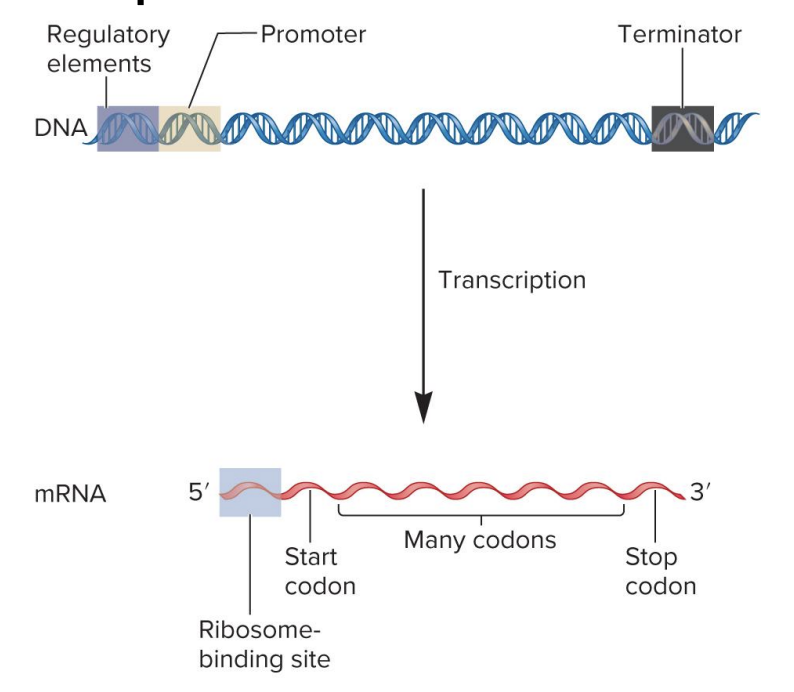
sequence organization to form a bacterial gene that codes an mRNA transcription
DNA
regulatory element
site for binding of regulatory proteins
regulatory proteins influence transcription rate
found in variety of locations
promotor
RNA polymerase binding site
signals beginning of transcription
terminator
signals end of transcription
mRNA
ribosome binding site
site for ribosome binding
translation begins near this site in mRNA
in eukaryotes, ribosome scans mRNA for start codon
start codon
specifies 1st amino acid in a polypeptide sequence
formylmethionine (bacteria)
methionine (eukaryotes)
codons
3 nucleotide sequences within mRNA that specify particular amino acids
codon sequence within mRNA determines the sequence of amino acids within a polypeptide
stop codon
specifies the end of a polypeptide synthesis
bacterial mRNA may be polycistronic (codes 2 or more polypeptides)
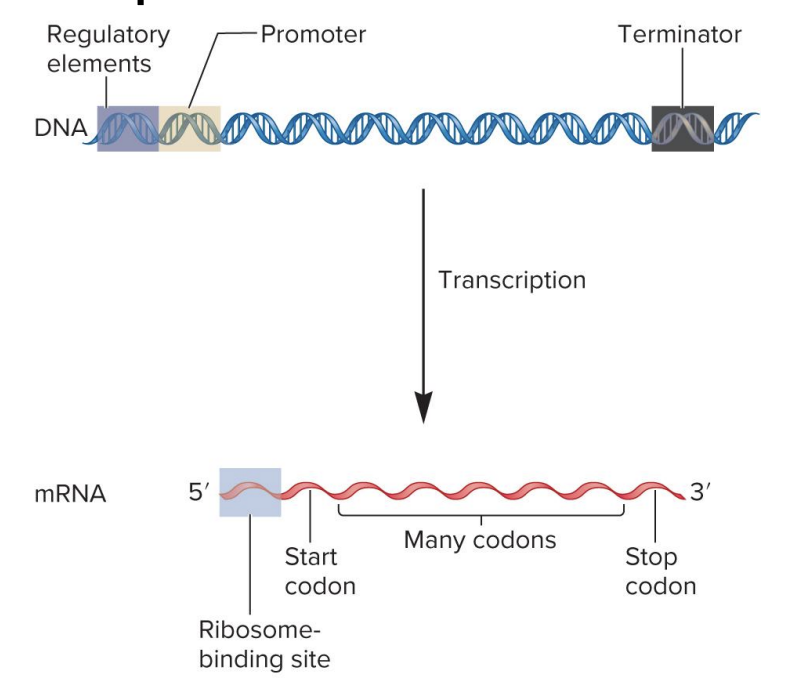
regulatory elements
specific DNA sequences that serve as binding sites for regulatory transcription factor proteins
these regulatory proteins (when bound) influence transcription rate
found in a variety of locations (upstream, downsream of gene or within introns)
DNA
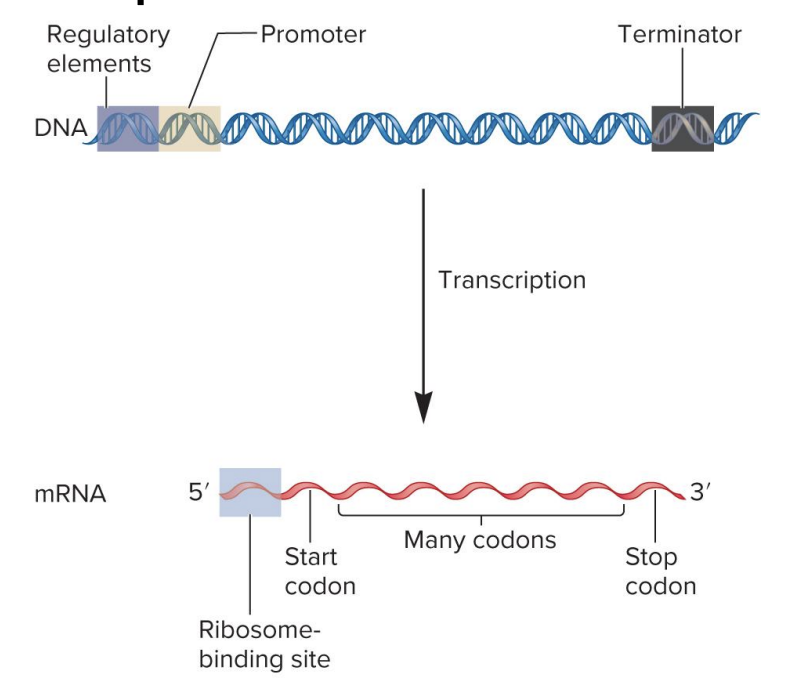
promotor
site for RNA polymerase binding
signals beginning of transcription
dictates where RNA polymerase should start synthesizing RNA
DNA sequence
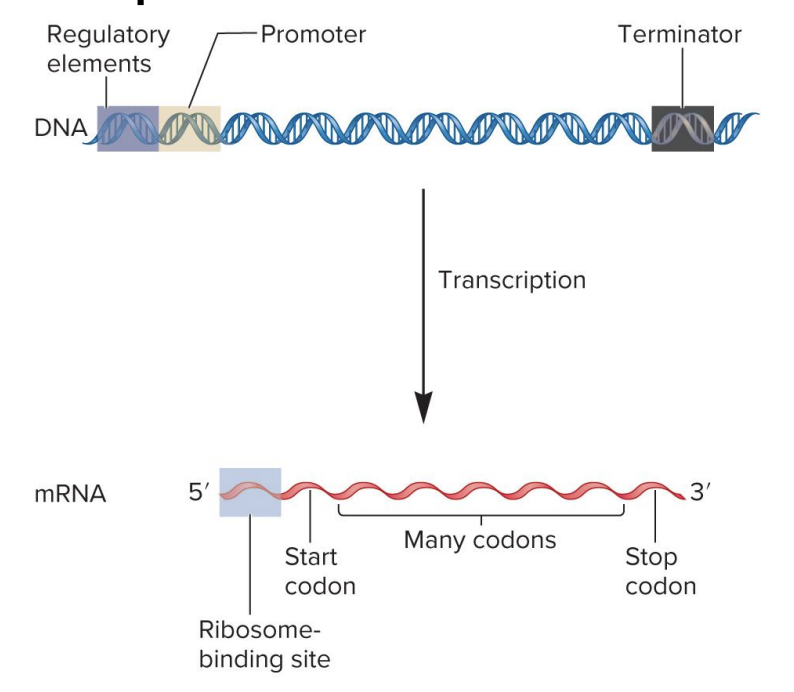
terminator
signals end of transcription
when RNA polymerase encounters ?, RNA polymerase detaches from DNA, releasing newly synthesized RNA molecule
DNA sequence
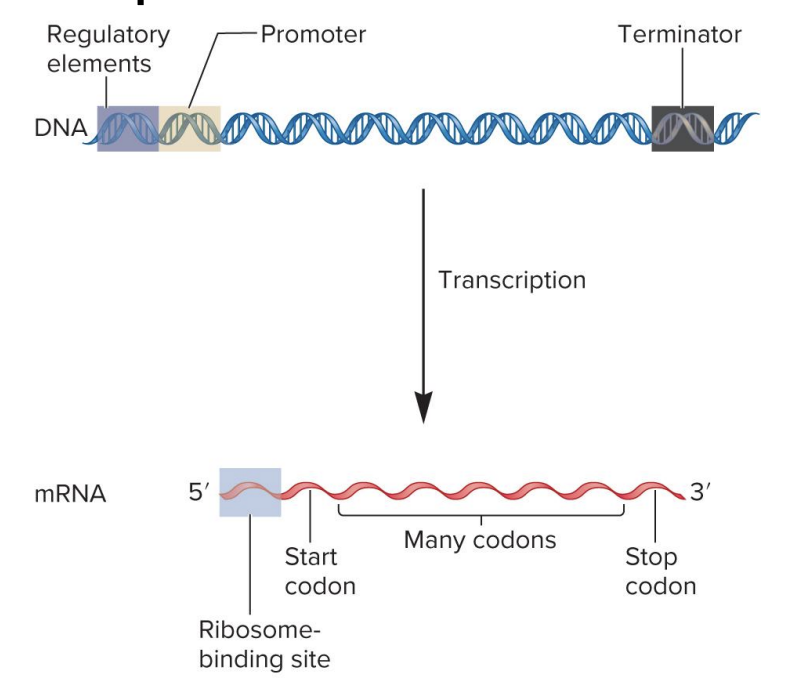
ribosome binding site
site for ribosome binding to mRNA in bacteria
translation begins near this site in the mRNA
in eukaryotes, the ribosome binds to a 7-methyguanosine cap in mRNA and ribosome scans the RNA for a start codon
mRNA
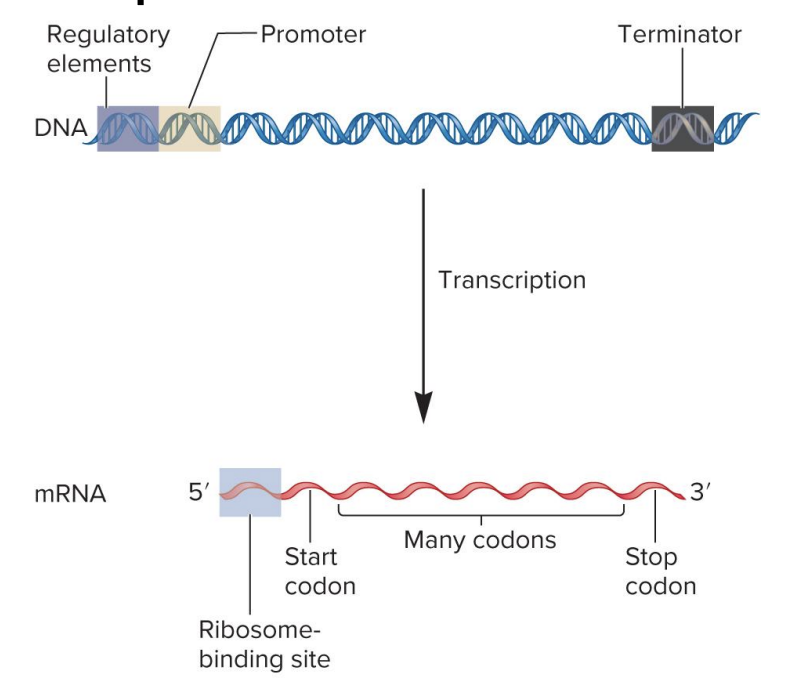
codons
3 nucleotide sequences within the mRNA that specify particular amino acids
? sequence within mRNA determines the sequence of amino acids within a polypeptide chain during translation
mRNA
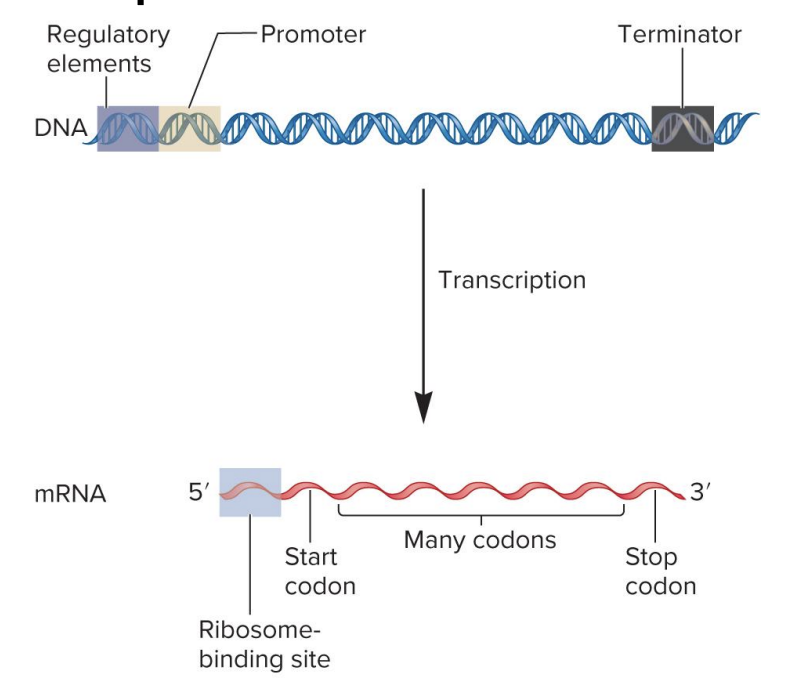
start codon
specifies the 1st amino acid in a polypeptide sequence (initiate translation)
formylmethionine (bacteria)
methionine (eukaryotes)
mRNA (AUG)
stop codon
specifies the end of polypeptide synthesis (translation)
mRNA
polycistronic
bacteria mRNA may code 2 or more polypeptides
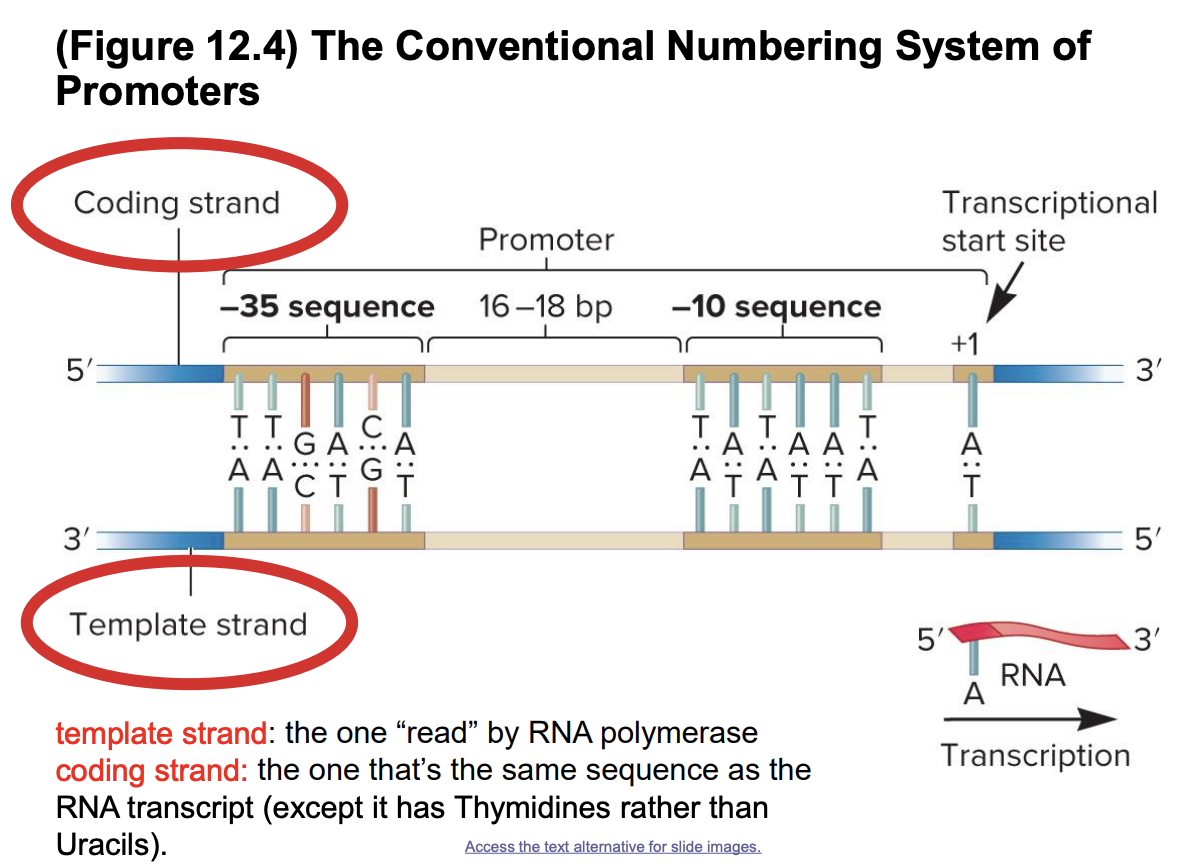
template/noncoding/antsense strand
DNA strand that is actually transcribed (used as template) for RNA synthesis during transcription
RNA transcript is complementary to this sequence
coding/sense/nontemplate strand
DNA strand not used as template during transcription (opposite to template strand)
base sequence in RNA is identical to the ? strand of DNA, except for the substitution of uracil in RNA for thymine in DNA
transcription factors
recognize the promotor and regulatory elements to control transcription rate
bind to enhancers (DNA), silencers (DNA)
proteins that bind to specific DNA sequences
mRNA
? sequences like ribosomal binding site and codons direct translation
ribosomal binding site in bacteria (or 5’ cap in eukaryotes) position ribosome correctly on mRNA
codons specify amino acids to be incorporated into growing polypeptide chain
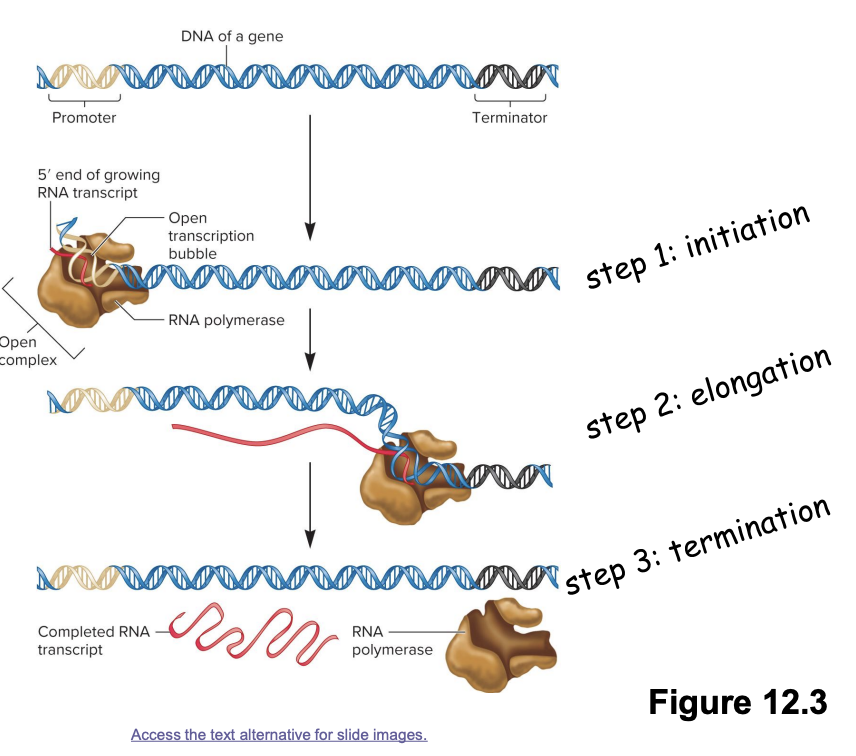
transcription stages
3 stages
Initiation
RNA polymerase binds promotor region on DNA→ initiate unwinding of DNA double helix
Elongation
RNA polymerase moves along DNA template strand, synthesizing complementary RNA molecule in 5’ to 3’ direction
Termination
RNA polymerase encounters terminator DNA sequence→ release RNA transcript
involve protein-DNA interactions
proteins like RNA polymerase and various transcription factors interact with DNA sequences (promotors, terminators, regulatory elements) to regulate gene expression
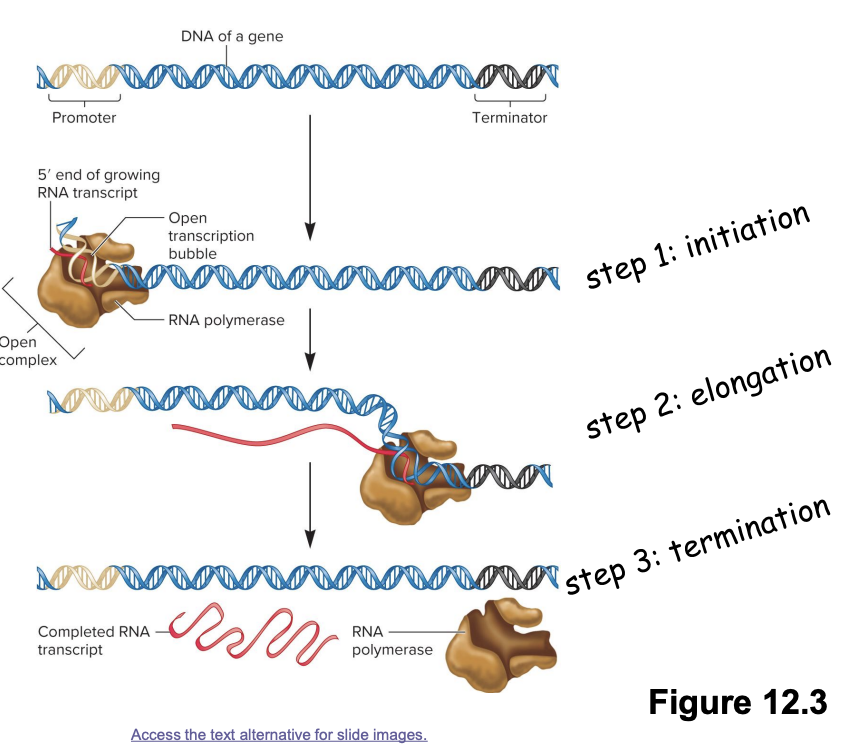
initiation
promotor functions as a recognition site for transcription factors
transcription factors enable RNA polymerase to bind to the promotor region of DNA
after binding, the DNA is denatured into an open transcription bubble (double helix→ open complex transcription bubble)
transcription stage

elongation/synthesis of RNA transcript
RNA polymerase slides 5’ to 3’ along DNA template strand in an open complex to synthesize complementary RNA
continuous addition of ribonucleotides to growing mRNA transcript
A with U
T with A
C with G
G with C
transcription stage
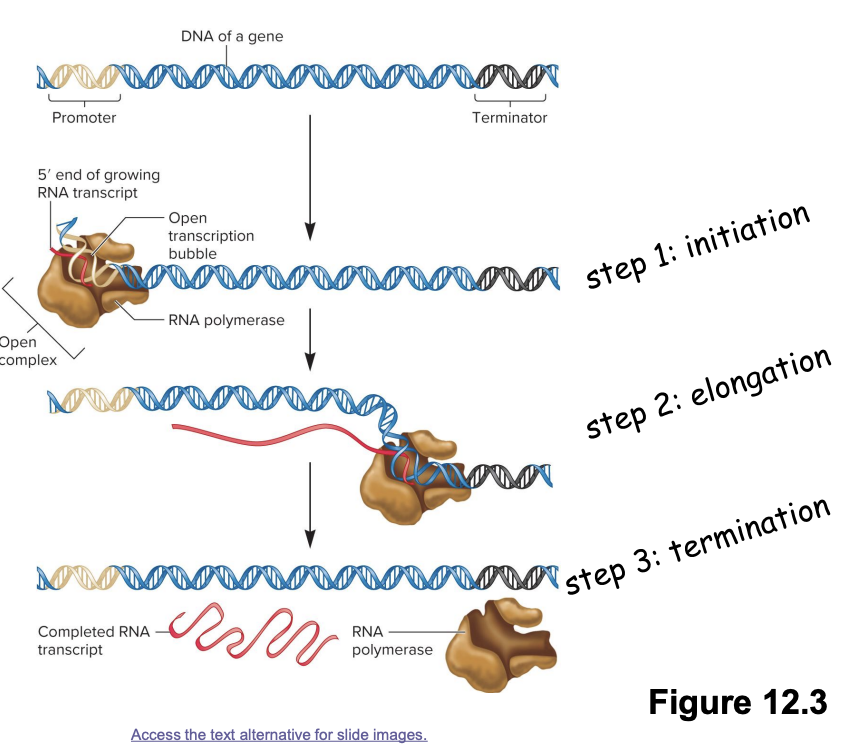
termination
a terminator DNA sequence is reached that causes RNA polymerase and the end RNA transcript to dissociate from the DNA template
transcription stage
bacteria
our molecular understanding of gene transcription came from early studies involving ? and ?phages
much of our knowledge coms from studies of E. coli
promoters
DNA sequences that “promote” gene expression
direct the exact location for transcription initiation
typically immediately upstream of site where transcription of a gene actually begins
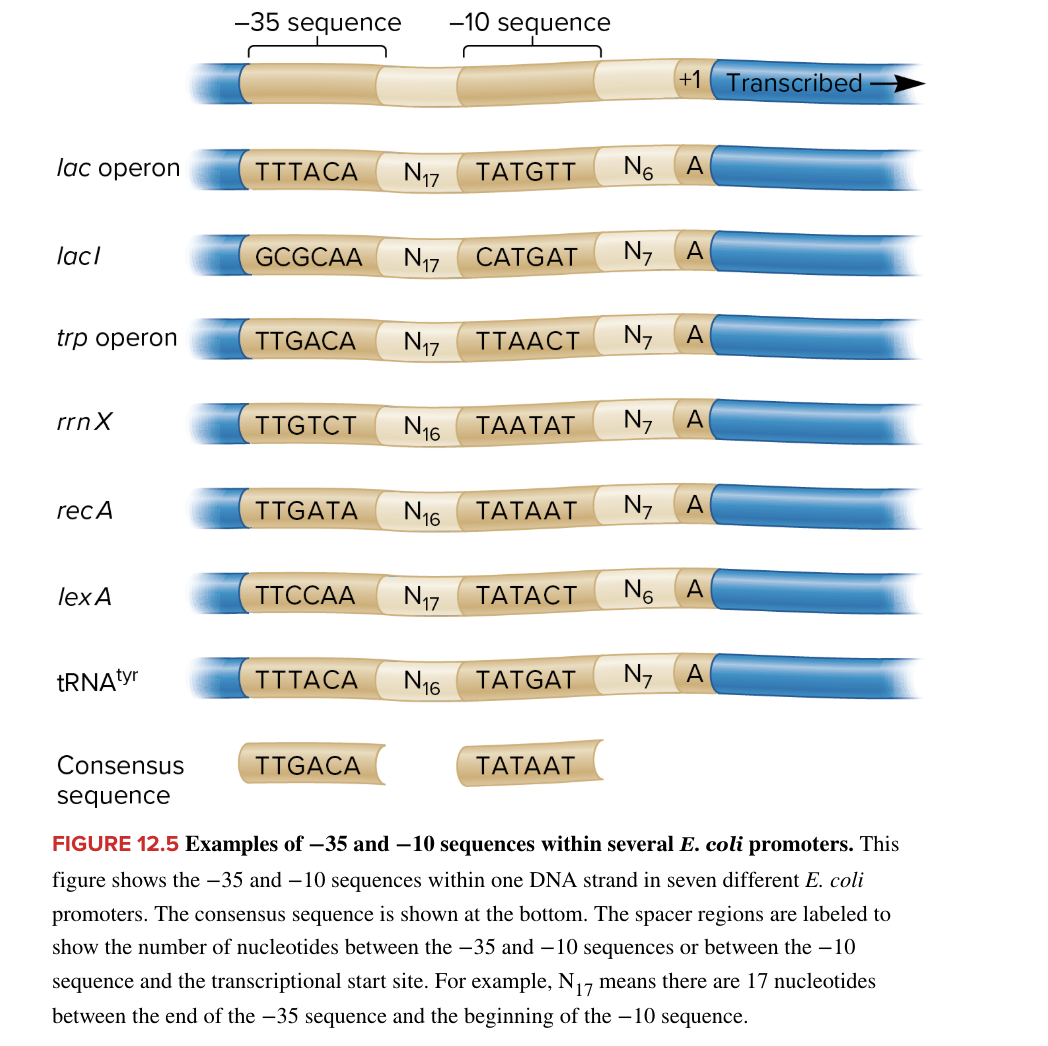
bases in a ? sequence are numbered in relation to the transcriptional start site (allow precise ID of regulatory elements)
no zero nucleotide in this numbering system
negative numbers ID bases preceding beginning of transcription (to the left)
vary at the -35 and -10 sequences
most common is consensus sequence
consensus sequence likely to result in high level of transcription
sequences that deviate from consensus sequence typically result in lower levels of transcription
template strand
the one “read” by RNA polymerase to synthesize a complementary RNA molecule
coding strand
the one that has the same sequence as the RNA transcript (except it has Thymidines rather than Uracils)
the DNA strand with same sequence as mRNA transcript (but with T instead of U)
not used as a template for RNA synthesis
consensus sequence
most common promotor sequence at the -35 and -10 sequences
likely to result in a high level of transcription
if sequence deviates from the ? sequence it typically results in lower levels of transcription
DNA
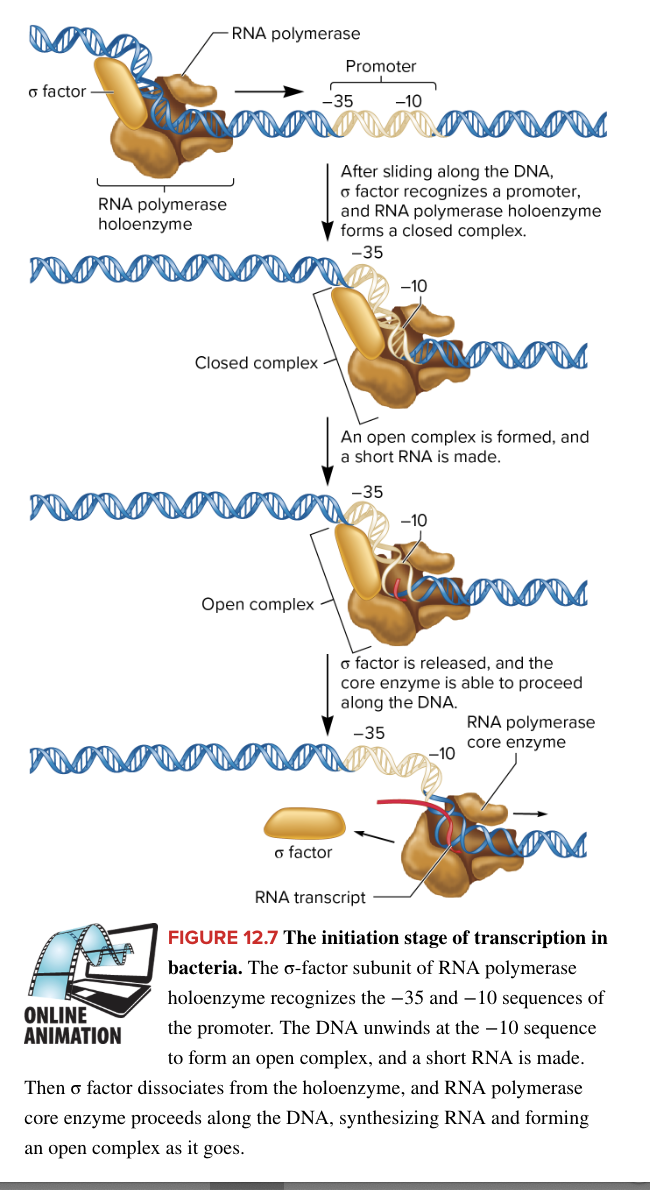
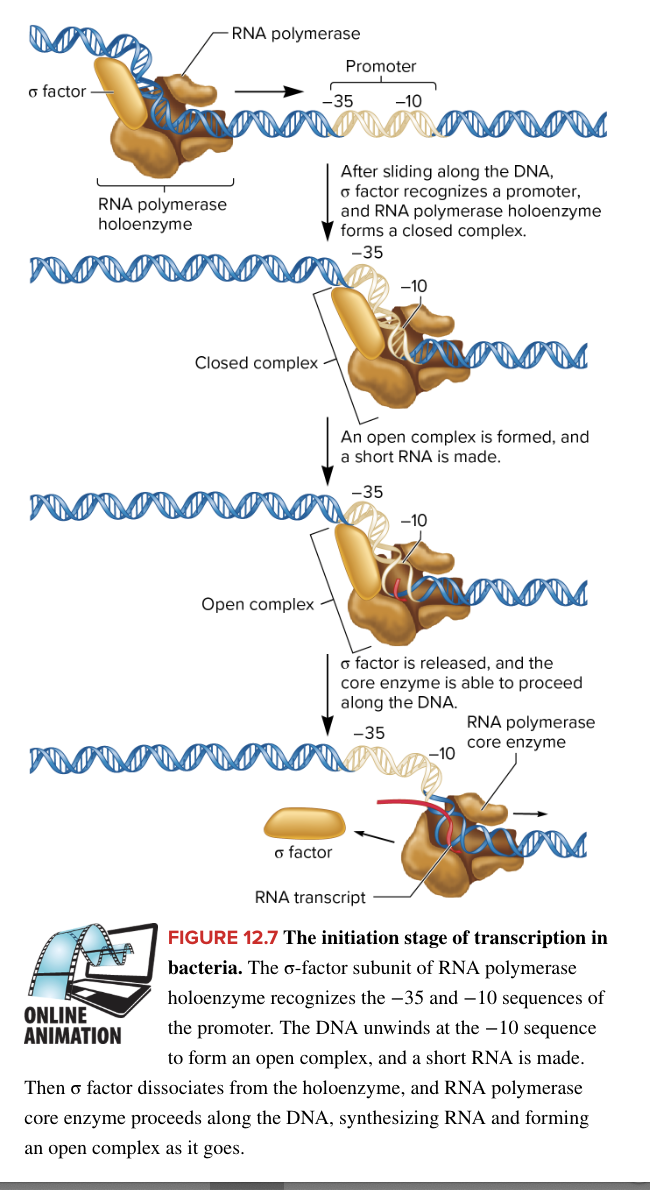
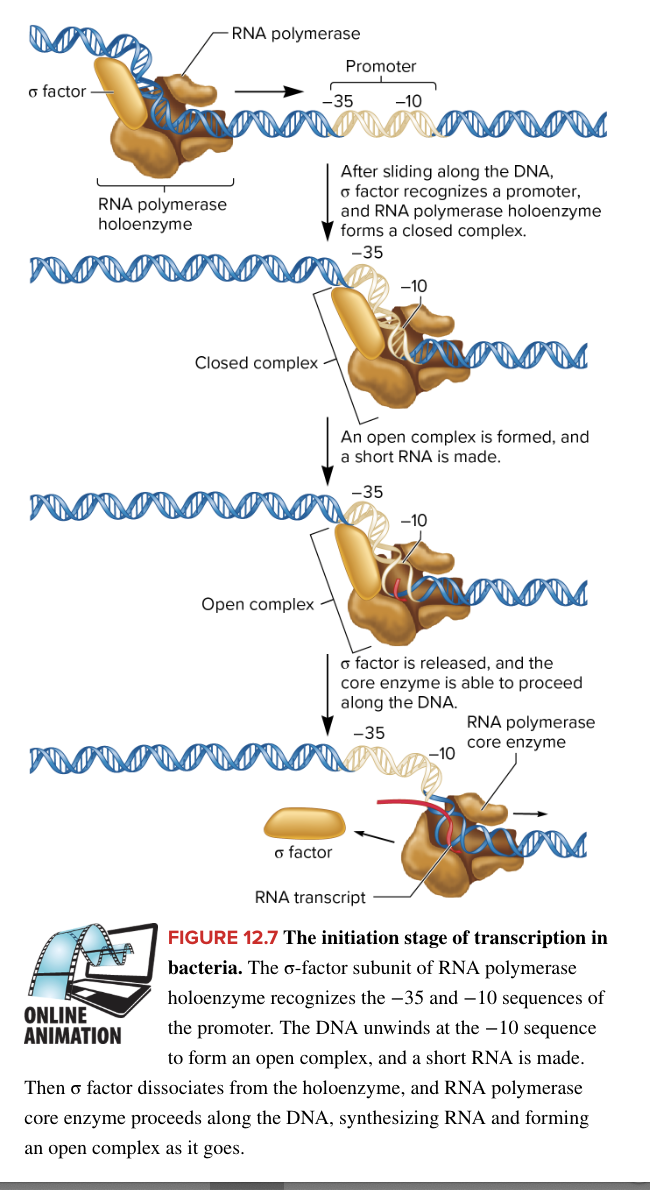
initiation of bacterial transcription
RNA polymerase catalyzes RNA synthesis
RNA polymerase holoenzyme binds loosely to the DNA and scans along the DNA until it encounters a promotor
in E. coli, RNA polymerase holoenzyme is composed of
core enzyme (5 subunits) responsible for transcription
sigma factor (1 subunit) protein helping RNA polymerase locate the promotor
these subunits play distinct functional roles
when RNA polymerase encounter a promotor, sigma factor recognizes both the -35 and -10 promotor sequences
a region within the sigma factor, containing a helix-turn-helix structure, facilitates tighter binding to the DNA (this binding forms the closed complex)
binding of RNA polymerase to promotor forms closed complex
then the TATAAT box in the -10 sequence is unwound, forming the open complex, and a short RNA strand is made
A-T bonds more easily separated, aiding in unwinding
sigma factor is released (after synthesizing the short RNA strand within the open complex)
core enzyme is able to proceed along the DNA marking the end of initiation and beginning of elongation where core enzyme now slides down DNA to synthesize an RNA strand
bacterial transcription
RNA polymerase
enzyme that catalyzes the synthesis of RNA in bacterial initation of transcription
bacterial transcription
core enzyme
part of RNA polymerase holoenzyme that does the transcription
5 subunits
bacterial transcription
sigma factor
protein that is a part of RNA polymerase holoenzyme, helps RNA polymerase find the promotor
1 subunit
recognizes the -35 and -10 promotor DNA sequences as RNA polymerase holoenzyme scans the DNA for a promotor
contains a region with a helix turn helix structure that helps the RNA polymerase bind tighter to the DNA promotor→ closed complex formation
? is released when the short RNA strand is synthesized within the open complex
bacterial transcription
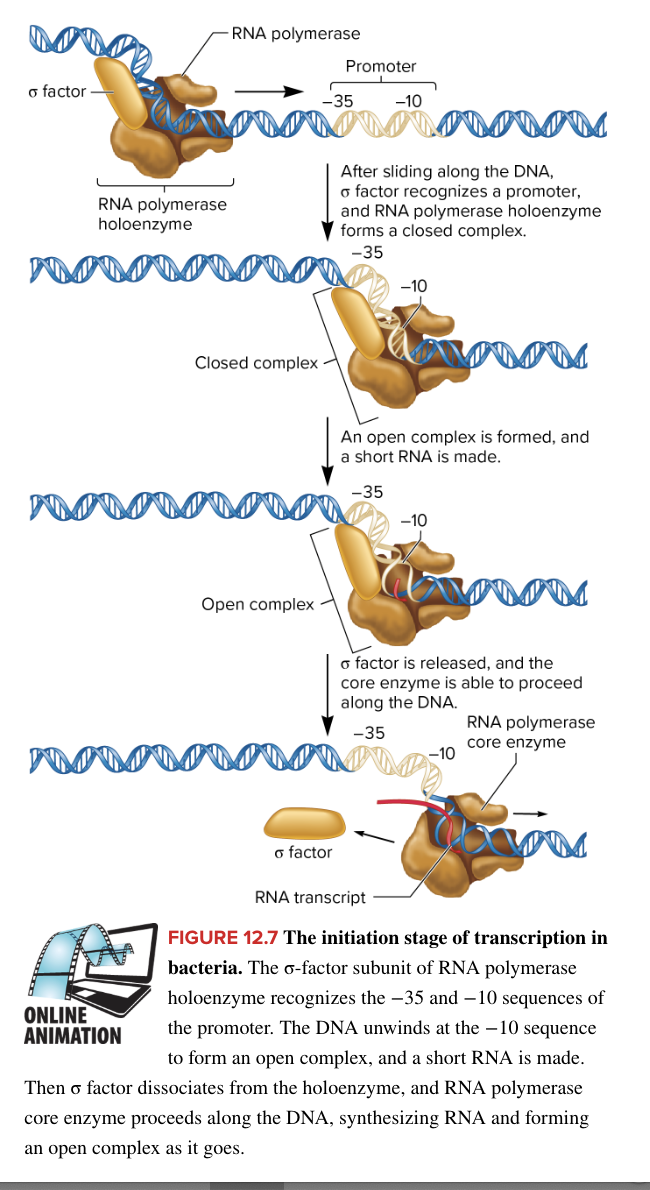
RNA polymerase holoenzyme
responsible for bacterial transcription, composed of
core enzyme
5 subunits
catalyzes RNA synthesis (does the transcription)
sigma factor
1 subunit
protein that helps RNA polymerase find the promotor
loosely binds to DNA and scans for a promoter. Upon encountering a promoter, the sigma factor facilitates binding, leading to the formation of a closed complex, which subsequently transitions into an open complex
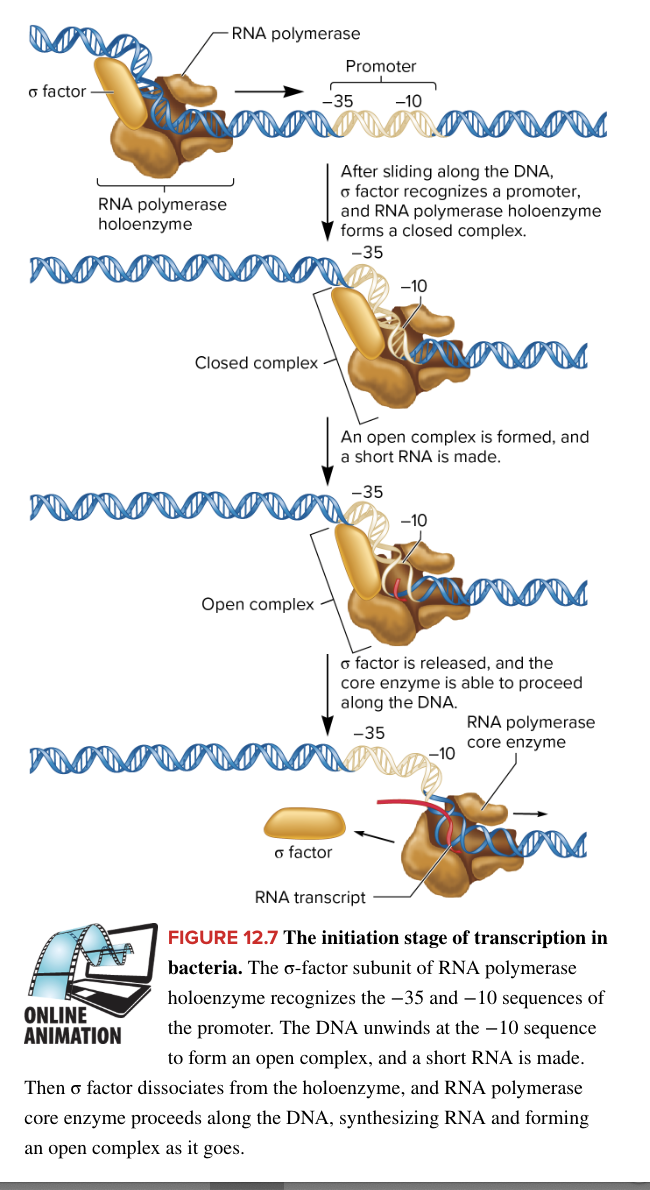
elongation in bacterial transcription
RNA transcript is synthesized
RNA polymerase slides along the DNA in a 3’ to 5’ direction, creating an open complex as it moves
RNA is synthesized in a 5’ to 3’ direction using nucleoside triphosphates as precursors
pyrophosphate is released
complementary rule is the same as the AT/GC rule except U is substituted for T in the RNA
each gene uses one DNA strand as a template, but the template strand used is not always the same
DNA strand used as template for RNA synthesis is the template or antisense strand
the opposite DNA strand is the coding strand
has the same base sequence as the RNA transcript except that T in DNA corresponds to U in RNA
a promotor specifies the direction of transcription
with regard to adjacent genes along a chromosome, some promoters direct transcription in one direction and others direct transcription in the opposite direction
in any case, the template strand is read in the 3’ to 5’ direction (direction of transcription)
synthesis of RNA transcript occurs in a 5’ to 3’ direction
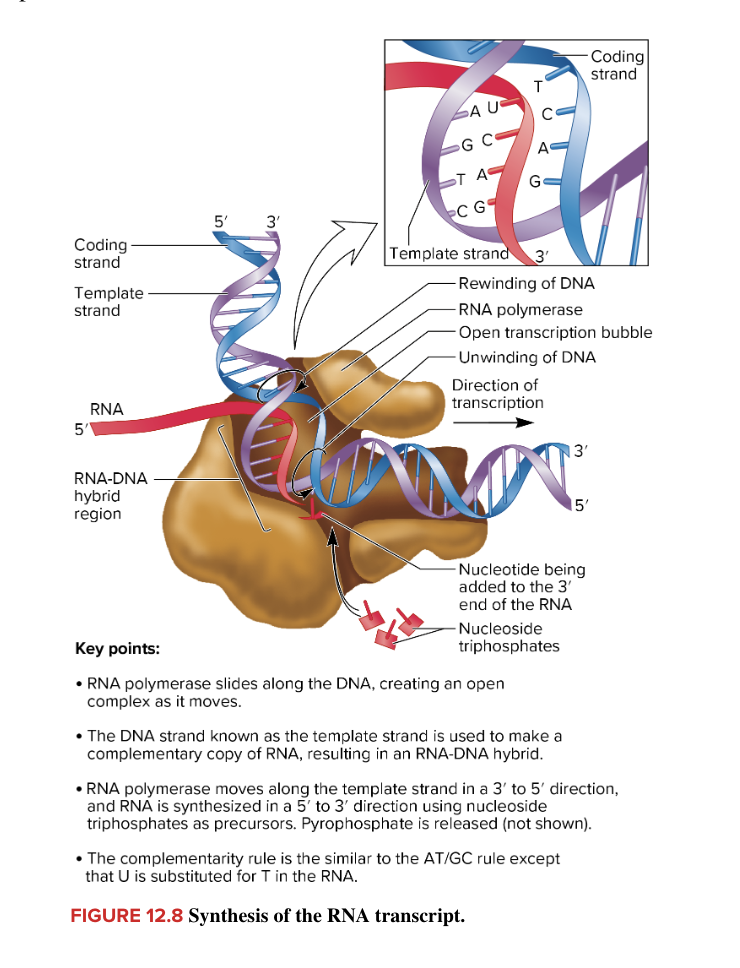
promoter
specifies the direction of transcription
for adjacent genes along a chromosome, some direct transcription in one direction and others direct transcription in the opposite direction
for transcription of multiple genes within a chromosome, the direction of transcription and the DNA strand used as a template can vary among genes
template strand is always read in the 3’ to 5’ direction (direction of transcription is 3’ to 5’)
RNA synthesis occurs in a 5’ to 3’ direction
in the figure, Genes A and B are transcribed L to R (promotor to terminator)
template strand is the bottom strand (bc direction of transcription is 3’ to 5’/read template strand from 3’ to 5’)
Gene A and B RNA is synthesized 5’ to 3’ to the right
Gene C is transcribed R to L (promotor to terminator))
template strand is top strand (bc direction of transcription is 3’ to 5’/read template strand from 3’ to 5’)
Gene C RNA is synthesized 5’ to 3’ to the left
always remember
direction of transcription/template strand reading is always 3’ to 5’
RNA transcript is always synthesized 5’ to 3’
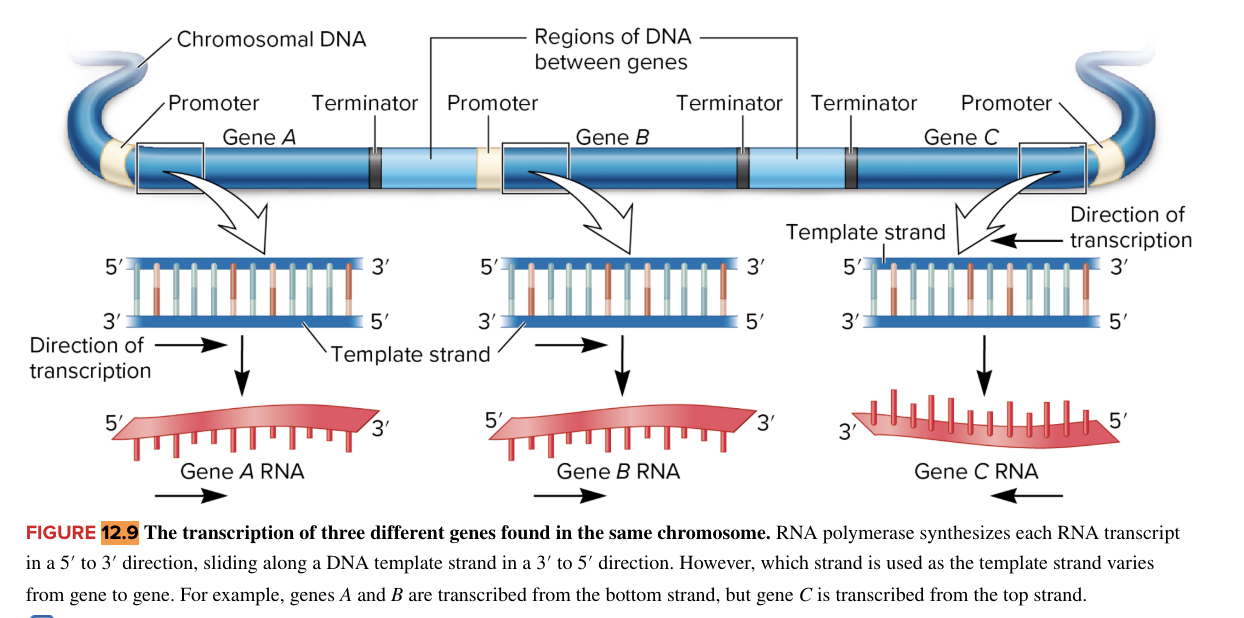
elongation in bacterial transcription
RNA pol slides along DNA 3’ to 5’, creating an open complex as it moves
DNA template strand used to make a complementary copy of RNA, resulting in a short RNA DNA hybrid
as RNA pol moves along template strand 3’ to 5’, RNA is synthesized 5’ to 3’ using nucleoside triphosphates as precursors
pyrophosphate is released
complementarity rule is similar to AT/GC rule except U is substituted for T in RNA
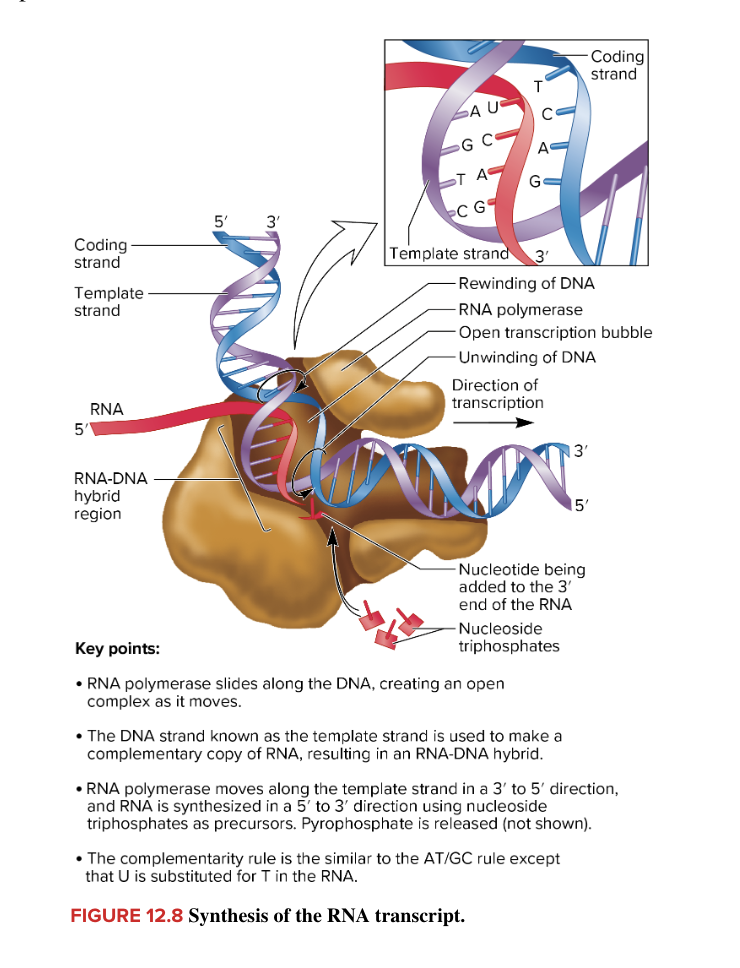
termination of bacterial transcription
end of RNA synthesis
occurs when short RNA-DNA hybrid of the open complex is forced to separate
this releases RNA polymerase and newly made RNA
E. coli has 2 diff mechanisms
rho-dependent
requires protein ρ (rho)
rho independent (intrinsic) termination
does not require ρ (rho) protein
facilitated by 2 sequences in RNA
stem loop structure upstream of uracil rich sequence
causes RNA polymerase to pause synthesis of RNA
uracil rich sequence at 3’ end of RNA
while RNA polymerase pauses, weakly bound U rich sequence can’t hold RNA-DNA hybrid together (leads to dissociation of the RNA pol and new RNA transcript from the DNA template)
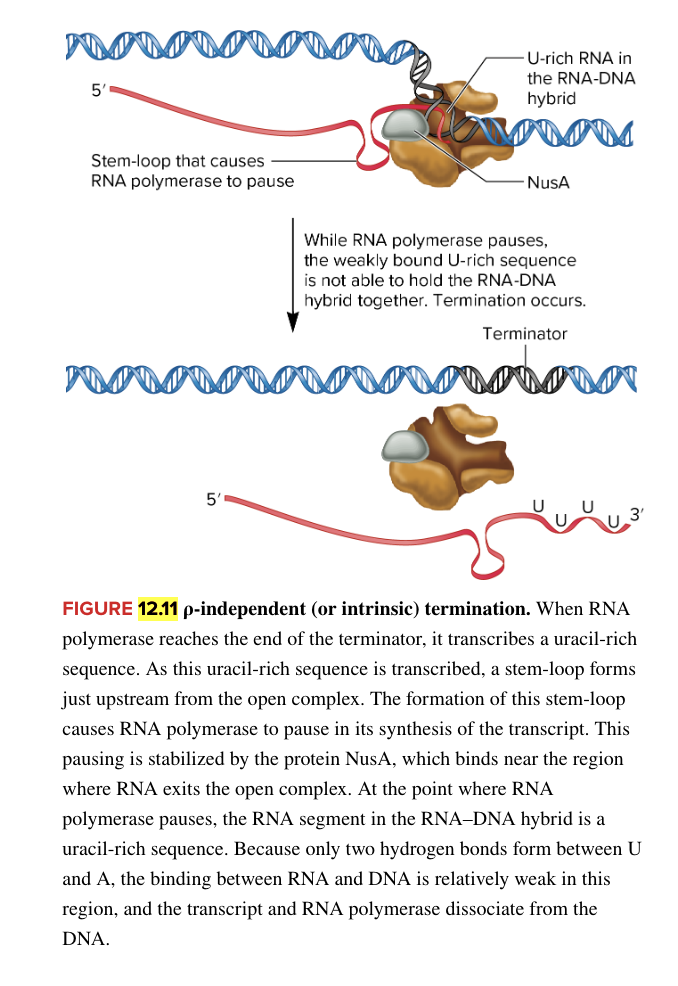
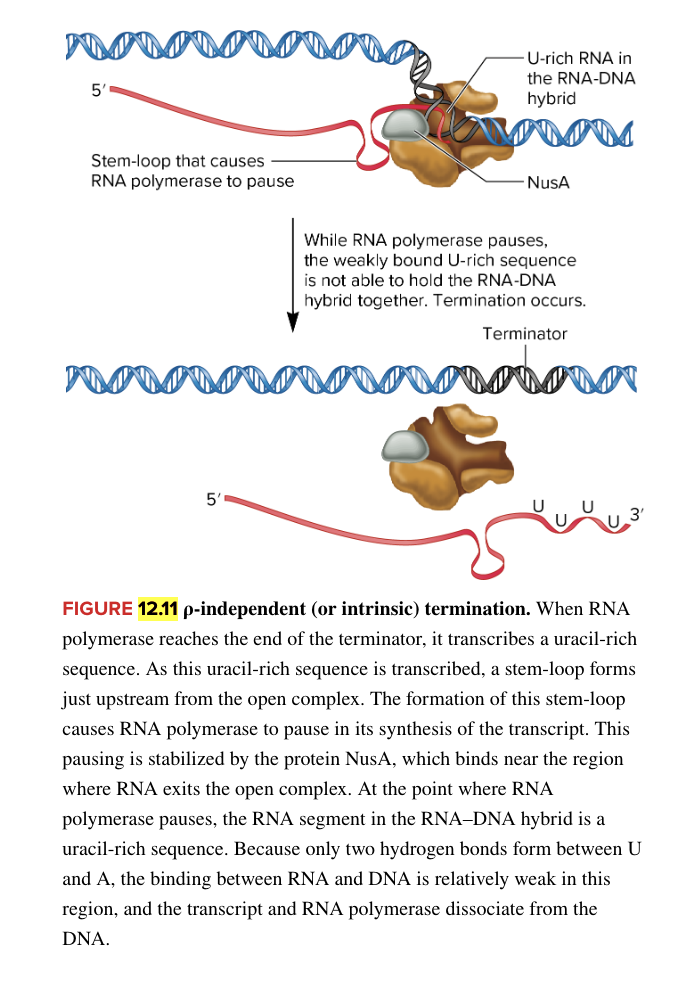
rho independent intrinsic termination
facilitated by 2 sequences in RNA
stem loop structure upstream of uracil rich sequence
causes RNA polymerase to pause synthesis of RNA
uracil rich sequence at 3’ end of RNA
while RNA polymerase pauses, weakly bound U rich sequence can’t hold RNA-DNA hybrid together (leads to dissociation of the RNA pol and new RNA transcript from the DNA template)
mechanism for termination in E. coli bacteria
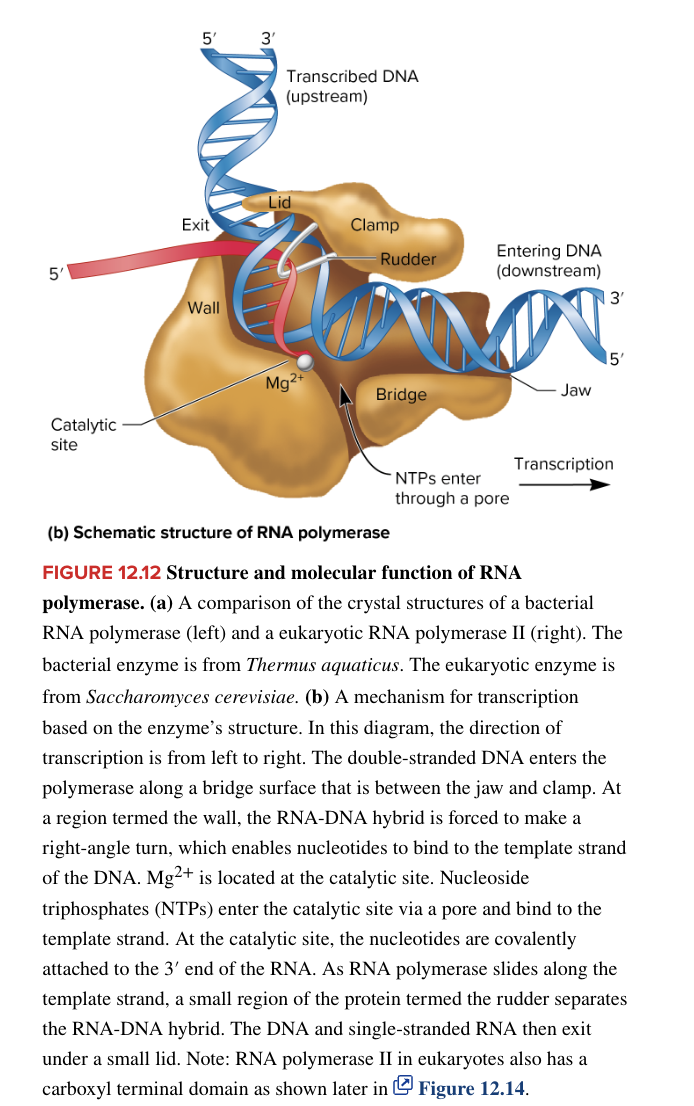
eukaryotic transcription
more complex bc
larger, more complex cells (organelles)
added cellular complexity means more genes that code proteins are required (more protein-coding genes)
multicellularity adds another level of regulation
express genes only in the correct cells at the proper time
eukaryotic RNA polymerases
nuclear DNA is transcribed by
RNA pol I
transcribes all rRNA genes (except for 5S rRNA)
RNA pol II
transcribes all protein-coding structural genes (basically synthesizes all mRNAs)
transcribes some snRNA genes needed for splicing
RNA pol III
transcribes all tRNA genes
transcribes the 5S rRNA gene
transcribes microRNA genes
all 3 are very similar structurally and are composed of many subunits
there is also remarkable similarity between bacterial RNA polymerase and its eukaryotic counterparts
eukaryotic RNA pol I
transcribes all rRNA genes (except for 5S rRNA)
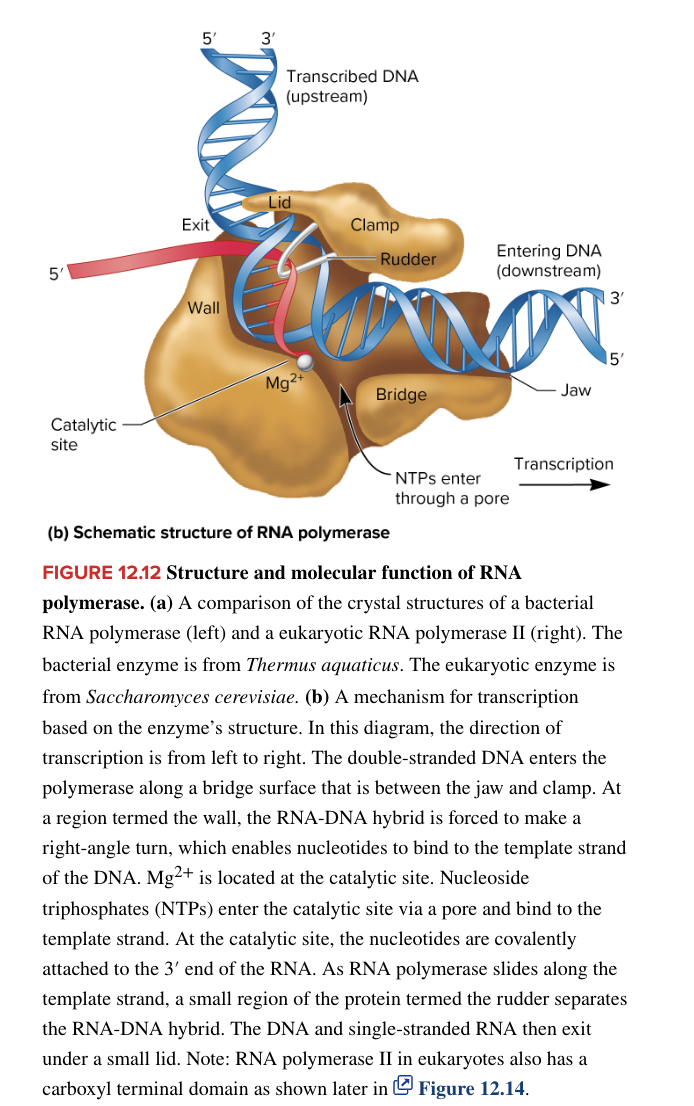
eukaryotic RNA pol II
transcribes all protein-coding structural genes (basically synthesizes all mRNAs)
transcribes some snRNA genes needed for splicing
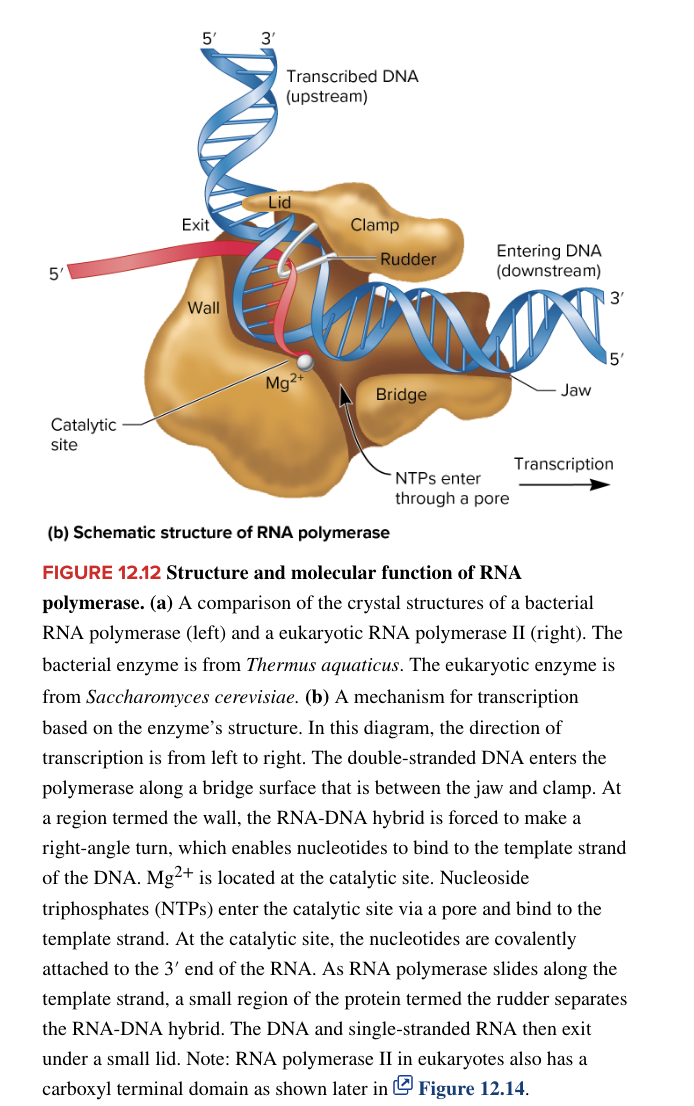
eukaryotic RNA pol III
transcribes all tRNA genes
transcribes the 5S rRNA gene
transcribes microRNA genes
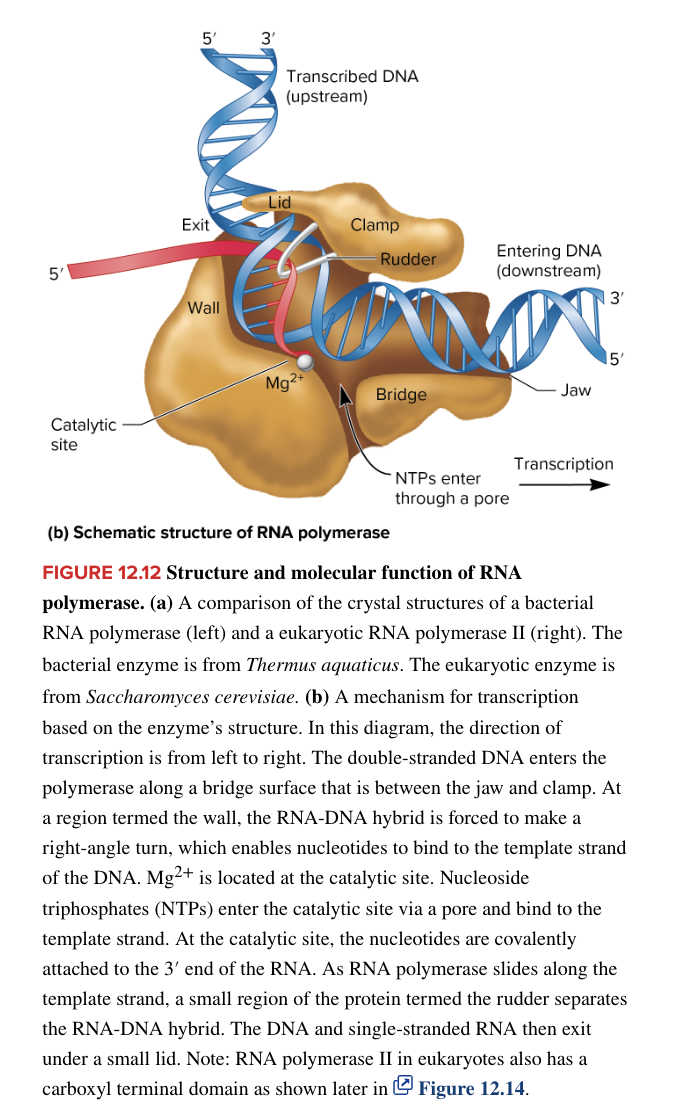
eukaryotic transcription initation
protein-coding genes are influenced by core promotor and regulatory elements
core promotor
relatively short DNA sequence necessary for transcription to occur
consists of TATA box, transcriptional start site, and 1 or more downstream promoter elements DPEs
TATA box not always present, but is important for determining the precise start point for transcription (so if it is missing, the transcription start site becomes undefined and transcription may start at a variety of different locations)
core promoter by itself produces a low level of transcription (basal transcription)
regulatory elements
enhancers
DNA segments, usually 50 bp to 1000 bp
contain 1 or more regulatory elements
vary widely in their locations but are often found in the -50 to -100 region
regulatory transcription factors
proteins that affect RNA pol’s ability to recognize core promoter and begin transcription
bind to regulatory elements and influence transcription rate
activators- recognize enhancers and stimulate transcription rate
repressors- bind enhancers, inhibit transcription
factors regulating gene transcription can be divided into 2 general types
cis acting elements
DNA sequences that exert their effect only over a particular gene
TATA box, enhancers containing regulatory elements
trans acting factors
proteins (general and regulatory transcription factors) that bind to cis-acting elements
3 protein categories are required for basal transcription to occur at the promotor
RNA polymerase II
6 general transcription factors (GTFs)
bc RNA polymerase can’t find promoters on its own
proteins that assist RNA pol II bind to promoter and initiate transcription
mediator (protein complex)
general transcription factors and RNA polymerase II assemble at the promoter of a gene, which often contains a TATA box
core promoter
relatively short DNA sequence necessary for transcription to occur
consists of TATA box, transcriptional start site, and 1 or more downstream promoter elements DPEs
TATA box not always present, but is important for determining the precise start point for transcription (so if it is missing, the transcription start site becomes undefined and transcription may start at a variety of different locations)
? by itself produces a low level of transcription (basal transcription)
eukaryotic transcription initiation
TATA box
DNA sequence found within eukaryotic core promoters that determines transcription starting site
important for determining the precise start point for transcription
if it is missing, the transcription start site becomes undefined and transcription may start at a variety of different locations
eukaryotic transcription initiation
basal transcription
in eukaryotes, a low level of transcription produced by the core promoter (when alone)
The binding of transcription factors to enhancer elements may increase transcription above this level.
3 protein categories required for this to occur at the promoter
RNA polymerase II
6 general transcription factors (GTFs)
bc RNA polymerase can’t find promoters on its own
proteins that assist RNA pol II bind to promoter and initiate transcription
mediator (protein complex)
general transcription factors and RNA polymerase II assemble at the promoter of a gene, which often contains a TATA box
enhancers
DNA segments, usually 50 bp to 1000 bp
contain 1 or more regulatory elements
vary widely in their locations but are often found in the -50 to -100 region
influences transcription in eukaryotes
regulatory transcription factors RTFs
proteins that affect RNA pol’s ability to recognize core promoter and begin transcription
bind to regulatory elements and influence transcription rate
activators- recognize enhancers and stimulate transcription rate
repressors- bind enhancers, inhibit transcription
eukaryotic transcription initiation
activators
regulatory proteins that recognize and bind to DNA sequences called enhancers
stimulate transcription rate (otherwise most eukaryotic genes have low levels of basal transcription)
repressors
proteins that bind to DNA sequences called enhancers
inhibit eukaryotic transcription rate
cis acting elements
DNA sequences that exert their effect only over a particular gene
TATA box, enhancers containing regulatory elements
regulate eukaryotic gene transcription
“next to”
may be located far away from core promoter, but always found within same chromosome as the genes they regulate.
trans acting factors
proteins (general and regulatory transcription factors) that bind to cis-acting elements
regulate eukaryotic gene transcription
“across from”
may be encoded by genes far away from genes they control, even on a diff chromosome
general transcription factors GTFs
proteins necessary to initiate basal transcription at the core promoter
proteins that assist RNA pol II bind to promoter and initiate transcription (bc RNA polymerase can’t find promoters on its own)
mediator
large protein complex
interacts with RNA pol II and various regulatory transcription factors
depending on its interactions with RTFs, may stimulate or inhibit RNA pol II
basal transcription reqs
6 diff general transcription factors GTFs, mediator (protein complex, RNA pol II
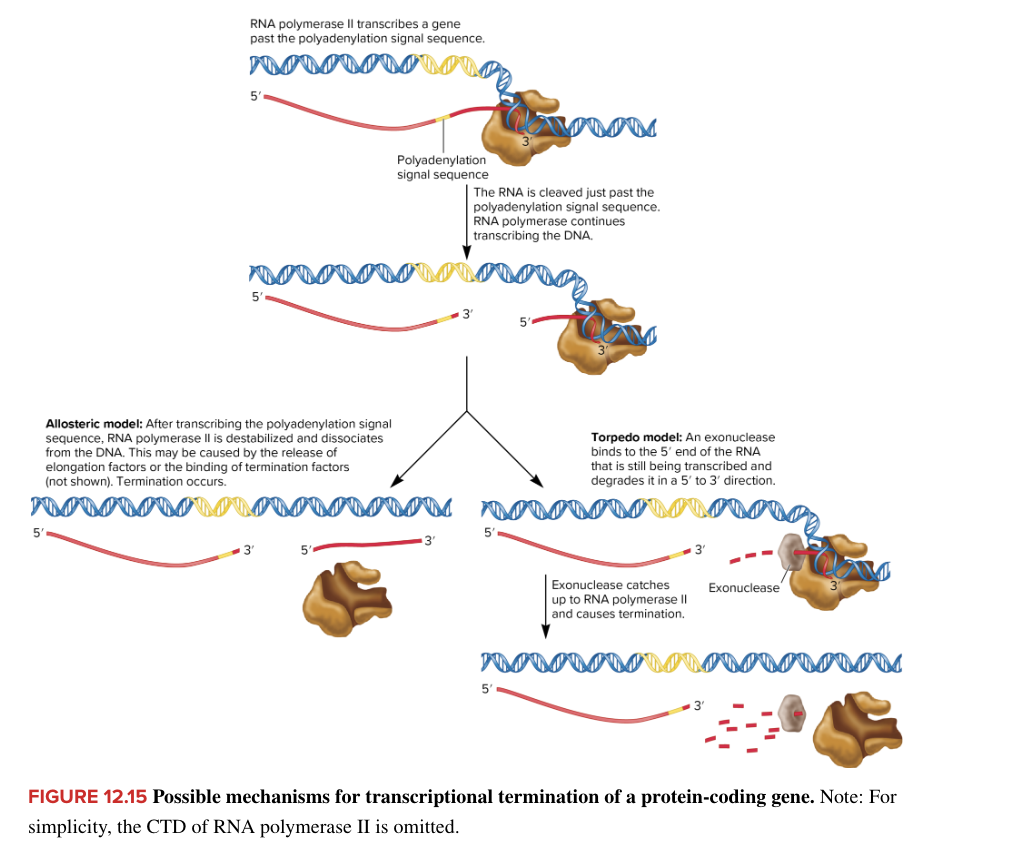
eukaryotic transcript termination
RNA pol II transcriptional termination
pre-mRNAs are modified by cleavage near 3’ end with subsequent attachment of a string of adenines to form a polyAtail
transcription terminates 500-2000 nucleotides downstream from polyA signal
2 models for termination (unclear which/if correct)
allosteric
torpedo
eukaryotic transcription elongation
RNA polymerase II: Moves along the DNA template strand, synthesizing RNA.
5' to 3' RNA synthesis: Builds the mRNA using ribonucleotide triphosphates.
Maintaining the open complex: Unwinds DNA ahead of the polymerase and rewinds it behind.
Proofreading: Corrects errors in the growing RNA strand.
Modification of the mRNA: As RNA polymerase II transcribes, the growing pre-mRNA undergoes modifications, including 5' capping (shortly after initiation) and 3' polyadenylation (adding a poly-A tail). These modifications are crucial for mRNA stability and processing.
Coordination with RNA processing: Couples transcription with modifications like splicing, ensuring the mRNA is ready for translation.
eukaryotic RNA modification
RNA transcripts can be modified by
chopping
splicing
capping
tailing
analysis of bacterial genes in the 1960s and 1970s revealed
DNA sequence in coding strand corresponds to nucleotide sequence in mRNA
codon sequence in mRNA provides instructions for amino acid sequence in the polypeptide
→ termed the colinearity of gene expression
analysis of eukaryotic protein-coding genes in the late 1970s revealed that they aren’t always colinear with their functional mRNAs
instead, coding sequences (exons) are often interrupted by intervening sequences (introns)
transcription produces pre-mRNA corresponding to entire gene sequence
introns are subsequently removed/excised
exons connected together or spliced
→ RNA splicing
common genetic phenomenon in eukaryotes
(occasionally occurs in bacteria as well)
RNA transcripts can also be modified by
processing of rRNA and tRNA transcripts to smaller functional pieces (chopping)
5’ capping
role in intron splicing, mRNA export from nucleus, and binding of mRNA to ribosome
3’ polyA tailing of mRNA transcripts
RNA stability and translation in eukaryotes
eukaryotic RNA modification
colinearity
the direct correspondence between the sequence of codons in the DNA coding strand and the amino acid sequence of a polypeptide.
in simple terms, the order of nucleotides in the DNA determines the order of amino acids in the protein
but eukaryotic protein coding genes aren’t always ?
contain numerous introns interrupting exons
RNA splicing necessary to remove introns from the pre-mRNA transcription and produce functional mRNA molecule
eukaryotic RNA modification
exons
region of RNA molecule that remains after splicing has removed the introns
in mRNA, contains the coding sequence of a polypeptide
eukaryotic RNA modification
intron intervening sequences
segment of RNA, specifically within a pre-mRNA transcript, that are removed during RNA splicing
don’t code for protein and are excised to produce a continuous coding sequence in mature mRNA
eukaryotic RNA modification
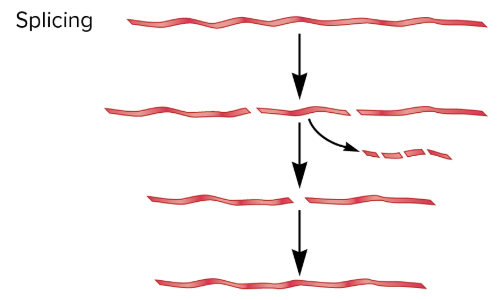
RNA splicing
process by which introns, non-coding segments of RNA within a pre-mRNA transcript, are removed, and the remaining exons, coding segments, are covalently joined together to form a continuous mature mRNA molecule
common in eukaryotes, occasionally occurs in bacteria
RNA modification
chopping
cleavage of a large rRNA or tRNA transcript into smaller pieces
one or more of the smaller pieces becomes a functional RNA molecule
transfer RNAs are also made as large precursors, and are cleaved at both the 5’ and 3’ ends to produce mature, functional tRNAs
prokaryotic and eukaryotic rRNAs and tRNAs
RNA modification

splicing
involves cleavage and joining of RNA molecules
RNA is cleaved at 2 sites, allowing an internal segment of RNA (intron) to be removed
after intron is removed, the two ends of the RNA are joined together
common among eukaryotic pre-mRNAs, occasionally with rRNAs, tRNAs, and a few bacterial RNAs
RNA modification
capping
attachment of a 7-methylguanosine cap to 5’ end of mRNA
cap plays a role in splicing of introns, mRNA export from nucleus, mRNA binding to ribosome
occurs on eukaryotic mRNAs
RNA modification
tailing
attachment of a string of adenine containing nucleotides to the 3’ end of mRNA at a site where the mRNA is cleaved
important for RNA stability and translation in eukaryotes
done on eukaryotic mRNAs and occasionally on bacterial RNAs
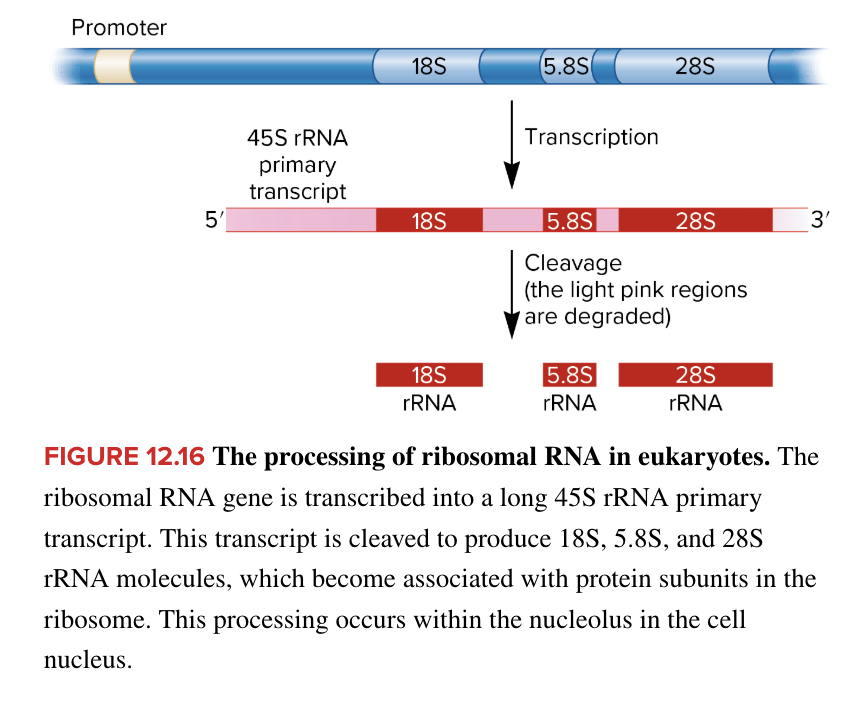
rRNA processing in eukaryotes
in eukaryotes, rRNA gene is transcribed into a long 45S rRNA primary transcript
transcript is cleaved to produce 18S, 5.8S, and 28S rRNA molecules which become associated with protein subunits to form functional ribosomes
occurs in nucleolus of cell nucleus
chopping (rRNA) RNA modification
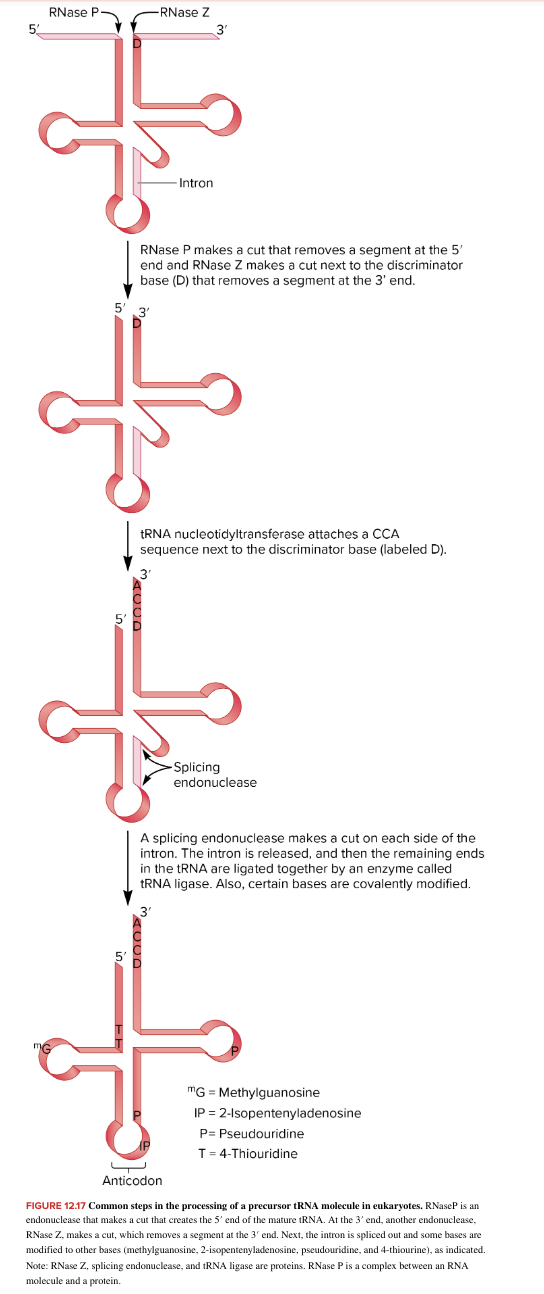
precursor tRNA processing
5’ End processing: RNaseP, an endonuclease, cleaves/cuts the precursor tRNA, creating the mature 5’ end
3’ End processing: RNase Z, another endonuclease, removes a segment from the 3’ end
Intron Removal: If present, an intron is spliced out.
Base Modifications: Some bases within the tRNA are modified to other bases, such as methylguanosine, 2-isopentenyladenosine, pseudouridine, and 4-thiouridine (convert from G,A,U,C to the ones listed)
note: RNase Z, splicing endonucleuase, tRNA ligase are proteins. RNase P is a complex btw an RNA molecule and a protein
chopping (rRNA) RNA modification (eukaryotes)
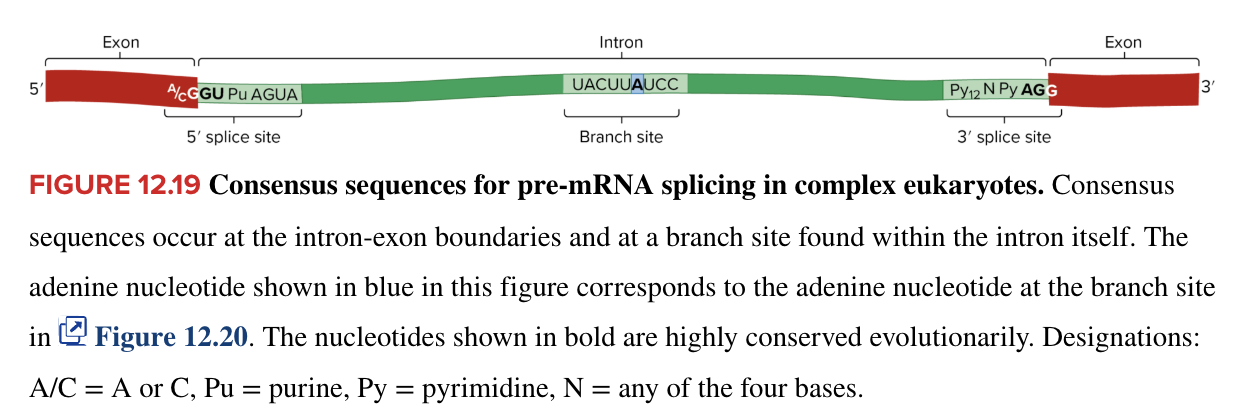
splicing
3 mechanisms have been ID’ed
all 3 cases involve
removal of the intron RNA from pre-mRNA transcript
intron RNA is defined by particular sequences within the intron and at the intron-exon boundaries
consensus sequences for pre-mRNA splicing in complex eukaryotes occur at the intron-exon boundaries and at a branch site within the intron itself
bases at these sites are highly conserved evolutionarily
covalent linkage of the exon RNA fragments to form mature mRNA molecule
eukaryotic RNA modification
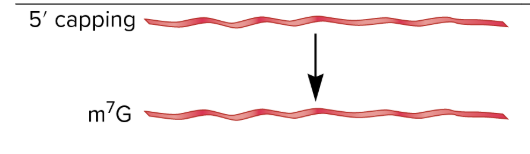
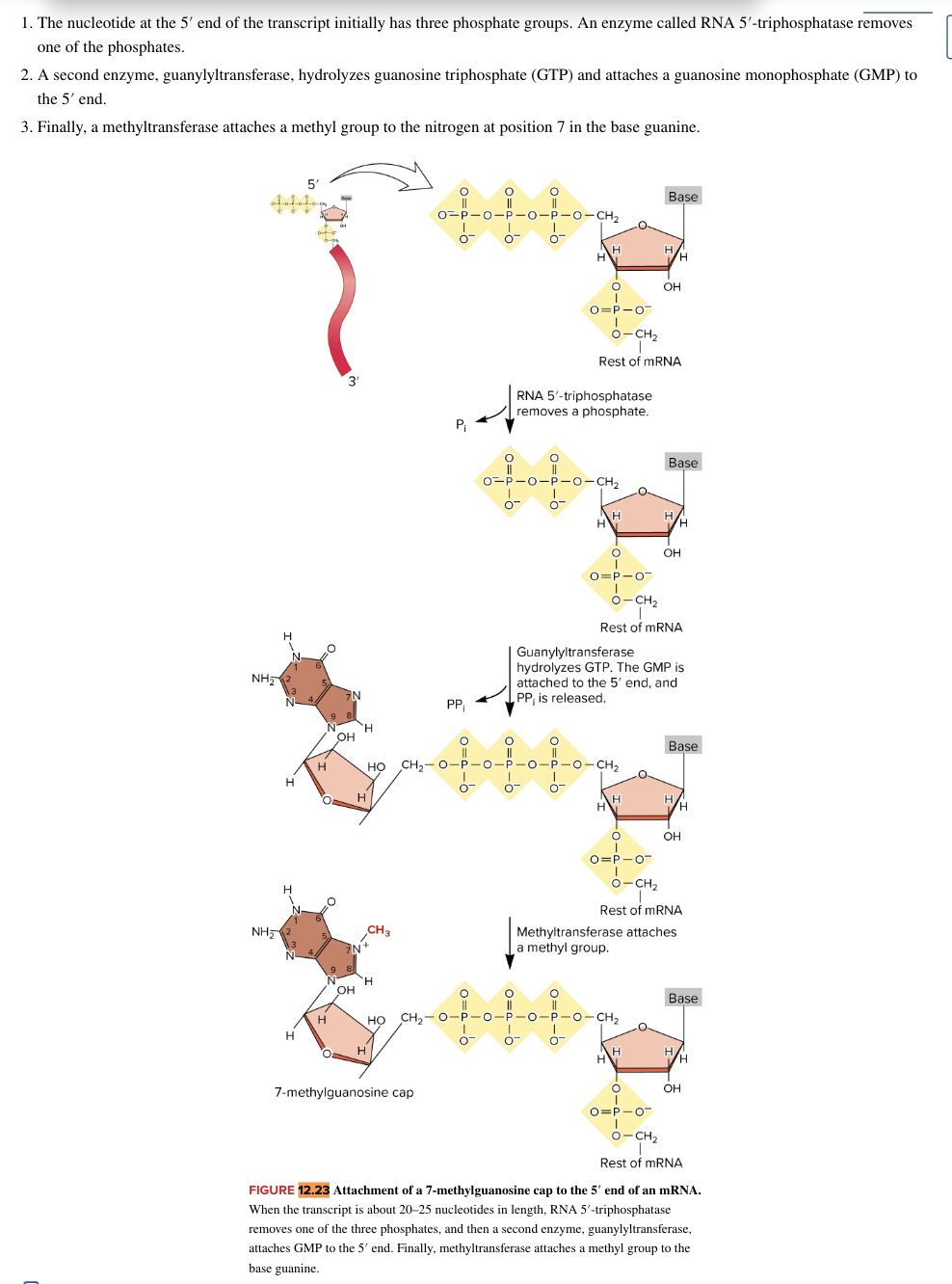
capping
most mature eukaryotic mRNAs have a 7 methyl guanosine covalently attached at their 5’ end
role of the cap
7 methylguanosine cap structure is recognized by cap-binding proteins
cap binding proteins play roles in
movement of some RNAs out of the nucleus
early stages of translation
splicing of introns
occurs as pre-mRNA is being synthesized by RNA pol II
usually when pre-mRNA transcript is only 20-25 bases long
3 step process
RNA 5’ triphosphatase removes one of the three phsophates
guanylyltransferase enzyme hydrolyzes GTP to GMP, which is attached to 5’ end
methyltransferase attaches a methyl group to the base guanine
eukaryotic RNA modification
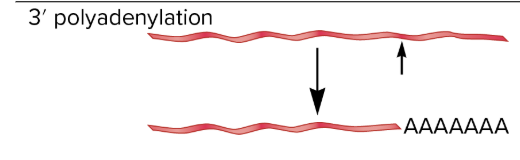
tailing/polyadenylation
most mature mRNAs have a string of adenine nucleotides at their 3’ ends
polyA tail
polyAtail is not coded in the gene sequence, but is added enzymatically after the gene is completely transcribed
an endonuclease cleaves pre-mRNA about 20 nucleotides downstream from 3’ end AAuAA sequence, making the pre-mRNA shorter at its 3’ end
poly A polymerase enzyme adds adenine nucleotides to the newly created 3’ end
eukaryotic RNA modification (although some bacterial RNAs are also polyadenylated but is degraded at degradosome)

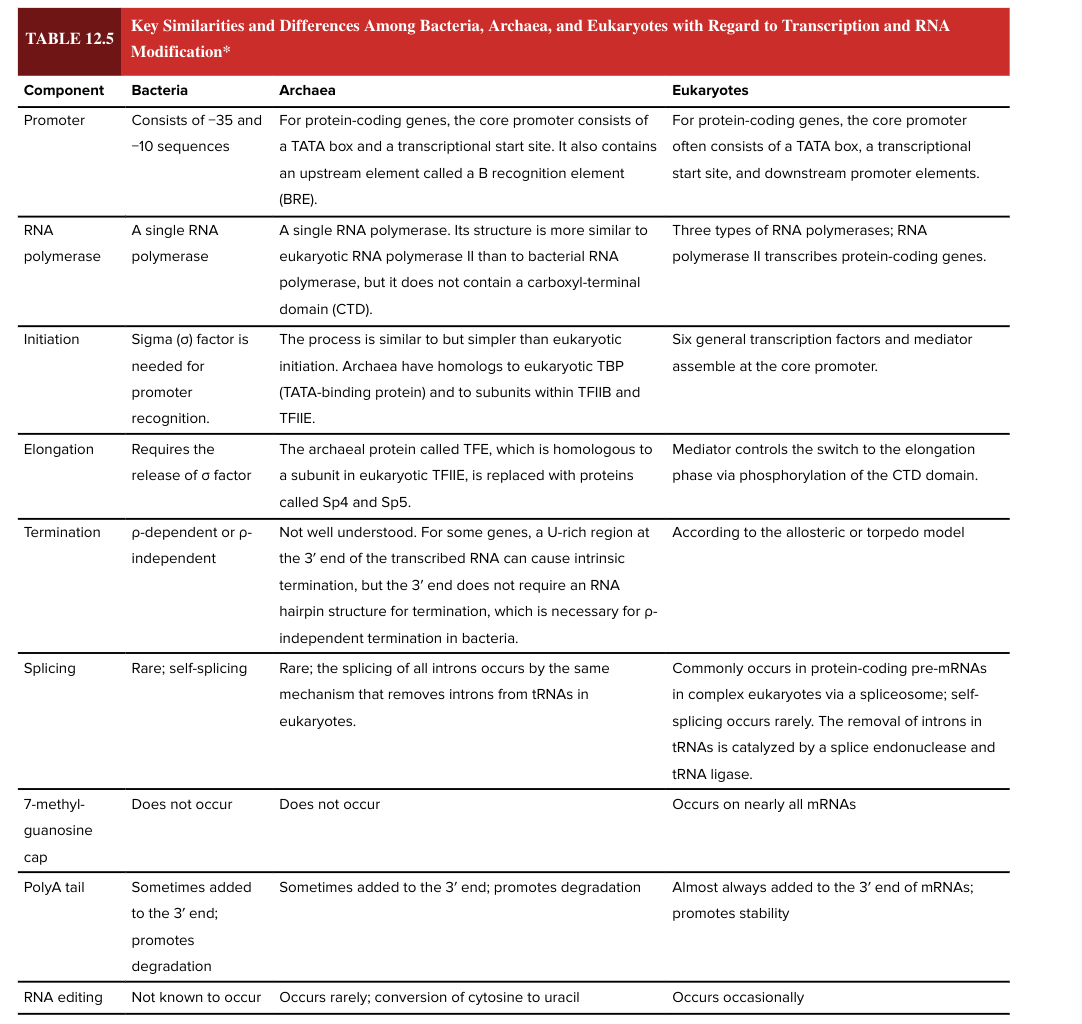
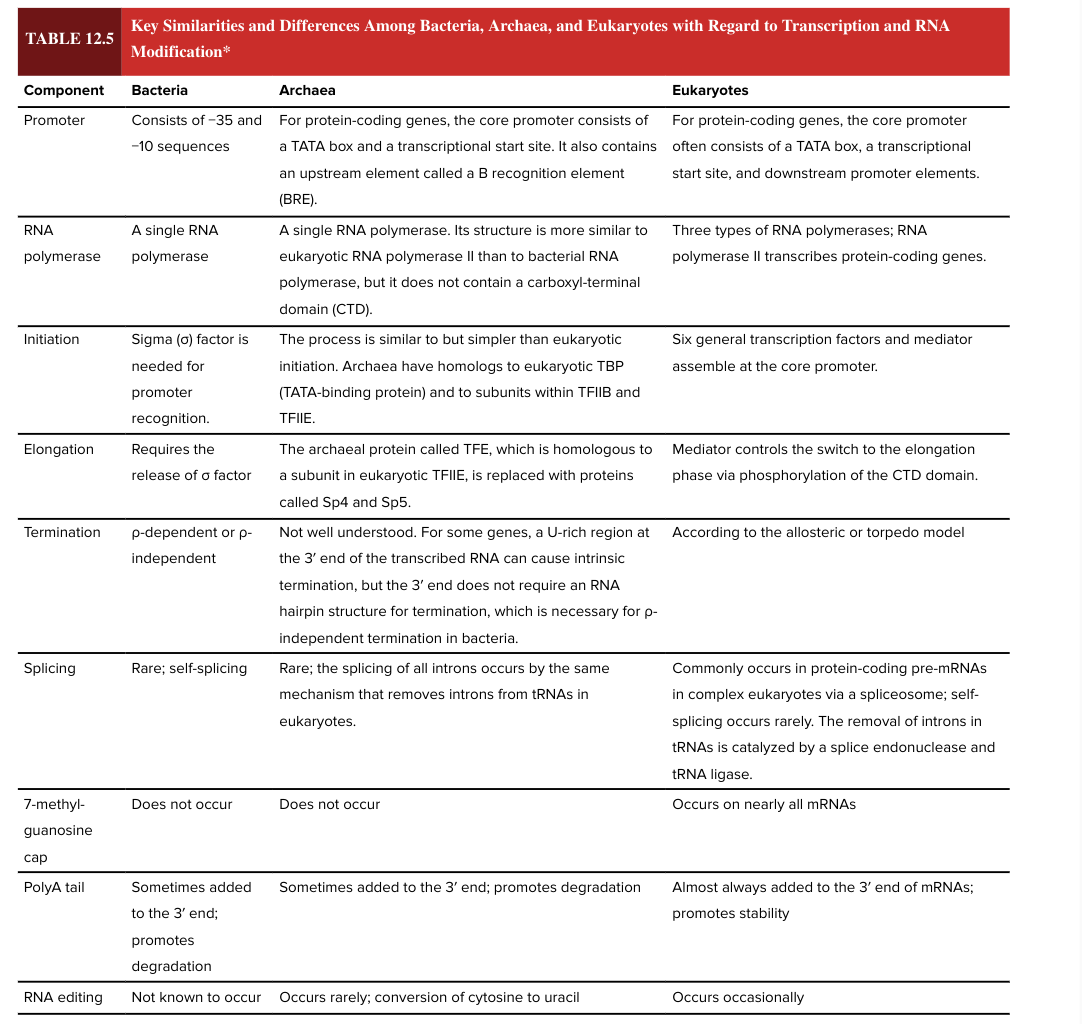
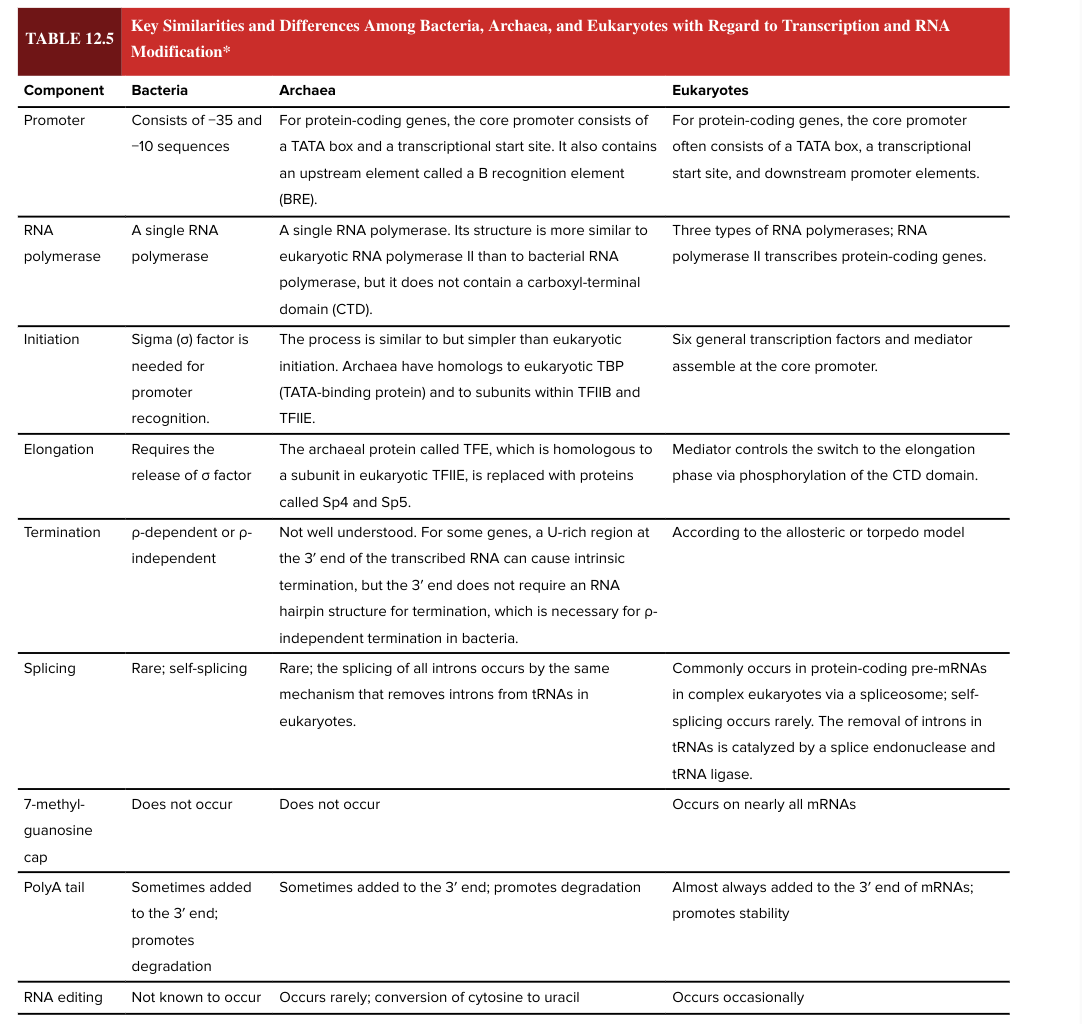
bacterial promoter
consists of -35 and -10 sequences
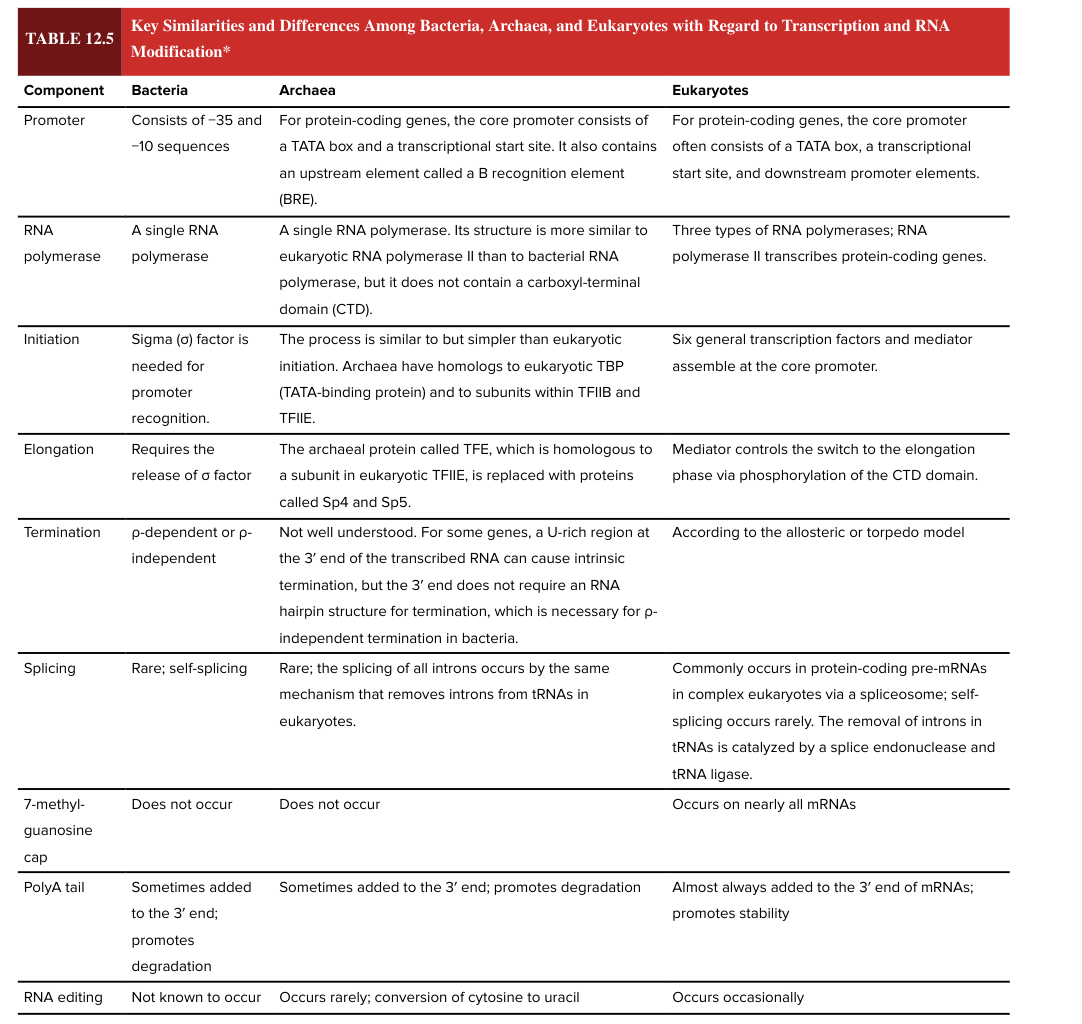
eukaryotic promotor
for protein coding genes, the core promoter often consists of a TATA box, a transcriptional start site, and downstream promoter elements
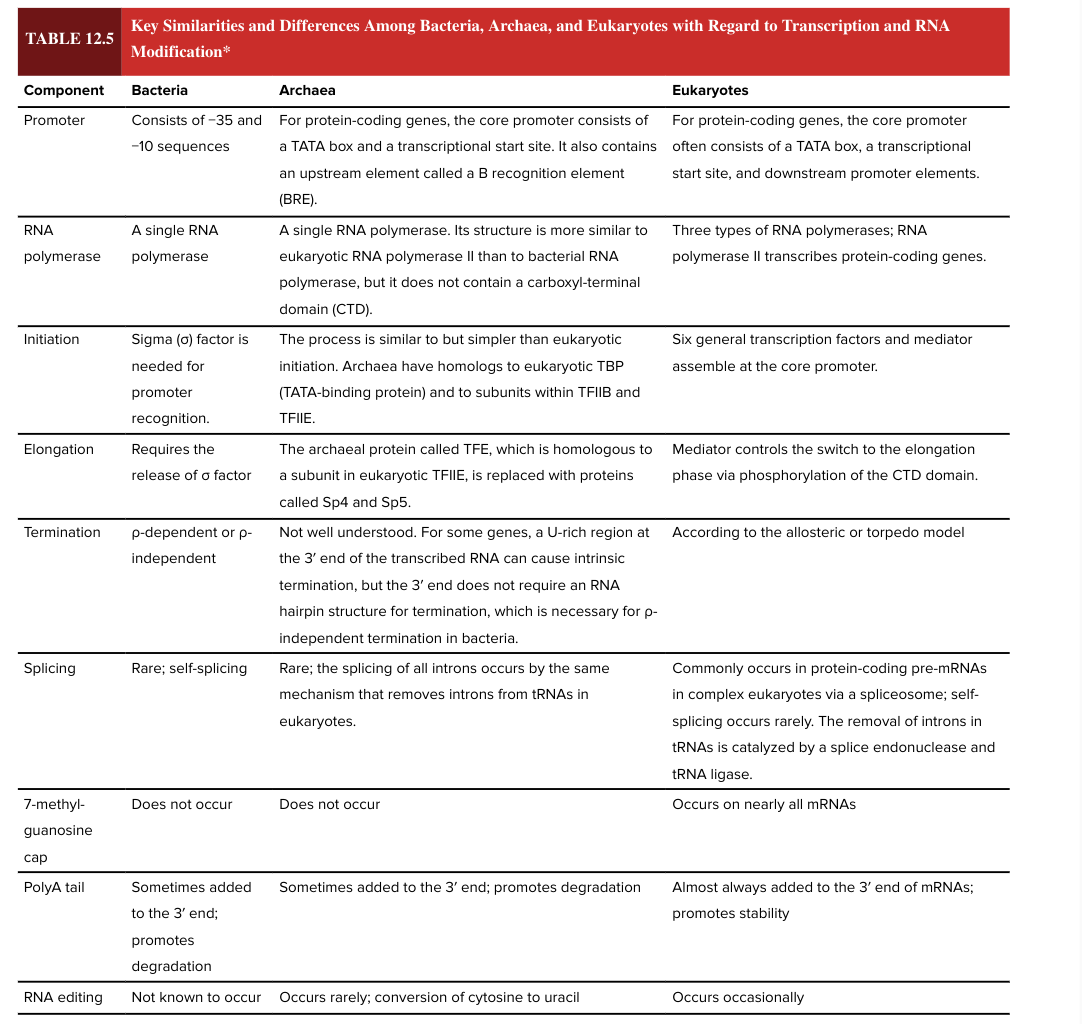
bacterial RNA polymerase
a single RNA polymerase
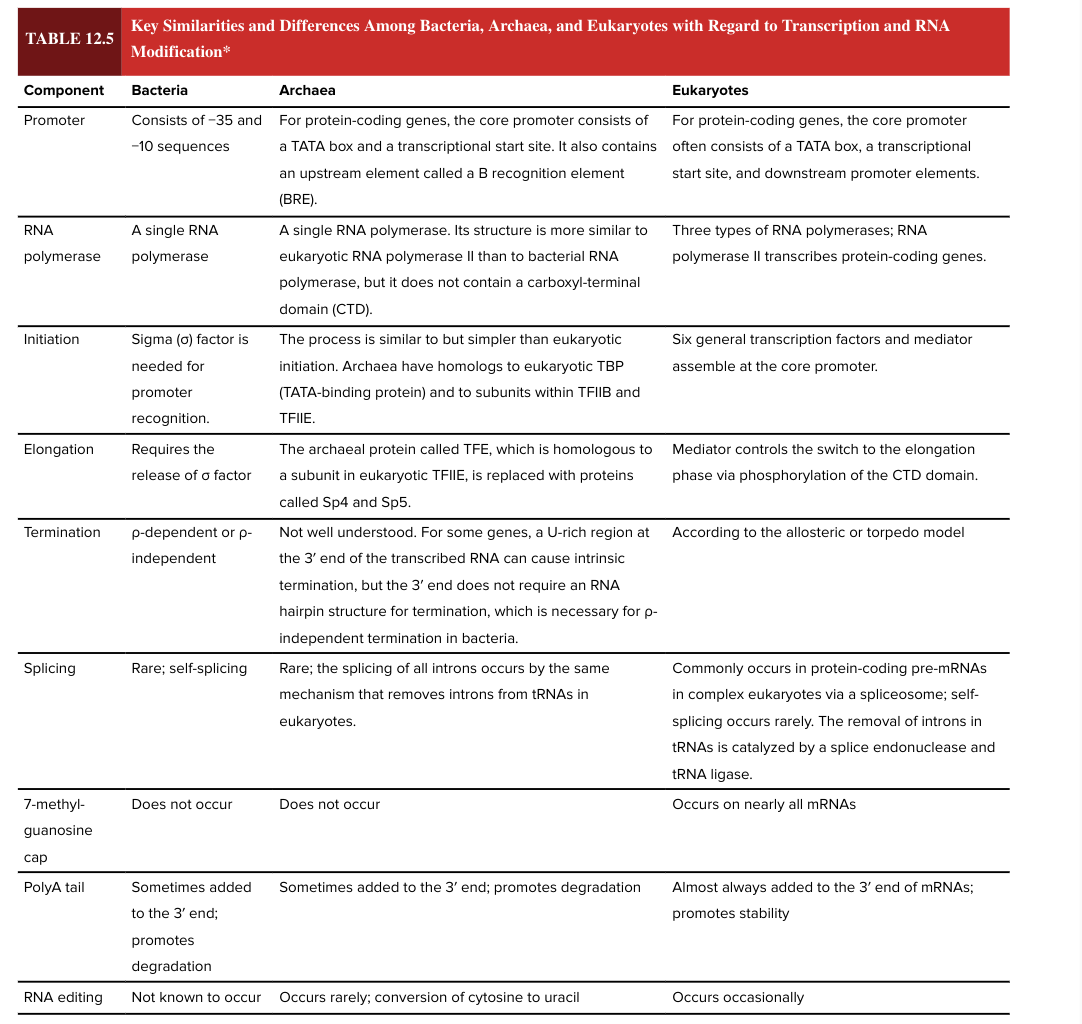
eukaryotic RNA polymerase
3 types
RNA pol II transcribes protein-coding genes
bacterial transcription initiation
sigma factor is needed for promoter recognition
eukaryotic transcription initiation
6 general transcription factors and mediator assemble at core promoter
bacterial transcription elongation
requires release of sigma factor
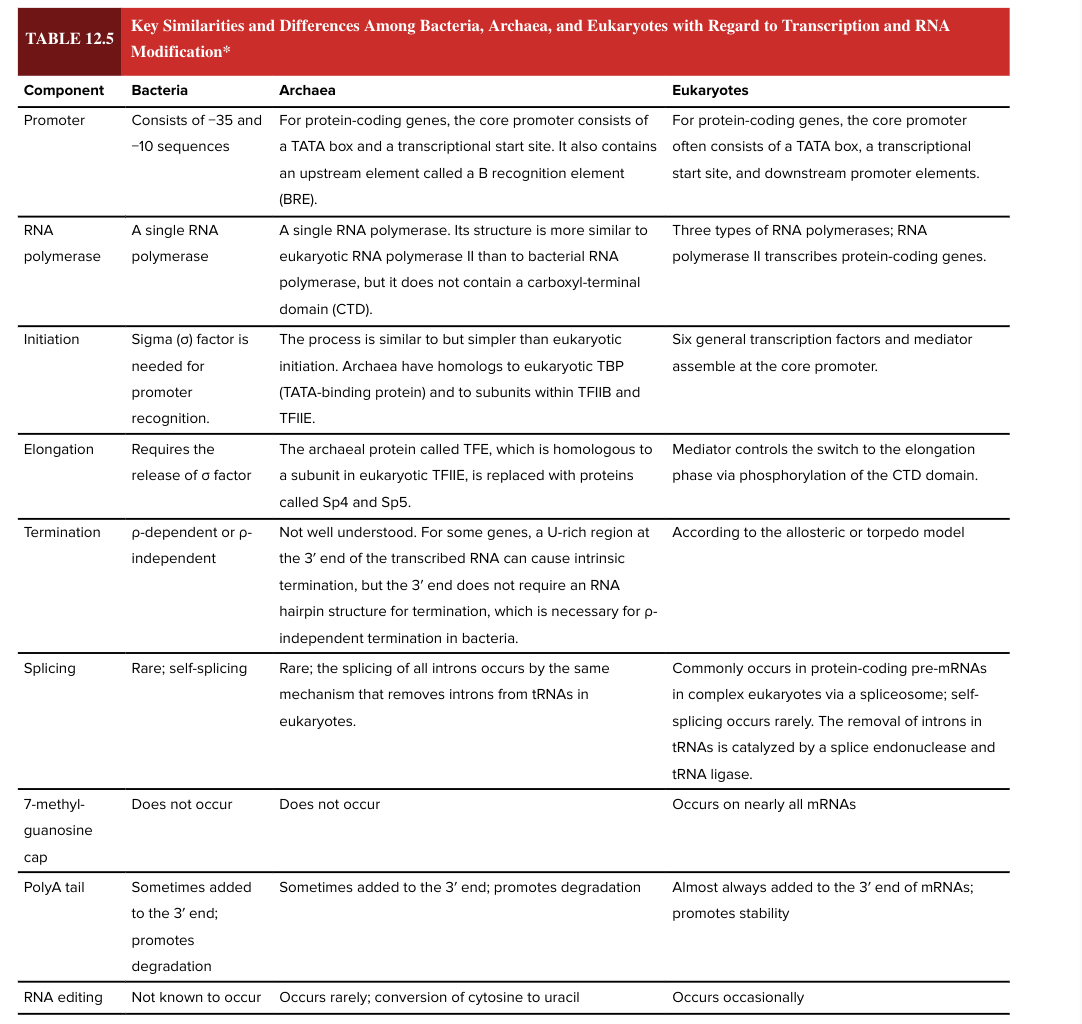
eukaryotic transcription elongation
mediator controls the switch to the elongation phase via phosphorylation of the CTD domain
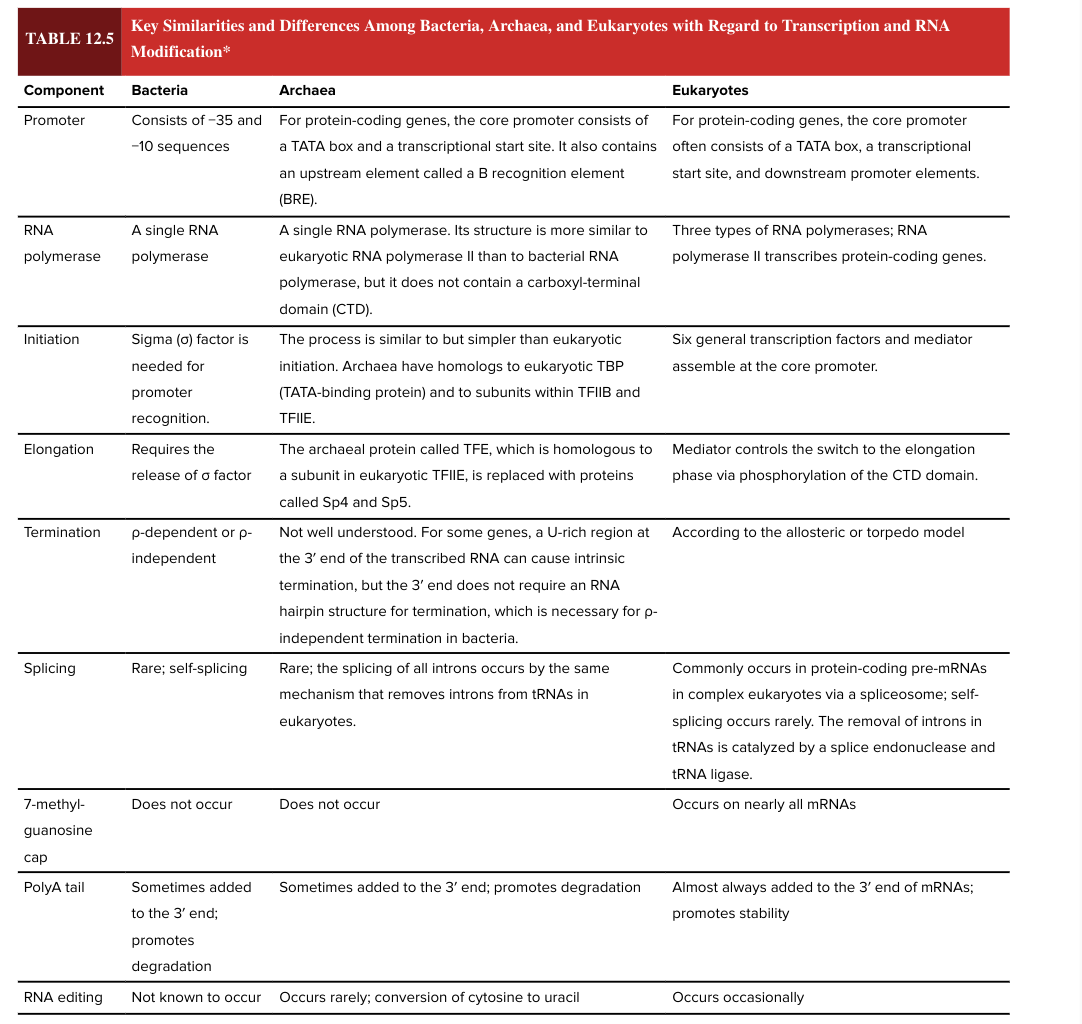
bacterial transcription termination
rho dependent or rho independent
eukaryotic transcription termination
according to the allosteric or torpedo model
bacterial splicing
rare; self-splicing
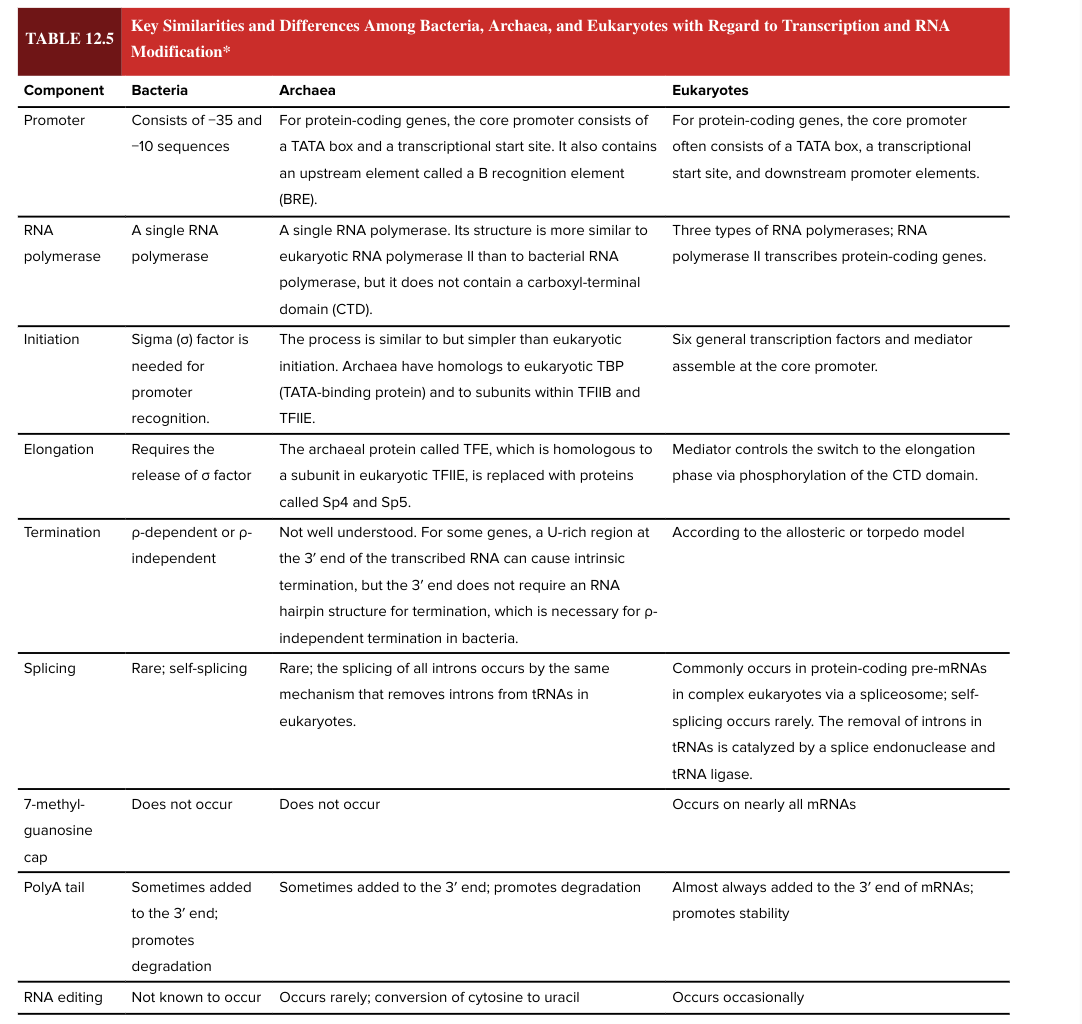
eukaryotic splicing
commonly occurs in protein-coding pre-mRNAs in complex ? via a spliceosome
self-splicing is rare
removal of introns in tRNAs is catalyzed by a splice endonuclease and tRNA ligase
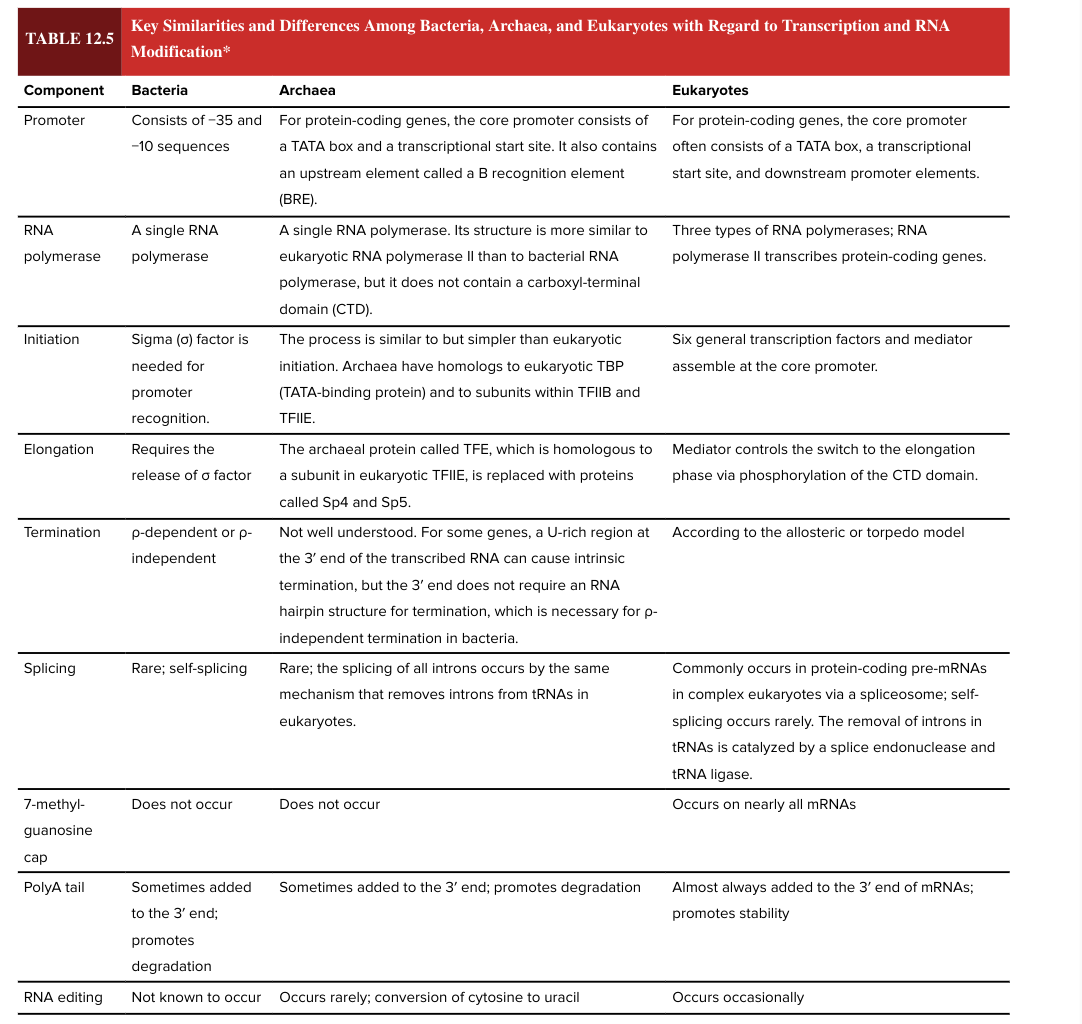
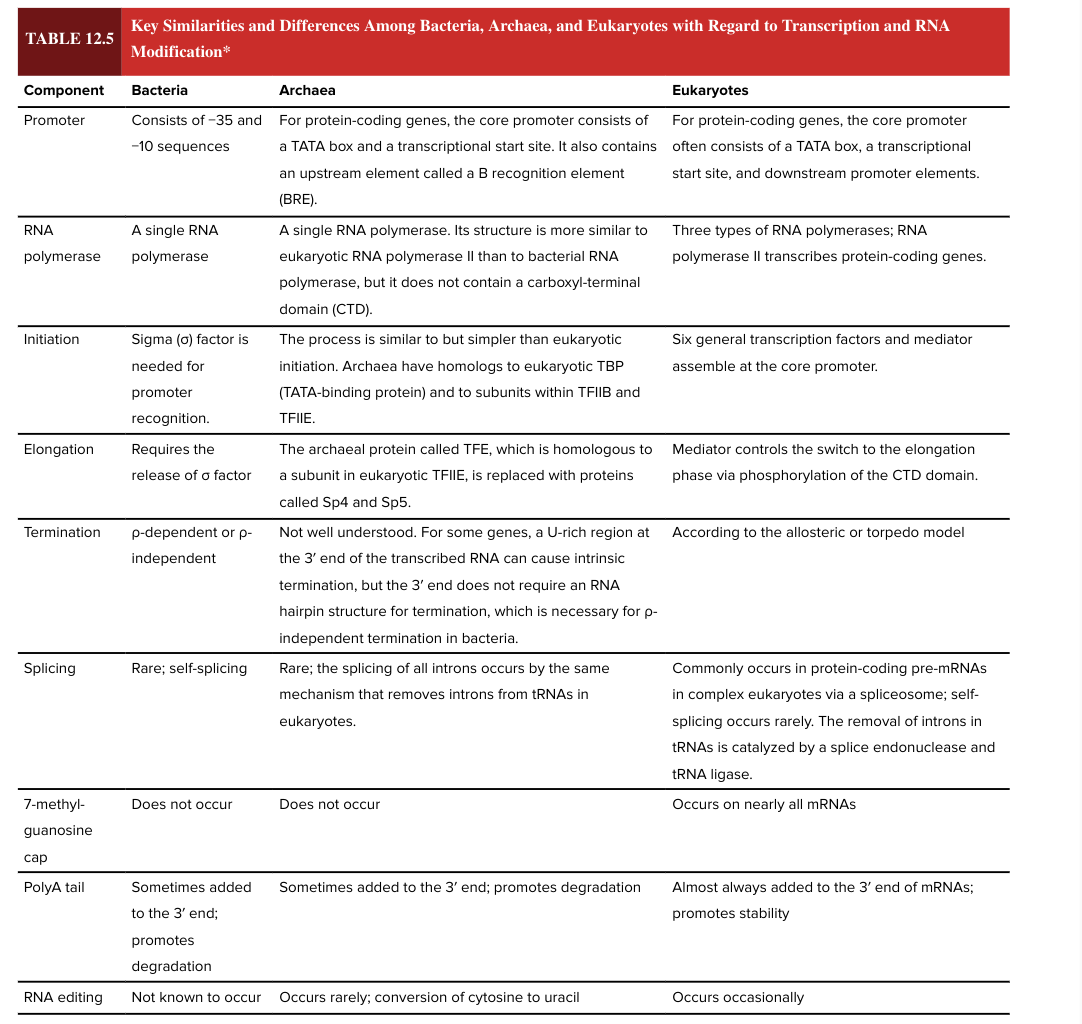
7 methylguanosine cap
occurs on nearly all mRNAs in eukaryotes
does NOT occur in bacteria
5’ end
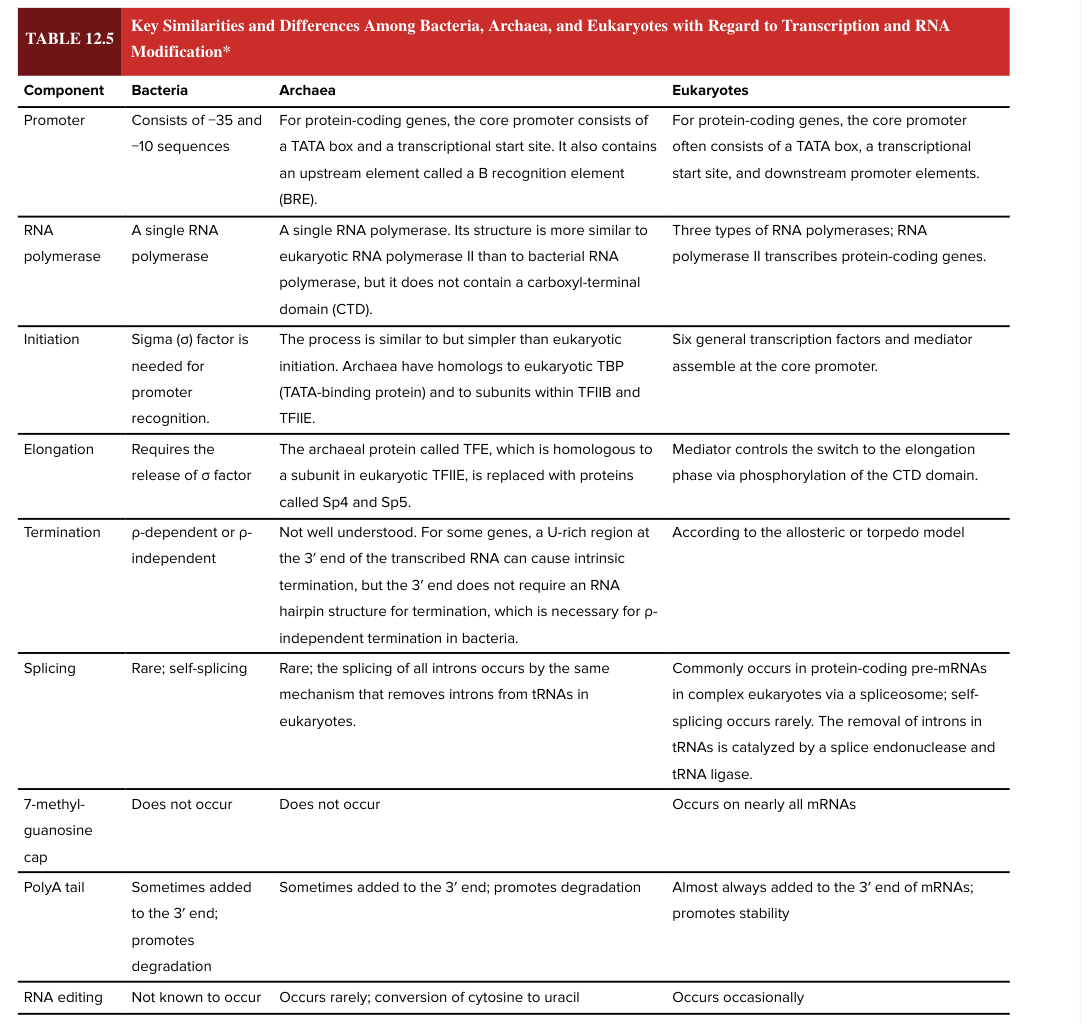
bacterial poly A tail
sometimes added to the 3’ end of the mRNA
promotes degradation (adding the poly A tail has opposite functional effect in bacteria vs eukaryotes)
eukaryotic poly A tail
almost always added to the 3’ end of mRNAs
promotes stability (adding the poly A tail has opposite functional effect in bacteria vs eukaryotes)
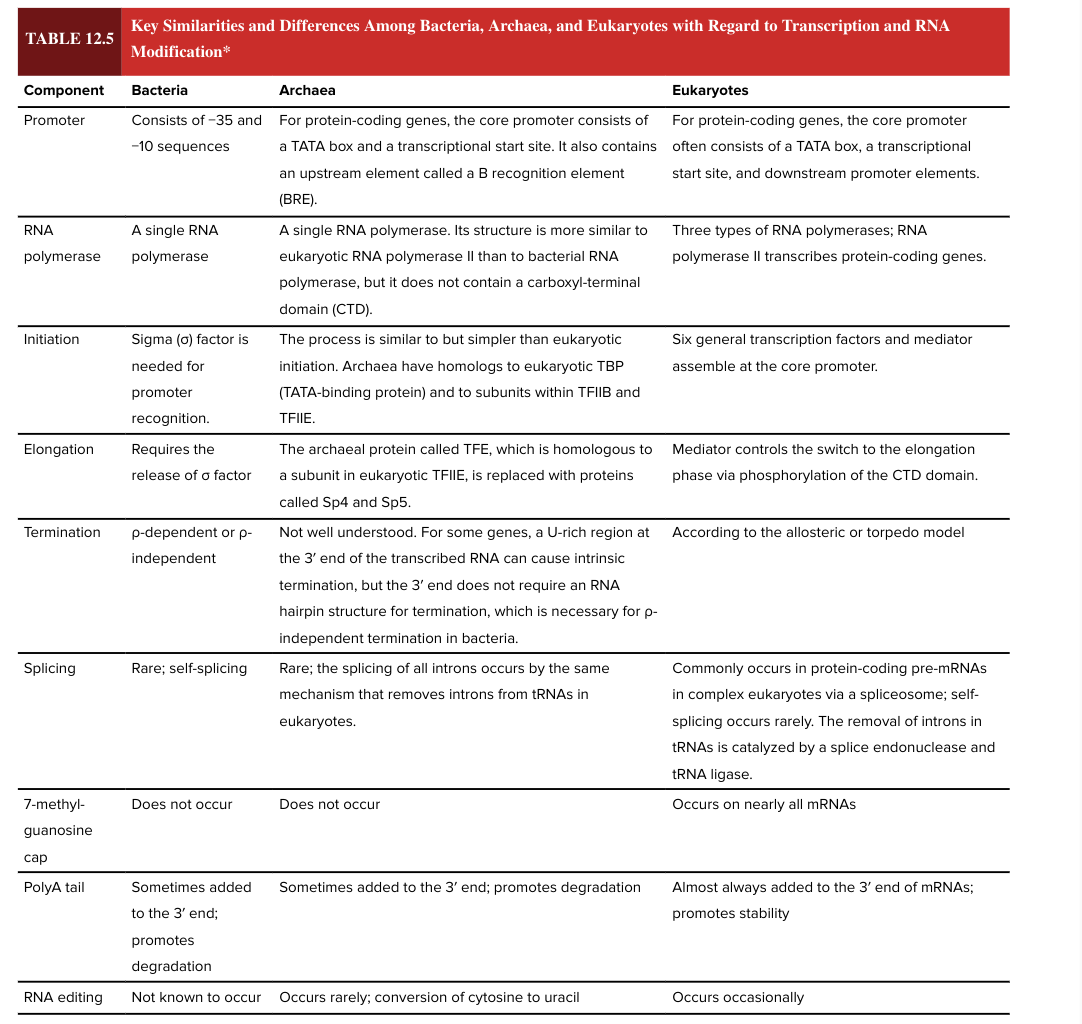
RNA editing
occurs occasionally in eukaryotes
not known to occur in bacteria