U3 AOS1
1/155
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
156 Terms
what are the four key organic biomacromolecules?
carbohydrates
nucleic acids
lipids
proteins
what are nucleic acids?
They are polymers (long chains of repeating units/monomers, in this case nucleotides). These monomers are joined together in a condensation (water is released) polymerisation reaction to form a polymer.
They are acidic
Two types are DNA and RNA
Role is to store and transmit hereditary information. The sequence of nucleotides in the DNA encodes the instructions on the synthesis of proteins.
difference between structure of DNA and RNA
RNA is single stranded and DNA is double stranded.
They both have a helical shape, however DNA is a double helix and RNA is a single helix. naturally spirals in a right-handed twist.
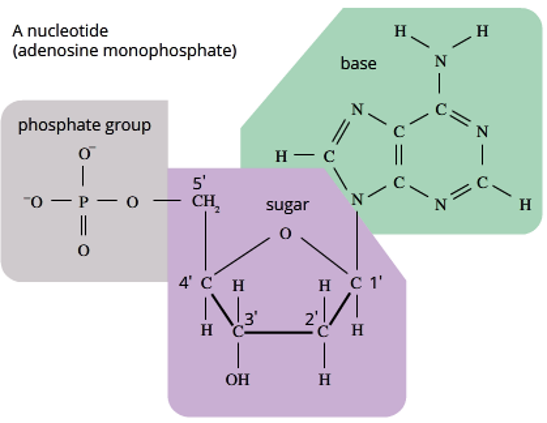
what does a nucleotide consist of?
-a phosphate group
-a five-carbon sugar (pentose)
-a nitrogenous base
difference between DNA and RNA nucleotides
DNA has a deoxyribose pentose sugar (missing oxygen) and RNA has a ribose pentose sugar (has an oxygen).
DNA has Thymine nitrogen base, whereas RNA has Uracil.
structure of nucleotides (P group and Sugar group)
The PHOSPHATE GROUP is made up of:
1 x Phosphorus atom (P)
4 x Oxygen atoms (O)
Can also have a Hydrogen atom making a hydroxyl group when the nucleotide is not yet part of polymer.
The SUGAR is a 5-Carbon Sugar and each Carbon is labelled with a number 1’, 2’, 3’, 4’, and 5’ = Number ‘Prime
1’ is always the carbon where the Nitrogenous base attached
3’ and 5’ are important in creating the Sugar-Phosphate backbone.
RNA has a hydroxyl (-OH) functional group at the 2’ position.
DNA has a hydrogen atom at the 2' position, as it lacks oxygen)
2 types of nitrogenous bases & complementary base pairing
Pyrimidines: (C, T and U) Single ring molecule
Purines: (A and G) Double ring molecule
Purines always bond with pyrimidines by forming hydrogen bonds.
A pairs with T/U (2 H bonds)
G pairs with C (3 H bonds)
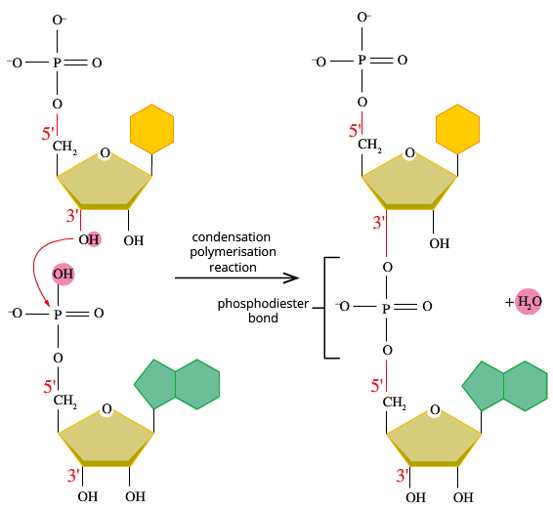
Joining nucleotides together (reaction & bond name, steps, final product)
-free nucleotides join through a condensation polymerisation reaction (water gets released)
-the bond formed between nucleotides is called a phosphodiester bond (between the sugar and phosphate of the nucleotides) There are oxygens connecting the nucleotides with a phosphorous atom in between.
STEPS:
1.The OH on the 3' carbon of pentose sugar bonds to where the OH on the phosphate group is on the next nucleotide
2.The H is lost from the OH on the 3’ Carbon and the OH on the Phosphate group is also lost
3.The lost H and OH join to form H2O, aka water
Once the nucleotides are joined together they form a sugar-phosphate backbone which has nitrogenous bases pointing out from it. If RNA, it will remain single stranded. If DNA< then it will bond with a complementary strand to become double stranded.
role of DNA & location in prokaryotes vs eukaryotes
DNA contains the instructions to make RNA (tRNA, rRNA, mRNA)
In eukaryotic organisms: DNA makes linear chromosomes, which are primarily found in the nucleus. However, mitochondria and chloroplasts also contain their own small amount of DNA, but these are circular.
In prokaryotic organisms: DNA makes circular chromosomes, just found in the cytosol as here are no organelles. Can also make plasmids, which are even smaller bits of circular DNA.
strands in relation to 5’ to 3’
these are the ends of the polynucleotide strand. 5’ is a free phosphate. 3’ is a free hydroxyl group.
a strand runs 5’ to 3’ direction, as nucleotides can only be added on the 3’ end.
the two complementary strands of DNA are antiparallel, running opposite to one another.
further DNA coiling (into chromatins)
To help package the DNA into the nucleus of eukaryotic cells, the DNA goes through additional coiling.
The DNA double helix wraps around proteins called Histones, this forms a Nucleosome.
The Nucleosomes then coil up to form a chromatin fiber.
The chromatin fibers then coil up and condense further to form a condensed chromosome (this last part will only happen at the start of mitosis and meiosis).
types of RNA
There are 3 main types of RNA:
-mRNA – messenger RNA
-tRNA – transfer RNA
-rRNA – ribosomal RNA
All of these types of RNA are produced (in the nucleus of eukaryotes) using the appropriate section of DNA as a template.
RNA is found in the nucleus and cytoplasm of Eukaryotes and the cytoplasm of Prokaryotes
mRNA
In the nucleus, the desired DNA sequence is transcribed into mRNA, so a portable copy of the DNA instructions for protein synthesis can exit the nucleus.
The mRNA carries the genetic message to ribosomes where it is translated into a particular protein.
tRNA
segment of RNA that has folded in on into into a 3D shape.
used to transport individual amino acids to the ribosome during protein synthesis, which are attached at the top.
the bottom section is the anticodon. tRNA molecules will carry different amino acids based on their 3-letter sequence at the anticodon.
rRNA
forms the ribosome (site of protein synthesis) by joining together with specific proteins.
primary function is in protein synthesis, as it binds to mRNA and tRNA to ensure that the codon sequence of the mRNA is translated accurately into amino acid sequence in proteins.
what is the genetic code? universal & degenerate
set of rules used by cells to translate the information encoded in nucleic acids. considered both universal and degenerate.
universal: all living things read the sequence of nitrogenous bases and interpret them the same way. meaning that all 3 base sequences will be interpreted for the same amino acid.
degenerate: there are multiple 3 letter combinations that code for the same amino acid. this redundancy is called degeneracy when talking about the genetic code.
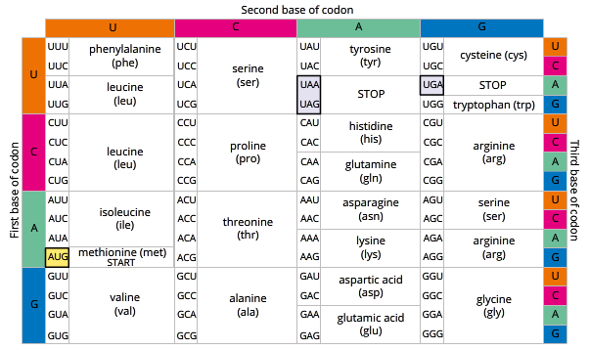
reading the genetic code
to read the genetic code the sequence of bases is grouped into 3 letter sets.
DNA - generally called triplets. mRNA- usually called codons.
each triplet/codon will then code for a specific amino acid.
4 codons that code for something other than an amino acid:
AUG – Start Codon (which codes for methionine after starting)
UAA, UAG & UGA – Stop Codons
levels of genetic code
-At it’s most basic level the Genetic Code is a sequence of nitrogenous bases in a nucleic acid.
-A Gene is a section of a nucleic acid where the sequence of bases contains the instructions for making a protein.
-A Chromosome is a whole nucleic acid molecule and can have many genes along the length of the molecule.
-A Genome is is the complete set of genes or genetic material of an individual cell or organism.
-The Proteome is the complete set of proteins expressed/ produced by the genome, but individual cells differ in their proteomes. by a single cell or organism in a particular environment.
The study of the structure, function and interaction of proteins of the proteome is called Proteomics
what is a gene?
a segment of DNA (a discrete locus) where the sequence of nucleotides code for a gene product which affect an organism’s treats by being expressed as a functional product or by regulation of gene expression.
typical gene is tens of thousands bases long
either regulatory or structural
types of genes
structural gene: codes for a specific protein that becomes part of the structure and functioning of an organism (eg. codes for enzymes, protein channels, rRNA, structural proteins). Code for proteins and RNA that are not involved in gene regulation, so they never code for a regulatory factor - protein.
regulatory gene: codes for transcription factors, which control transcription and hence the expression/action of a target gene. these can either be activators, increasing expression , turning the gene on. or repressors, reducing expression and turning the gene off. these regulatory genes may only affect some genes in certain cells, at a certain stage of development, under certain conditions. but it means that the gene is able to control the expression of one or more other genes.
regulated gene: genes which are controlled by transcription factors. eg. could be structural genes that are being controlled by regulatory genes. Means their expression can vary depending on the presence or absence of transcription factor.
constitutive gene: “house keeping genes” they are transcribed continually, and are always turned on.
2 main sections of a gene (regions)
the flanking regions: non-coding regions on either side of a coding region.
the coding region: contain the information for making a protein
up / down stream regions
flanking before coding region - upstream region (near 5’):
segment of DNA that hormones attach to. contains the DNA code with the instructions where to begin the transcription process.
flanking after coding region - downstream region (near 3’):
contains the DNA code with the instructions where to Stop the transcription process.
promoter
located in the upstream region. found in both eukaryotes and prokaryotes.
it identifies the DNA strand to be transcribed and is where RNA polymerase binds for transcription. it promotes transcription of the sequence after it and also identifies the direction of transcription. in eukaryotes it is usually the sequence TATAAA (aka TATA box)
can also contain regulatory regions for gene expression.
operator
located upstream only in prokaryotes, after the promoter.
operator comes before a gene, and is where repressor proteins can bind to so that transcription of the following gene is stopped and prevented from being expressed.
if the repressor is inactivated, RNA polymerase can transcribe the genes.
start and stop codons
AUG is always the star codon and codes for the amino acid methionine after
UAA, UAG and UGA are all stop codons. They don’t code for amino acids and when read, terminate peptide synthesis.
coding region (of eukaryotes)
includes both exons and introns in its DNA sequence.
exons- are expressed by the cell and will be both transcribed and translated. they code for the synthesis of a protein.
introns- are interruptions. are non-coding regions which are transcribed but spliced/cut out before translation. they therefore, do not contribute to the synthesis of a protein.
general steps of gene expression (polypeptide synthesis)
A gene is expressed when its polypeptide is synthesised, converted into a protein and the protein is fully functional.
For a gene to be expressed it must go through 3 processes:
1 Transcription [DNA to pre-mRNA] “same language (nucleic acid)”
2 RNA processing [pre-mRNA to mRNA]
Post-transcriptional Modification
3 Translation [mRNA to Polypeptide/Protein] “different language (protein)”
DNA coding vs template strand
coding strand: contains genetic instructions for the desired protein. needs to be read in the 5’ to 3’ direction. cannot be copied directly as the mRNA would contain the complementary base sequence and not the original one.
template strand: is the non-coding sequence. Is copied in mRNA, and will be the same sequence as the coding strand as the template is complementary to the coding strand. Is transcribed in the 3’ to 5’ direction so it will produce an mRNA strand with the correct 5’ to 3’ direction.
transcription (what is it, the 3 phases)
process by which the information in the coding strand of DNA is copied into a new molecule of pre-mRNA by transcribing the template strand, uses RNA polymerase as the main transcription enzyme.
1 Initiation: RNA polymerase binds to the promoter sequence and unwinds and unzips the DNA (with the help of helicase)
2 Elongation: RNA polymerase reads and synthesis a complementary pre-mRNA strand from the DNA template according to base pairing rules. (U complimenting A instead of T)
3 Termination: RNA polymerase stops transcription when it reaches the stop codon. It then detaches, the pre-mRNA gets released and the DNA reforms.
mRNA editing / RNA processing (3 steps)
occurs after the primary transcript is formed (pre-mRNA), which needs to be processed before becoming the final mature mRNA that is sent to the ribosome for protein synthesis.
steps:
1- addition of a 5’ cap on the 5’ end (protects mRNA from degradation, and is recognised by initiation factors involved in proteins synthesis to help initiate translation by ribosomes)
2 - addition of a poly-A tail on the 3’ end (a string of approx. 200 A nucleotides, protects mRNA from degradation)
3 - splicing (removal) of the introns, so that only the coding (exon) sections are left for the final mRNA
IMPORTANT NOTE
mRNA Editing/RNA Processing ONLY occurs in Eukaryotes. Prokaryotic genes don’t have introns and the mRNA generally doesn’t need a 5’ cap or Poly-A tail added.
alternative splicing
in this splicing process, particular exons of a gene may be included or excluded from the final mRNA produced, meaning the exons are joined in different combinations. this leads to different mRNA strands.
this process during gene expression allows a single gene to code for multiple proteins with different amino acid sequences.
translation (what is it, the 3 phases)
process by which the mRNA strand is translated into the encoded sequence of amino acids to form a polypeptide. it occurs outside the nucleus at a ribosome, either free in the cytoplasm or attached to the rough ER.
1 Initiation: Ribosome attaches to the 5’ end of the mature mRNA and starts to read at the start codon (AUG)
2 Elongation: tRNA carries a specific amino acid to the ribosome and has an anticodon that is complimentary to the codon on the mRNA strand being read. The amino acid it is carrying is joined by a peptide bond to other amino acids. Amino acids are added sequentially as the ribosome moves along the mRNA reading each codon to make up the polypeptide chain.
3 Termination: ribosome stops reading the mRNA at a stop codon, and the polypeptide is then released, ready for post-translation modification.
post-translational modification
The human genome comprises about 25,000 genes. The proteome comprises more than 1,000,000 proteins. Therefore, on average, each gene contains the coded instructions for making 40 proteins or more! This is possible because of post-translational modification.
Once a polypeptide is synthesised, it may be: • methylated (CH3 – added). • acetylated (CH3CO – added). • phosphorylated (phosphate added). • glycosylated (sugar added). • lipidated (lipid added).
Each of these not only adds compounds to the protein, but can affect protein folding.
In addition: • proteases may cut the polypeptide into several smaller polypeptides. • chaperone proteins fold the polypeptide. • polypeptides may also be combined with other polypeptides into a functional protein with a quaternary structure
what is gene regulation? why do we need it?
the regulation of gene expression (usually of protein synthesis) so that only the right amount of proteins are synthesised by a given cell.
gene regulation can occur at many points along the process of transcription, RNA processing and translation.
if you don’t regulate genes, they’d be active in every cell all the time. meaning cells would be expressing things they don’t need to, at times they don’t need to be expressed. wasting energy and resources, and there would be no specialised cells.
turning genes on & off
-genes are turned on and off in different patterns for different cell types during development, which is what differentiates cells. (eg. brain cell from liver cell)
-some genes are active your whole life, some only for a certain period of time.
-each cell expresses or turns on only a fraction of its genes, depending on that cells function
-some genes do not express themselves in a person’s phenotype until adulthood eg. such as in Huntington’s disease
benefits of gene regulation
allows cells to react quickly to changes in their environments, as they can change what genes are being expressed in response to environmental change.
be controlling the level of transcription, cells can conserve necessary energy & resources early on. determining the amount of protein product made by a gene at any given time.
transcription factors
proteins that help turn specific genes “on” or “off” by binding to nearby DNA
Groups of transcription factor binding sites called enhancers and silencers can turn a gene on/off in specific parts of the body.
Activators boost a gene's transcription. (attach to enhancer)
Repressors decrease transcription. (attach to silencer)
“switched on” genes (differ between..?)
are active and are producing mRNA.
differ between:
cells of different tissues
of same tissues at diff stages of development
cells under diff environmental conditions (eg. drug exposure)
abnormal cells (eg.. cancerous cells)
what are operons
are mainly in prokaryotes only
the grouping of genes that code for proteins/enzymes in the same biochemical pathway, meaning they are located next to each other on the chromosome. it also includes the regulatory DNA sequences for that set of genes (promoter and operator.)
operon is controlled by a regulator gene far upstream, which produces a small protein molecule called a repressor. The repressor binds to the operator gene and prevents it from initiating the synthesis of the protein called for by the operon.
trp operon
tryptophan operon - found in E.coli bacteria
when there isn’t enough tryptophan in the environment, this pathway allow the bacteria to produce tryptophan itself (an amino acid)
The trp operon is regulated by the trp repressor. When bound to trp, the trp repressor blocks expression of the operon by binding to the operator. When not bound, operon is expressed, as the repressor is unable to bind.
This acts as a negative feedback, as once the Tryptophan levels increase enough, they will activate the trp repressor protein and “Switch Off” the trp Operon.
(turned "on") when tryptophan levels are low and (turned "off") when they are high.
trp operon - structure
1- A promoter (RNA polymerase binding site)
2- An operator (binding site for a repressor protein). Tryptophan acts as a co-repressor when it binds to the trp repressor to make it active.
3- five genes that encode enzymes needed for tryptophan biosynthesis (runs trp E - trp A)
Together these 5 enzymes made by these 5 genes will go on to produce Tryptophan, thus increasing the available levels of Tryptophan in the bacterial cell.
high tryptophan levels
the tryptophan present will bind to the trp repressor protein, creating a change in shape so that it is now able to attach to the trp operator. (active repressor protein) Once attached it acts as a physical block, stopping RNA polymerase from being able to move along the DNA and transcribe the genes.
low tryptophan levels
there is little to no tryptophan that can bind to the trp repressor protein. without tryptophan binding, it becomes inactive. meaning RNA polymerase is free to move along the DNA and transcribe the trp genes. The genes then get transcribed and translated, into tryptophan.
what is regulation via attenuation?
another way the trp operon can be regulated, and happens when tryptophan levels are high. This occurs when there are no free trp in the cell, but there is still some attached to tRNA molecules. (meaning there is no reason ot make more because it’s a waste of energy)
-attentuation prevents transcription from being completed properly, so you never end up with your final proteins.
(repressor proteins regulate gene expression by stopping any transcription)
leader section and attenuator region
leader section is the untranslated region that comes after the operator.
the attenuator region comes after the leader section. it can stick to itself forming different structures that can then affect transcription & translation. What structure the “attenuator” region forms is different depending on the speed of translation, and how quickly mRNA moves through the ribosome. 3 & 4 if quickly. 2 & 3 if slower.
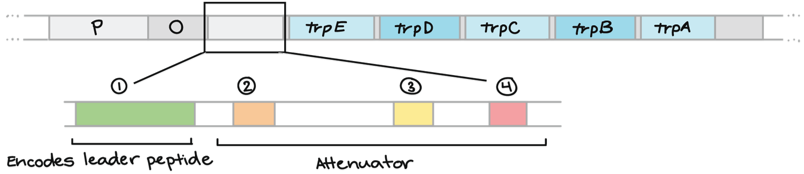
attenuation process in detail
in prokaryotic cells, ribosomes don’t have to wait for the mRNA to finish being transcribed before it begins to translate it (as there are no organelles separating the ribosome from the DNA)
As the ribosome starts translating the “leader” part of the polypeptide, it requires needs several tryptophan amino acids to get started.
high trp: ribosome moves quickly along the leader as many tryptophan-carrying tRNA are available. the attenuator region forms a terminator hairpin, which make RNA polymerase detach and ends transcription.
low trp: ribosome will stall at the trp codons (waiting for trp-carrying tRNA) = slower translation. non-terminator hairpin forms, which prevents formation of the terminator and transcription of the trp operon continues.
what do proteins do?
DNA codes for a variety of proteins, which carry out a wide range of functions. Such as growth and repair, metabolic processes etc. Nearly every function of a living organism depends on proteins.
proteins are made of? how is it joined together? how are they broken apart?
proteins are polymers, made up of amino acid monomers.
polypeptides are created through the joining together of amino acids at the ribosome (which facilitates the reaction)
Amino acids are connected through condensation polymerisation reactions, in which an ―OH is lost from the carboxyl group of one amino acid along with a hydrogen from the amino group of a second, forming a molecule of water and leaving the two amino acids linked via a peptide bond. (Aka, dehydration reaction)
Opposite of a condensation reaction is a hydrolysis reaction, in which water is required to break amino acids apart.
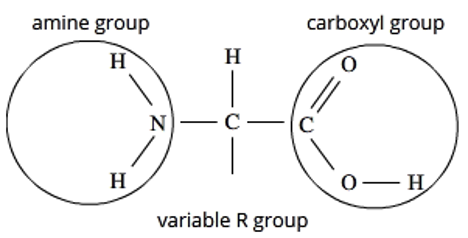
amino acid structure
central carbon + singular Hydrogen
an amino group on one end
a carboxyl group on the other end (acid)
a variable R group/side chain, which is what distinguishes each amino acid from the other.
essential vs non essential amino acids
There are 20 key amino acids found in all organisms
There are a couple of other amino acids found in some other organisms.
Not all of these amino acids can be made within a cell and instead have to obtained from an organisms diet. These are called “Essential” amino acids because it is essential that an organisms diet has them.
Nonessential means that our bodies can produce the amino acid, even if we do not get it from the food we eat.
Example: tryptophan is essential for us. But for e-coli it is not because it can be produced by itself.
levels of protein structure
Once a polypeptide is formed it then folds into a 3D shape. It is this 3D shape that dictates its function. (how it interacts with other molecules and enzymes etc)
There are 4 levels of organization for protein structure:
-Primary
-Secondary
-Tertiary
-Quaternary
All proteins have the first three.
primary structure of protein
the sequence/order of amino acids in the protein chain.
secondary structure (2 forms)
coiling and folding of the protein, caused by weak hydrogen bonds between amino acids in the chain.
alpha helices
beta pleated sheets
tertiary structure
refers to the overall 3D shape of the polypeptide, which is what determines its function.
It is generally caused by the R groups of the amino acids, either being attracted or repelled from one another. (eg. 2 hydrophilic R groups attract one another)
is held together by hydrogen bonds, ionic bonds, or covalent disulfide crosslinks.
Enzymes (called chaperonins) helps fold the peptide chain into its tertiary structure.
how do chaperonins help fold other proteins?
chaperonins are proteins that have a shape of a hollow cylinder with a cap.
1- an unfolded polypeptide enters the cylinder
2- the cap attaches, which causes the cylinder to change shape and create a hydrophilic environment for polypeptide folding. (facilitating)
3- the cap comes off and the correctly folded protein is released
Quaternary structure
occurs when two or more polypeptide chains interact to form a protein. each being referred to as a subunit of the protein.
protein structural classifications (2 main types)
classified based on the proteins overall 3D structure.
Fibrous proteins: elongated and insoluble. function is usually structural.
Globular proteins: compactly folded and coiled into a spherical overall shape. Function as enzymes and hormones.
shape of proteins relation to function
The structure of a protein is directly connected to it’s function.
This means for a protein to perform its function properly it also needs to be structured correctly. Any change in a proteins structure can affect reduce a proteins function or stop it functioning entirely. These are known as prions.
Factors that affect protein structure (+ denaturation)
-change in the amino acid sequence
-extreme temperature or pH (which affect the protein structure by breaking bonds at the secondary, tertiary or quaternary levels. which disrupts its shape causing the protein to denature as it has been unfolded. → this could lead to either the protein refolding incorrectly or it remaining unfolded.)
protein secretory pathway (reason)
The way in which proteins are secreted outside the cell. This pathway involves various different organelles that produce, fold, modify, and package proteins, eventually exporting them from the cell via the process of exocytosis.
An important process which allows for cells to communicate with other cells elsewhere in the body.
secretory pathway steps
nucleus – ribosome – endoplasmic reticulum – vesicle – Golgi apparatus - vesicle - plasma membrane - secreted
Steps:
1.mRNA from the nucleus after transcription and processing moves to the ribosomes on the Rough ER and are translated to produce Proteins.
2.Proteins move into the Rough ER.
3.Protein exits Rough ER in a Vesicle and travels to the Golgi Apparatus and binds to it.
4.Protein Travels through the Golgi Apparatus.
5.Protein enters a Secretory vesicle and leaves Golgi Apparatus.
6.Secretory vesicle binds to the cell membrane and secretes protein via exocytosis.
The role of rough ER (SP)
A series of connected flattened plasma membrane sacs studded with ribosomes. located near the nucleus.
The ribosomes synthesis the protein.
The rough er is then responsible for folding the protein (providing the environment needed by the protein to fold it correctly) Once folded, the protein is packaged into a transport vesicle and send to the golgi apparatus.
Role of Golgi apparatus (SP)
A series of flattened stacked pouches, different to the ER because the membranes aren’t all connected.
Responsible for further modifying and packaging the protein. eg. adding or removing chemical groups such as lipids, sugar molecules etc.
Then the proteins are often packing into secretory vesicles for export.
Role of vesicles (SP- 2 types)
transport vesicle: transports proteins WITHIN the cell. buds off the Rough ER with newly folded protein inside. travels to golgi body and fuses with the golgi membrane, releasing the protein into its lumen.
secretory vesicle: transports proteins OUT OF the cell. buds off the golgi apparatus with the finalised protein inside. travels through the cytoplasm’s to the cell membrane. fuses with the cell membrane and releases the protein out of the cell through exocytosis.
bulk transport (2 forms)
A type of active transport (requires energy)
transports lots of things at once, and uses vesicles in order to move molecules in and out of the cell.
Exocytosis and Endocytosis are 2 key forms of bulk transport.
Exocytosis
the process by which the contents of a vesicle are released from a cell. It’s a form of bulk transport.
Involves the vesicle fusing with the plasma membrane and releasing its contents to the outside of the cell, which is possible due to the fluid nature of the plasma membrane, which allows it to fuse with the vesicle.
Exocytosis is not just proteins ! Can also be used in the export of waste products to ensure that toxins do not build up within the intracellular environment.
endocytosis
opposite of exocytosis. bulk transport.
involves engulfment of molecules by extensions of the plasma membrane, importing them into the cell.
mitochondria role (SP)
site of ATP synthesis
provides energy required to move the vesicles and modify the proteins
name some types of proteins
structural (provide structure and structure eg. collagen)
enzymes (speed up chem reactions… —ase)
contractile & motor (important for muscle/cellular movement)
immunoglobin (defends body against diseases eg. antibodies)
hormones (regulate activities within the body eg. insulin)
receptors (respond to external and internal stimuli)
transport (carry molecules eg. channel proteins)
storage (store metal ions and amino acids)
what is activation energy? how do catalysts work?
activation energy is the amount of energy required by chemical reactions to get started. some require more than others.
a catalyst is something that speeds up or triggers a chemical reaction by lowering the activation energy of the reaction.
catalysts are not consumed by the reaction, and can therefore be reused
usually catalyse reactions in both directions
what are enzymes (structure, use, binding)
are soluble (hydrophilic properties) globular proteins with tertiary or quaternary structure which act as biological catalysts. most names of enzymes end with “ase”
They are specific with what they can interact with and the molecule that they act on is called a substrate. The area of the enzyme that the substrate binds to is called the “Active Site”. Also known as “substrate binding site”
The shape of an enzyme is critical to its function – particularly the shape of its active site. The shape of the active site needs to complement the shape of the substrate or they won’t be able to bind with each other. When the substrate binds to the active site, it forms an enzyme-substrate complex
2 mechanisms of enzyme binding with substrate.
lock and key: substrate molecules have the right shape to fit an enzyme (like a puzzle piece)
induced fit: the interaction of the substrate with the enzyme changes the shape of the enzyme to produce the right fit (like molding into shape)
why are enzymes regulated?
It is important that enzymes are regulated as otherwise reactions in the cell would happen too quick or too slow. This would result in either too much product or not enough.
The factors that affect the rate of enzyme efficiency (hint, they are proteins)
-Temperature
-pH
-Concentration of substrate and product
-Inhibitors & Feedback Inhibition
-Phosphorylation
-Cofactors and Coenzymes
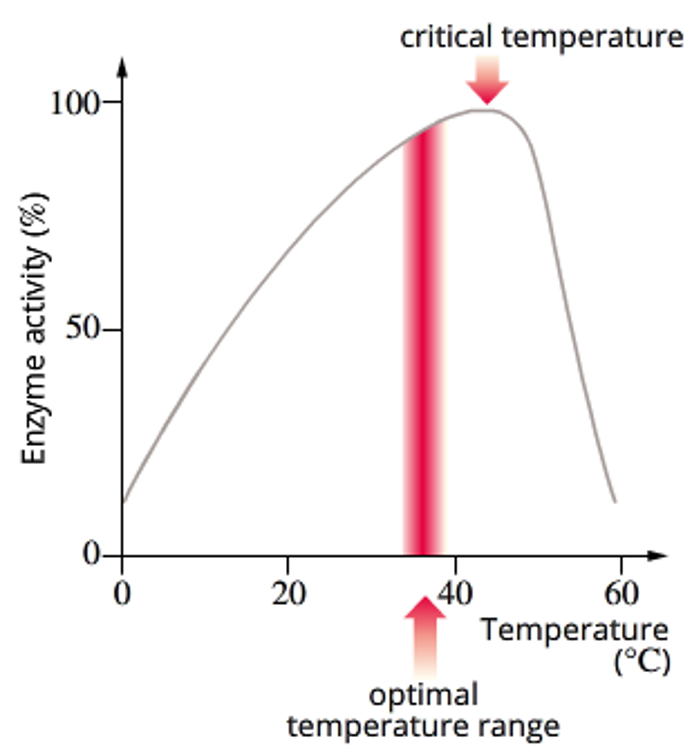
temperature - increasing enzyme rate of reaction, over heating
As temperature increases, particles move around more quickly, so substrates will collide more often with enzymes, increasing the chance of binding to the active site. So generally, as temperature increases, rate of reaction increases.
However, there is a limit to how much an enzyme can be heated up before it’s bonds begin to break and it denatures, losing it’s functional shape. This would result in the change in the active site’s shape, meaning the substrate can no longer bind to it. So rate of reaction would drop off abruptly.
Most enzymes have an optimum temp range where the enzyme activity is the greatest, and the temperature at which an enzyme starts to denature is called its critical temperature.
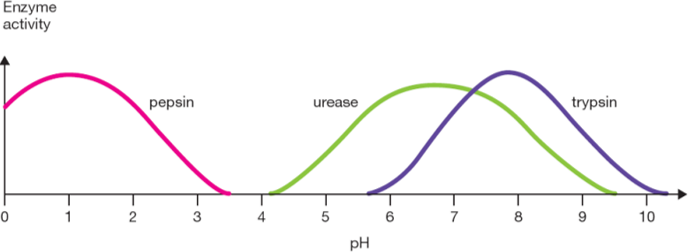
pH - enzymes
each enzyme has an optimum pH. Enzymes each have a varying range of pH environments which they can work in. (eg. enzyme in our stomach, optimum pH 1.5)
Change in pH affects the amino acids making up the enzyme
As a solution becomes more basic, amino acids lose hydrogen ions (react with OH-)
As a solution becomes more acidic, amino acids gain hydrogen ions (react with H3O+)
This affects the secondary and tertiary structure of the enzyme, as amino acids may change their attraction to each other and begin to unfold. So extreme pH will denature an enzyme.
concentration of substrate, product or enzymes
The rate of the reaction can be adjusted by increasing or decreasing the concentration of substrate, product or enzyme:
increasing substate: increased rate & product until the enzyme is saturated, then the rate will not continue to increase.
increasing product: decreased rate, as too much product can cause the reaction to be reversed. This is a way cells can regulate their enzyme production – the product of the reaction itself inhibits enzyme function, because the cell doesn’t need to make any more.
increasing enzyme: increased rate until all the substrate is depleted.
enzyme inhibition (2 types)
some molecules can interact with the enzyme and cause it to be inhibited.
competitive inhibitors: Competes with the substrate to bind to the active site. Therefore it has to have the same or very similar shape to the substrate. It forms an enzyme-inhibitor complex, and directly blocks the active site.
non-competitive inhibitors: binds elsewhere on the enzyme and causes the enzyme and its active site to change shape, preventing the substrate from being able to bind
reversibility of enzyme inhibition
reversible (only competitive, but not all): the inhibitor weakly interacts with the active site of the enzyme, decreasing the frequency at which the substate can bind to the active site. can be reversed by increasing the substrate concentration, as it will tend to displace the inhibitor bound to the enzyme.
irreversible: inhibitor binds strongly either at the active site or another region. causes a permanent block of the active site or a conformational change so that the active site is no longer complementary. meaning, non-competitive inhibitors are usually irreversible.
Irreversible inhibitors form a covalent bond with part of the enzyme – causing a permanent change to the enzyme. Reversible inhibitors bind to an enzyme non-covalently.
feedback inhibition in enzymes
where the final product made by the enzyme can bind to and inhibit the function of the original enzyme, which can stop an enzyme from continuing to make a product that there is a lot of. This regulates the amount of product produced by the cell.
phosphorylation
the binding of a phosphate group to the enzyme, which can either activate or deactivate the enzyme. dephosphorylation reverses this affect.
Coenzymes & Cofactors
Additional molecules which can enable enzymes to properly function and may be required in order to catalyse a reaction. May sit in the active site to help the enzyme work best.
2 groups:
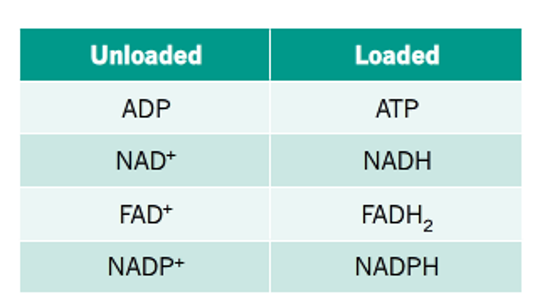
Coenzymes (organic): usually derived from vitamins. usually donate phosphates, hydrogens, electrons or protons. eg. NAD+ and NADP+
Cofactors (inorganic): metallic cations (eg. Mg2+, Fe, Zn)
coenzyme (re)cycling
many reactions catalysed by enzymes involve the transfer of chemical groups, particularly phosphate groups.
this calls for the use of coenzyme intermediates, which can be reloaded/refreshed multiple times and used again after donating these chemical groups.
enzymes regulating biochemical pathways
Many biological processes are a sequence of chemical reactions where each reaction is catalysed by a different enzyme
Therefore, biological processes in a cell may be regulated by genes because they have to code for the multiple enzymes requires in a process, so it relies on the genes being transcribed and processed.
metabolism & (its 2 types)
Metabolism is the sum total of biochemical reactions that occur in a cell.
These are either reactions involve the 'building up' or 'breaking down' of molecules. Two terms that describe this are:
Anabolism: anabolic reactions are where there is a building up of larger molecules from smaller molecules. (usually requires energy- endergonic)
Catabolism: catabolic reactions are where there is a breaking down of macromolecules into smaller molecules. (usually produces energy- exergonic)
enzyme action in catabolic reactions
The substrate binds to the active site of the enzyme, because their shapes are almost perfectly (but not quite) complementary. Upon binding, the enzyme changes shape, which stresses the bonds in the substrate, helping it to catabolise into two products
enzyme action in anabolic reactions
Anabolic enzymes work by aligning substrates and using an energy source (such as ATP) to transfer energy to the substrates to create a bond.
Upon binding, the enzyme changes shape, aligning the substrates, making it easier for a bond to form between them to form a product
what does DNA Manipulation allow scientists to do?
understand the function of genes (by turning them on and off in a chromosome)
compare genetic information between species
solve crimes
paternity testing and cases
product gm plants and animals (to improve crop and animal breeds)
what are the 3 branches of DNA manipulation
tools (molecules that rearrange segments of DNA)
techniques
application
what are DNA manipulation tools (3 key molecules)
these are different enzymes used to copy, cut and glue segments of DNA (manipulate DNA)
3 key types of molecules:
-polymerases to synthesis
-endonucleases to cut
-ligases to join
not to be confused with “equipment” eg. micropipettes and test tubes
polymerases (what they do, 3 key)
are enzymes that join together monomers to form polymer chains … “Polymer” + “-ase”
key polymerases:
DNA polymerase: reads DNA to make DNA (eg. to add nucleotides to the existing DNA strand or to repair damaged DNA strands.)
Reverse transcriptase: uses single strand of RNA to make DNA
RNA polymerase: reads DNA to make (m)RNA
DNA Polymerases
Create a complementary DNA strand to a template DNA strand- running from 3’ to 5’ by joining together nucleotides (deoxy-nucleotides) in a 5' to 3' direction according to the base-pairing rules..
Used normally in cells to replicate DNA
Requires a primer to initiate replication on the strand being synthesized, Usually a short nucleic acid sequence that provides a starting point for DNA synthesis.
Taq Polymerase (a type of DNA polymerase)
A DNA polymerase that is used to amplify DNA in the laboratory (used in PCR).
It’s originally from Thermophilic (heat loving) bacteria.
This makes it very heat resistant and robust enzyme. It is very heat resistant.
DNA polymerase - reverse transcriptase (use in producing cDNA)
Type of DNA polymerase enzyme that transcribes a complementary DNA strand to a template RNA strand. Is the reverse to transcription.
Works by joining together nucleotides (deoxy-nucleotides) in a 5' to 3' direction according to the base-pairing rules. E.g. u in RNA into a in DNA
It is useful for producing cDNA (complementary DNA) from mRNA. There are no introns as it is made from mRNA that has undergone processing, which is useful in prokaryotes.
RNA polymerase
creates a complementary RNA strand to a template DNA. synthesises mRNA, rRNA and tRNA.
dosen’t require a primer to bind to the DNA, because it usually has a promoter
Endonucleases (their restriction sites)
They are restriction enzymes that cut DNA into fragments at a specific sequence of bases (restriction site), which is unique to that endonuclease. More than one endonuclease can be used to cut the DNA.
The sequences are also palindromic (can be read the same forwards on one strand and backwards on the other).
The length of the restriction site is dependent on the enzyme, some 4 bases, some 6 etc. Statistically speaking 4 base cutters will cut DNA more frequently than 6 base cutters.
Endonuclease examples (Hind? & Eco??)
Hind3 and EcoR5
-found naturally in bacteria, and are named after these.
what are the 2 types of ends produced by endonucleases?
blunt ends (only cuts through the sugar-phosphate backbone)
OR
sticky ends (cuts through both the sugar-phosphate backbone and hydrogen bonds between nucleotides in restriction site)
difference between cutting circular vs linear DNA
When DNA is cut/digested by a restriction enzyme it is cut into fragments:
If the DNA is Circular (like a plasmid) to begin with then
# of fragments = # Restriction Sites cut at
Meaning that the number of restriction sites cut, is the number of fragments made. As in one cut opens up the circular DNA, leaving it as one, and another cut would make it two.
If the DNA is Linear (like our chromosomes) to begin with then
# of fragments = # Restriction Sites cut at + 1
As DNA is already a fragment to begin with because it is not closed.
Because the DNA is Double Stranded each fragment will also be made up of double stranded DNA.
how is fragment size measured?
measured in base pairs. a pair being 2 complementary nucleotides joined together across the 2 strands of DNA.
if a nucleotide is unjointed, it is not counted in the fragments size, as only paired bases are counted.
This means that DNA cut with blunt ends will have all of its bases paired and counted.