Systeembiologie: Genomics, Transcriptomics, and Proteomics
1/183
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
184 Terms
What is the primary goal of Systeembiologie?
To understand the entire picture of biological systems, including cells and organisms, by comprehending all components such as metabolites and proteins.
What is the order of biological omics fields in Systeembiologie?
Genomics → Transcriptomics → Proteomics → Metabolomics.
What does the genome consist of?
The genetic material of an organism, which can be DNA or RNA in viruses, composed of coding and non-coding material.
How can bacterial gene quantities vary?
Bacteria of the same species can have different amounts of genes.
What is a eukaryotic model organism mentioned in the notes?
Yeast cell, which has the first sequenced eukaryotic genome with 16 chromosomes.
What determines the reproduction of eukaryotic cells?
Whether the cells are diploid or haploid; two haploid cells can fuse to form a diploid cell.
What are Open Reading Frames (ORFs)?
Sequences in DNA that can potentially code for proteins, identified by searching for start and stop codons.
What are the start and stop codons in DNA?
Start codon: ATG; Stop codons: TAA, TAG, TGA.
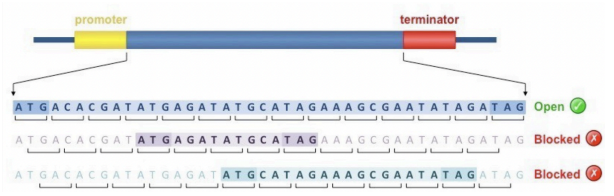
How many reading frames are there in total?
There are 6 reading frames: 3 on the forward strand and 3 on the reverse strand.
What does the systematic name YALo24C represent?
Y: Yeast, A: Chromosome 1, L: Left arm, o24: 24th open reading frame from the centromere, C: Crick strand (reverse strand).
What is Sanger sequencing?
A method used for sequencing small DNA fragments of about 1000 nucleotides, requiring plasmids or PCR.
What is a key advantage of Illumina sequencing?
It can produce 1 TB of DNA sequences in 6 days.
What is a disadvantage of Illumina sequencing?
It generates very small fragments of 35-450 nucleotides.
What is a notable feature of Pacific Bioscience sequencing?
It produces very long fragments up to 5000 nucleotides, making assembly easier.
What is a disadvantage of Pacific Bioscience sequencing?
It is very expensive.
What is Nanopore sequencing (MinION)?
A method that produces very long fragments (> 500 kb) but has relatively more sequencing errors.
What is shotgun sequencing?
A method where DNA is cut into small pieces, cloned into plasmids, and then sequenced.
What is the significance of coverage in genome sequencing?
The more times a genome is sequenced, the better the quality; typically, 20-40 times coverage is normal.
What types of sequences are difficult to assemble?
Repeats in DNA, such as rDNA, transposons, and viral sequences.
What is NOVO assembly?
A combination of short and long read sequencing methods to close gaps in DNA sequences.
What is the purpose of structural genome annotation?
To identify genes and their start-stop and intron-exon structure.
What is functional genome annotation?
The process of identifying the function of predicted genes.
Why is finding genes in eukaryotes more challenging than in prokaryotes?
Eukaryotic genomes are more complex, with introns and regulatory elements that complicate gene identification.
What is transcriptomics?
The analysis of the difference in mRNA gene expression at the transcriptional level.
What are introns and exons?
Introns are non-coding regions of DNA, while exons are coding regions that are expressed.
What are alternative start sites and alternative splicing?
Alternative start sites refer to different locations where transcription can begin, while alternative splicing allows for the generation of multiple mRNA variants from a single gene.
What are microORFs (μORFs)?
MicroORFs are small open reading frames that can encode peptides and are involved in regulating gene expression.
How is the relative level of gene expression calculated?
By measuring the amount of mRNA present under two different conditions.
What are three methods to analyze gene expression?
Affymetrix Microarrays, RNA sequencing, and design of experiment with data analysis.
What is Northern hybridization?
A technique for separating mRNA by size and charge, transferring it to a membrane, and hybridizing it with labeled DNA from a known gene.
What is quantitative reverse transcriptase PCR?
A method that converts mRNA to cDNA and then uses quantitative PCR with primers designed for known genes.
What are the types of gene arrays?
Macro-arrays and micro-arrays, with macro-arrays having DNA from different genes spotted on a filter and micro-arrays containing thousands of spots for individual genes.
What is RNA sequencing (RNAseq)?
A method that converts RNA to cDNA and sequences it, allowing for the discovery of new genes without prior genome sequence.
What is the principle of microarray technology?
It involves using complementary probes of specific genes on a microchip, where labeled cDNA from samples is hybridized to these probes.
What happens to cDNA that does not hybridize to oligonucleotide probes?
It indicates that the specific gene is not expressed in the sample.
What is hybridization in the context of microarrays?
The binding of cDNA to oligonucleotides due to complementary interactions, involving hydrogen bonds.
What are DNA arrays?
Microchips that contain oligonucleotide probes at each spot, designed based on genomic sequences.
What is the significance of mismatch probes in Affymetrix Microarrays?
They help identify false positives by comparing hybridization of perfect probes with mismatch probes.
How many genes can be tested using Affymetrix Microarrays?
Up to 500,000 genes can be tested.
What is the protocol for transcriptome analysis?
It includes sample preparation, RNA/mRNA isolation, labeling the sample, and purification.
Why is RNA purity critical in transcriptome analysis?
Impurities like cellular proteins and lipids can cause cDNA to bind to multiple probes, affecting results.
What labeling agent is mentioned for RNA/cDNA in the protocol?
Biotin is used for labeling in Affymetrix.
What is the purpose of purifying the sample in transcriptome analysis?
To remove unnecessary molecules that could interfere with the analysis.
What is the role of oligonucleotides in microarrays?
They are synthetic probes manufactured based on genomic sequences and predictions.
What is the primary advantage of RNA sequencing over Micro-Arrays?
RNA sequencing is less biased, does not require hybridizations, and can detect novel transcripts.
What are some systematic errors that can affect gene expression data?
- Poor RNA quality 2. Ineffective RNA labeling 3. Pipetting mistakes.
What are housekeeping genes and their role in gene expression studies?
Housekeeping genes are consistently expressed at the same level regardless of growth conditions, used to correct systematic errors in gene expression data.
How is the amount of RNA expressed quantified in RNA sequencing?
It is quantified as TPM-value (transcripts per Million transcripts).
What does Fold Change (FC) represent in gene expression analysis?
Fold Change is the average expression of a condition divided by the average expression of another condition.
What statistical test is used to determine the significance of Fold Change results?
A T-test, where a p-value < 0.01 indicates significant results.
What percentage of genes in a genome are functionally annotated?
About 30% of genes are functionally annotated, mostly inferred from computational analysis.
What is Gene Ontology (GO) and its purpose?
GO is a hierarchical classification system for proteins that provides a framework for functional annotation of genes.
What are the three main categories of Gene Ontology?
- Molecular function 2. Biological process 3. Cellular compartment.
What is the purpose of GO Enrichment analysis?
To interpret and analyze data from transcriptomics and proteomics experiments, identifying affected processes or classes of proteins.
Define metabolism in the context of living cells.
Metabolism is the total synthesis and breakdown of compounds within a living cell.
What are the two main types of metabolic processes?
- Catabolism (breaking down compounds) 2. Anabolism (building up compounds).
What is the first stage of cellular respiration?
Glycolysis, which breaks glucose into two pyruvate compounds.
What is the role of the pyruvate dehydrogenase complex in cellular respiration?
It converts pyruvate into Acetyl-CoA and CO2.
What is the function of the Citric Acid Cycle in cellular respiration?
It completes the breakdown of glucose to CO2.
What is the significance of RNA sequencing in detecting gene expression?
It allows for the detection of transcription start/termination sites, intron/exon boundaries, and splice variants.
Why is RNA sequencing preferred over Micro-Arrays in research?
It does not require genome sequence or good annotation, making it less biased.
What does it mean if a gene is inferred from computational analysis?
It means that the gene's function is predicted based on similarities with known genes from other organisms.
How can one protein belong to multiple GO terms?
Proteins can participate in various biological processes and molecular functions, allowing for multiple classifications.
What is the relationship between nodes in Gene Ontology?
Nodes are connected by directed edges that define hierarchical parent-child relationships.
What is the outcome of catabolic processes?
They break down large molecules into smaller ones, releasing energy.
What is the outcome of anabolic processes?
They build larger molecules from smaller ones, consuming energy in the process.
What is the maximum amount of ATP produced per one glucose molecule during glycolysis, TCA and oxidative phosphorylation?
About 36 ATP.
What is the end product of glycolysis?
2 pyruvate (C3) molecules.
Where does glycolysis occur in the cell?
In the cytoplasm.
What is the role of the pyruvate dehydrogenase complex?
It converts pyruvate (C3) into an acetyl group (C2) and CO2 (C1) in the mitochondrion.
How many ATP are produced from one pyruvate during the citric acid cycle?
15 ATP, resulting in 30 ATP per glucose.
What is the main function of oxidative phosphorylation?
To generate ATP by transferring electrons through the electron transport chain and reducing O2 to H2O.
What is produced as a result of the proton gradient created during oxidative phosphorylation?
ATP, as protons flow back through ATP synthase.
What process allows ATP production without oxygen?
Fermentation and anaerobic respiration.
What is the difference between aerobic and anaerobic conditions in glycolysis?
Glycolysis can produce ATP with or without O2.
What are the two types of fermentation mentioned?
Alcohol fermentation and lactate fermentation.
What does yeast produce when glucose levels are high?
Ethanol.
What is the diauxic shift in yeast?
When all glucose is consumed, yeast shifts to growing on ethanol.
What is the glyoxylate cycle?
A pathway similar to the TCA cycle, essential for growth on C2 carbon sources, involving isocitrate lyase and malate synthase.
What is a proteome?
The entire complement of proteins in a biological organism or system at a given time.
Why are there more proteins than genes in an organism?
Due to alternative splicing and posttranscriptional modifications.
What does the transcriptome measure?
mRNA levels and information carriers.
How can the transcriptome be amplified?
By PCR.
What is the main technique used for protein separation in proteome analysis?
2D gel electrophoresis.
What does the first dimension of 2D gel electrophoresis separate proteins by?
Their isoelectric point.
What does the second dimension of 2D gel electrophoresis separate proteins by?
Their molecular weight.
What is the isoelectric point (pI) of a protein?
The pH level at which the net charge of the protein is zero.
What is the role of SDS in SDS-PAGE?
To add a negative charge to proteins, ensuring they have the same charge density.
What does DIGE stand for and what is its purpose?
Difference Gel Electrophoresis, used to label protein samples with fluorophores for comparative analysis.
What is the purpose of electrophoresis in protein analysis?
To separate proteins from each other for identification.
What two specific features are used to separate proteins during electrophoresis?
Molecular weight and iso-electric point.
What does IEF stand for and how does it separate proteins?
Iso-Electric Focusing; it separates proteins according to their isoelectric point (pI).
What does SDS-PAGE stand for and what is its function?
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis; it separates proteins according to their length/size.
What are the two main methods of mass spectrometry (MS) mentioned?
Peptide Mass Fingerprinting and Tandem Mass Spectrometry (MS/MS).
What is the principle behind mass spectrometry?
It involves the ionization of target molecules in a vacuum and the accurate measurement of the mass of the resulting ions.
What are the stages of mass spectrometry?
- Ionization, 2. Acceleration, 3. Deflection, 4. Detection.
What happens during the ionization stage of mass spectrometry?
Target molecules are ionized by knocking off electrons, resulting in positive ions.
How are ions accelerated in mass spectrometry?
Positive ions are repelled from a positively charged ionization chamber and pass through slits to form a focused beam.
What determines the deflection of ions in mass spectrometry?
The mass of the ions and the number of positive charges they carry.
What is the output of mass spectrometry represented by?
X-axis: mass/charge ratio; Y-axis: abundance of ions.