1. control of eukaryote transcription
1/71
Earn XP
Description and Tags
Eukaryotic cells
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
72 Terms
1.1 what are the different RNA pol involved in the transcription
RNA pol I : transcribes ribosomal RNA ex. 18S RNA
RNA pol II : transcribes mRNA & different RNA (sRNA)
RNA pol III : transcribes tRNA and sRNA,miRNA
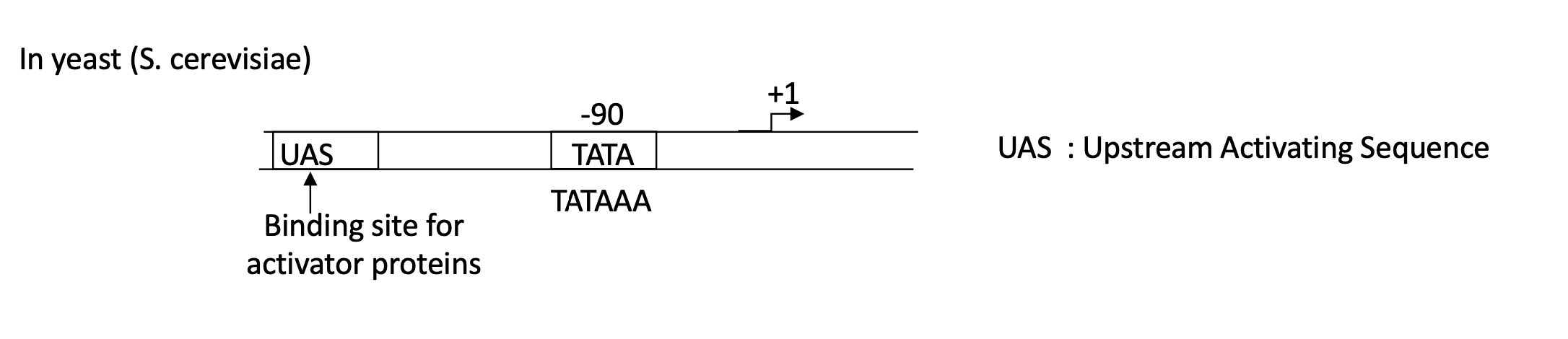
1.1 what is the required DNA sequences for efficient initiation of transcription in yeast
Promoter w/ :
UAS = Upstream Activation Sequence : binding site for activator proteins
TATA box at -90
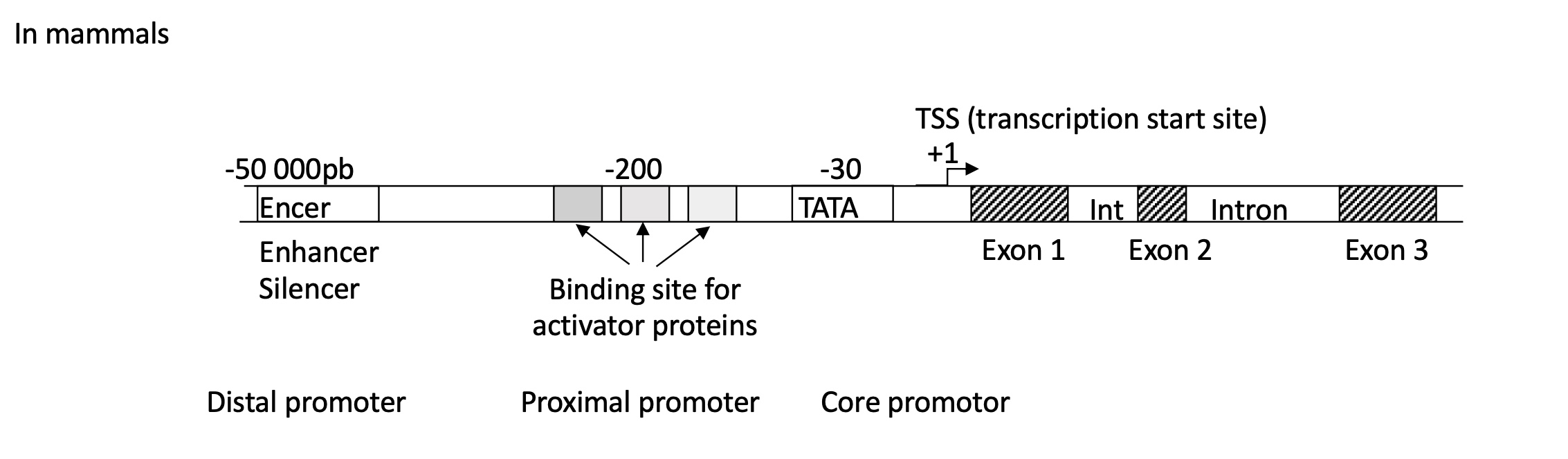
1.1 what is the required DNA sequences for efficient initiation of transcription in mammals
Promoter w/ :
distal : Enhancer/Silencer binding site
proximal : multiple binding sites for activator proteins
core : TATA box at -30
after TSS/+1 there is the presence of introns (non coding sequences) and exons (coding sequences)
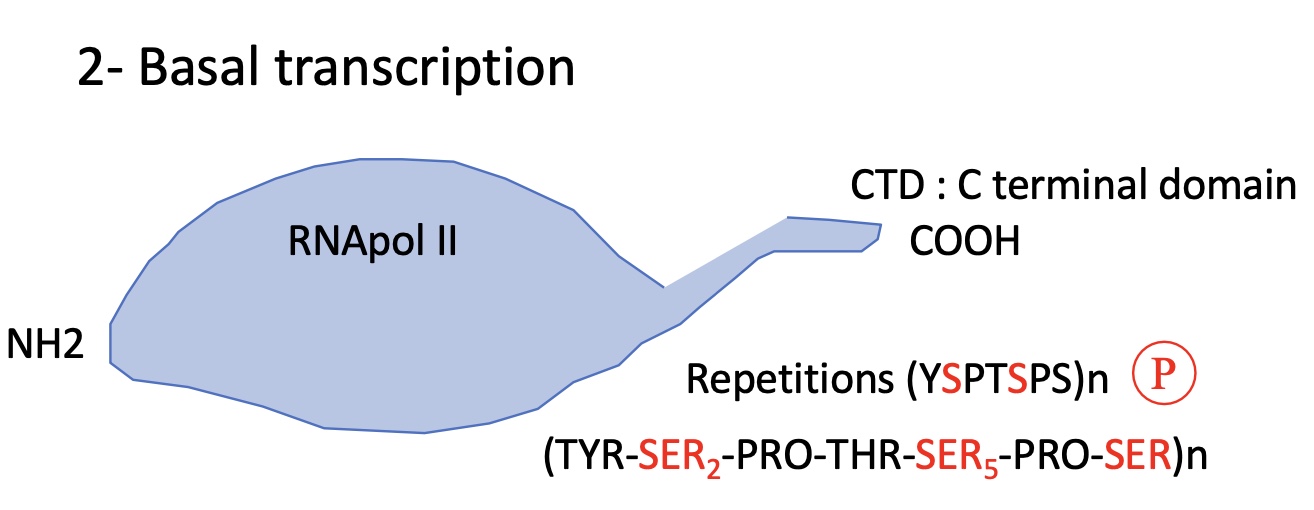
1.2 what is required structure of RNA pol II for minimum transcription/ basal transcription
RNA pol II has 12 subunits with 2 bigger sub units at the C terminal domain (CTD)
The CTD has a sequence repetition where the SER 2 & 5 are subject to phosphorylation —> can impact transcription
nb of repetitions depends on the species and complexity
1.2 regarding the CTD of RNA pol II how does its’ phosphorylation affect its binding to promoter
Hypo-phosphorylated —> RNA pol II binds to the promoter
Hyper-phosphorylated —> RNA poll II released from promoter and begins elongation
1.2 An experiment was carried out to test basal transcription, what was the conclusion ?
that DNA fragments (TATA, TSS etc.) + RNA pol II + nuclear extracts there was minimal transcription
wasn’t the case when nuclear extracts were absent
proteins in the nucleus are involved in the activation of transcription = Transcription Factors
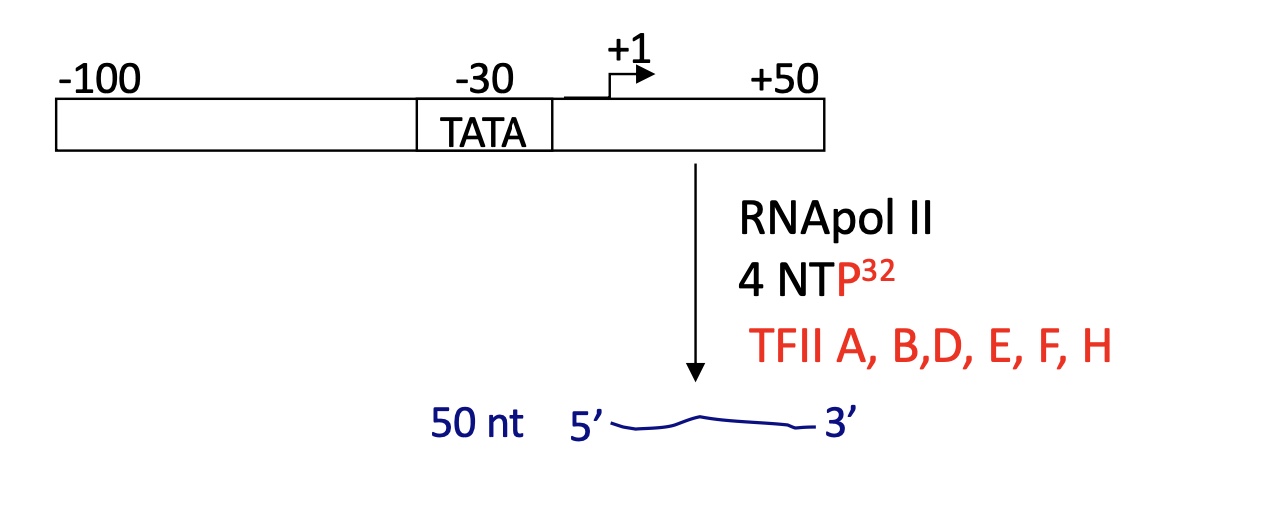
1.2 How are the transcription factors TF assembled to initiate transcription
TFIID binds to TATA box ( and other sequences but for a lesser affinity)
TFII A and B bind to TFIID
TFIIF helps the fixation of RNA pol II to the TFII B/D/A
TFII E & F and TFIIH binds to the RNA pol II
TFIIH = kinase that phosphorylates SER 2 & 5 on the CTD
==> Pre-Initiation complex PIC
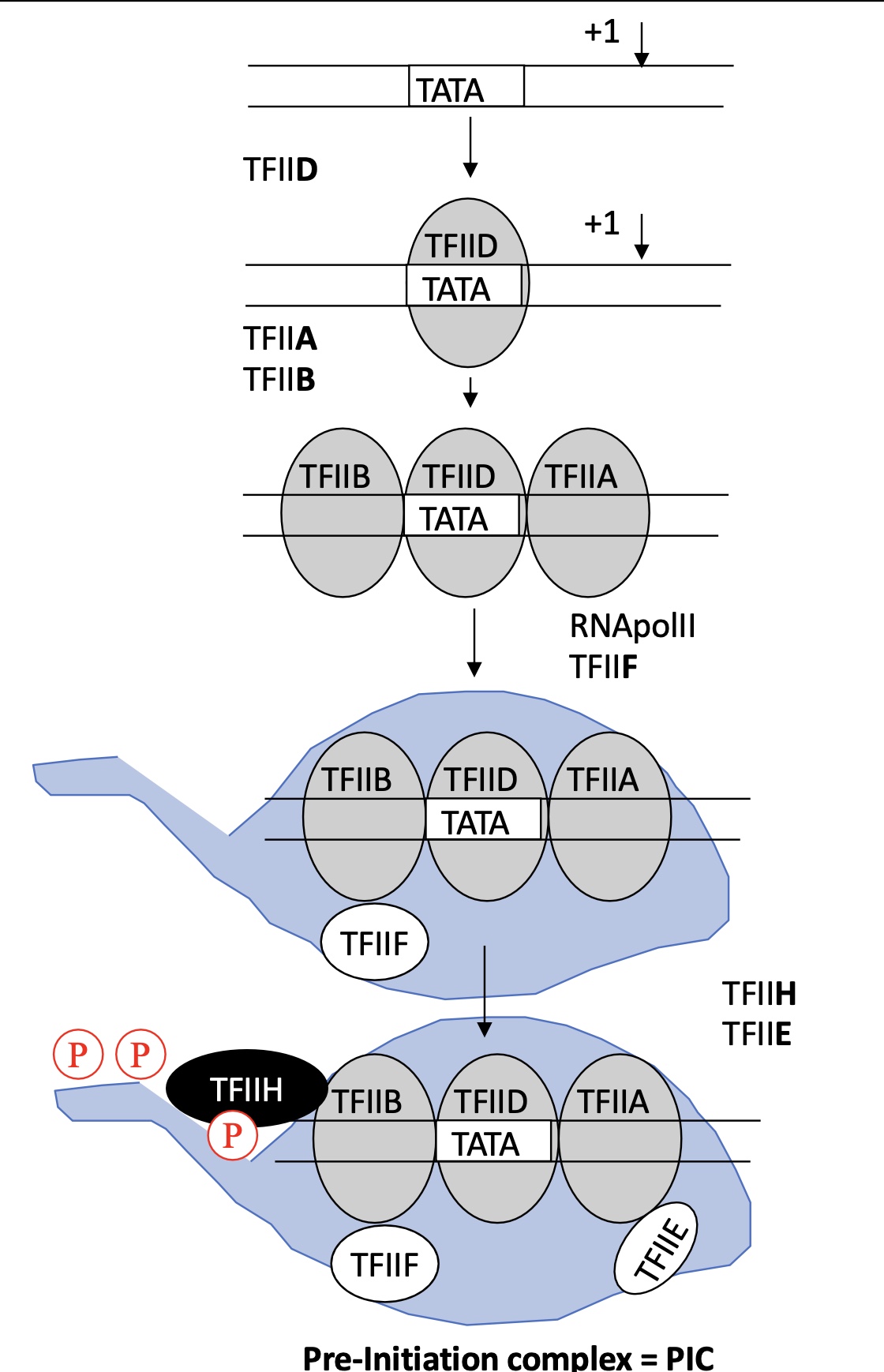
1.3 how does the mediator help in the activation of transcription
it helps install PIC :
recrute TFs
interacts w/ repressors o block RNA pol II , it can block :
fixation and action of activator
action and recrutement of mediator
condense chromatin
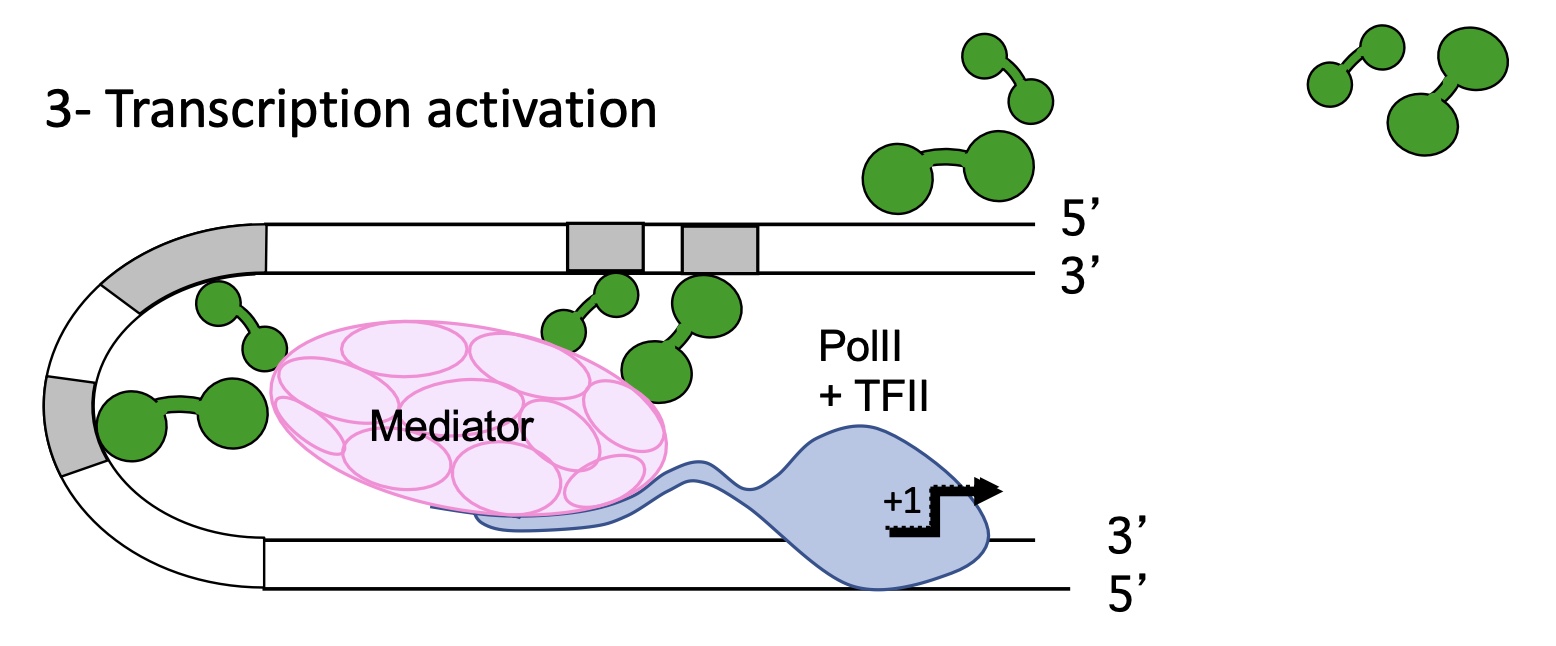
1.4.1 what is the structure of chromatin
organised in nucleosomes :
histones wrapped around by DNA
histone tailed
linker DNA that connects the 2 nucleosomes
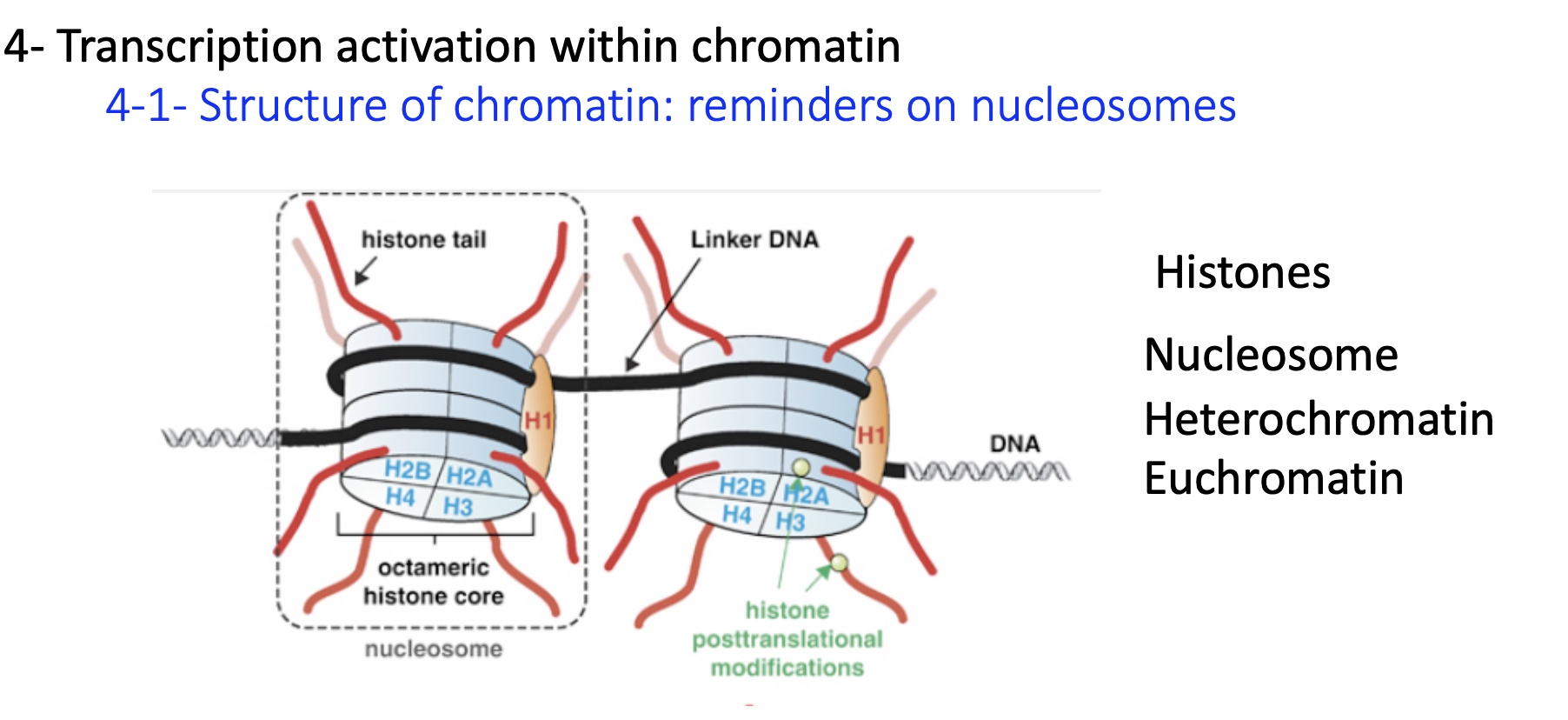
1.4.1 difference in chromatin condensation and activity
heterochromatin = condensed and transcriptionally inactive
euchromatin = loosely packed and transcriptionally active
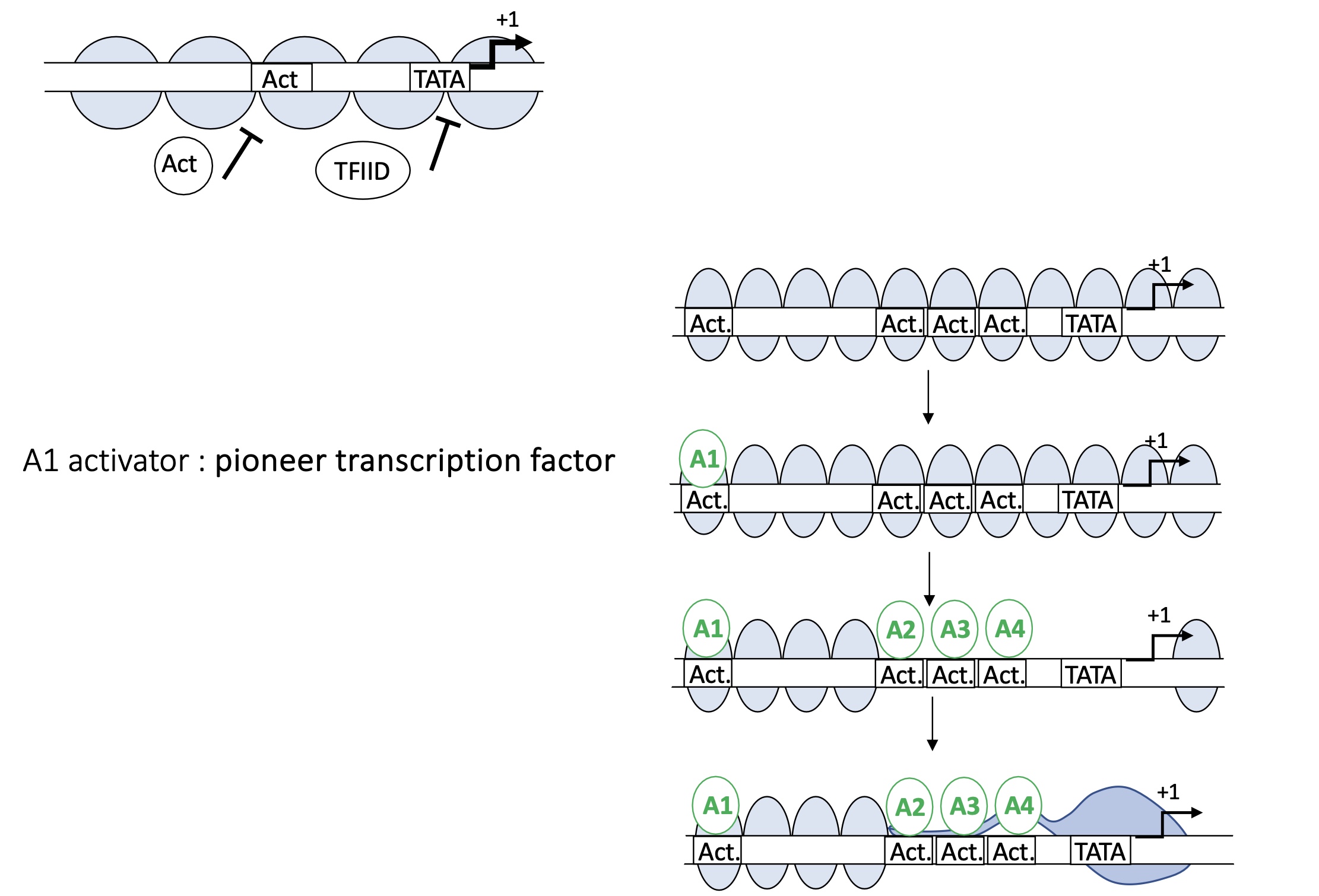
1.4.1 how do proteins such as TFII and activators bind to chromatin
Pioneer ACT will bind to the ACT binding sit on the DNA in between the nucleosomes
ejection or displacement of nucleosomes
more ACT binding sites available
TATA box available
TFIID can bind to TATA box and form PIC
1.4.2 what are examples of epigenetic marks
DNA methylation
Post translational modification of histones
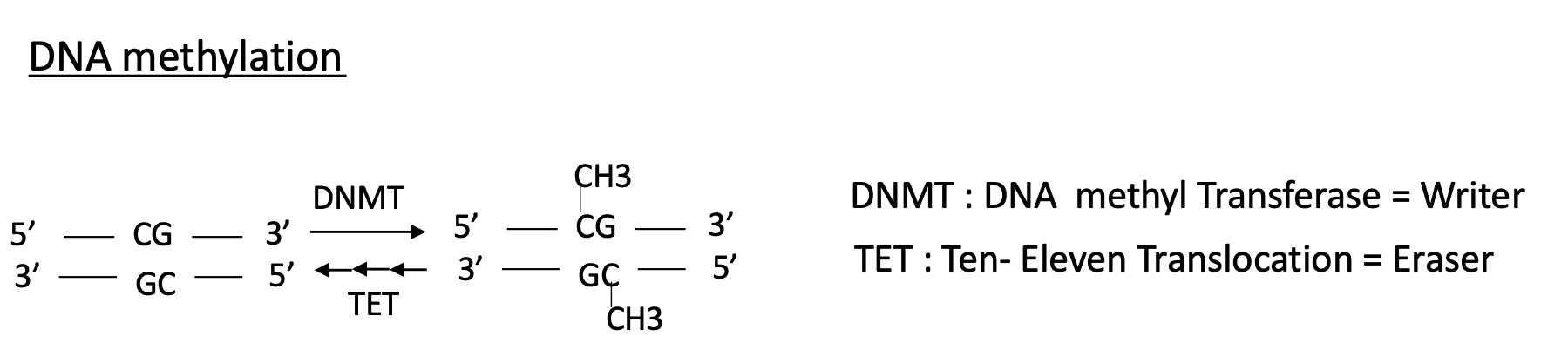
1.4.2 how is the methylation of DNA controlled
a DNMT DNA methyl transferase will methylate a CG s DNMT = writer
a TET ten-eleven translocation will remove the methylation so TET = Eraser
1.4.2 what is methylation usually related to? activation or repression of transcription?
methylation is mostly related to the inhibition of transcription

1.4.2 what are the types of post translational modification histones can undergo ?
on the Nt histone tail :
phosphorylation on the Ser
Methylation of Lys (K) or Arg(R)
acetylation on K or R
on the Ct histone head :
ubiquitination on a K
sumoylation
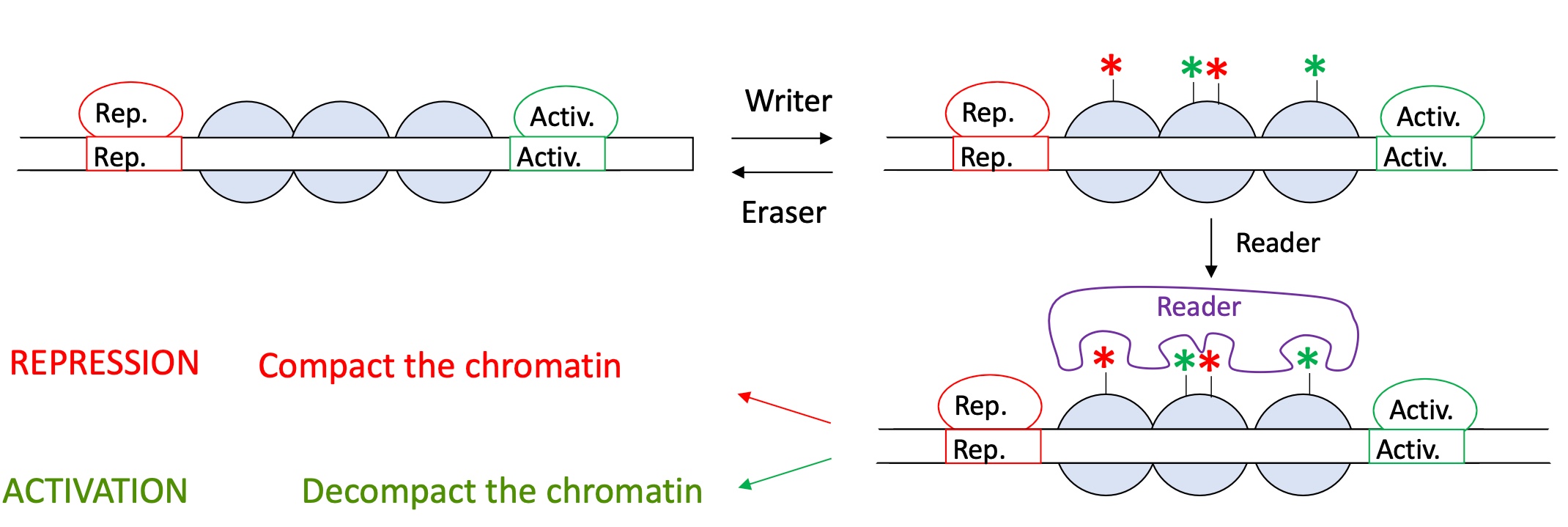
1.4.2 how does histone post translational modification impact the activity of transcription
certain modicifaction done by the writer on the nucleosomes recruit a read that will determine the repression or activation of transcription
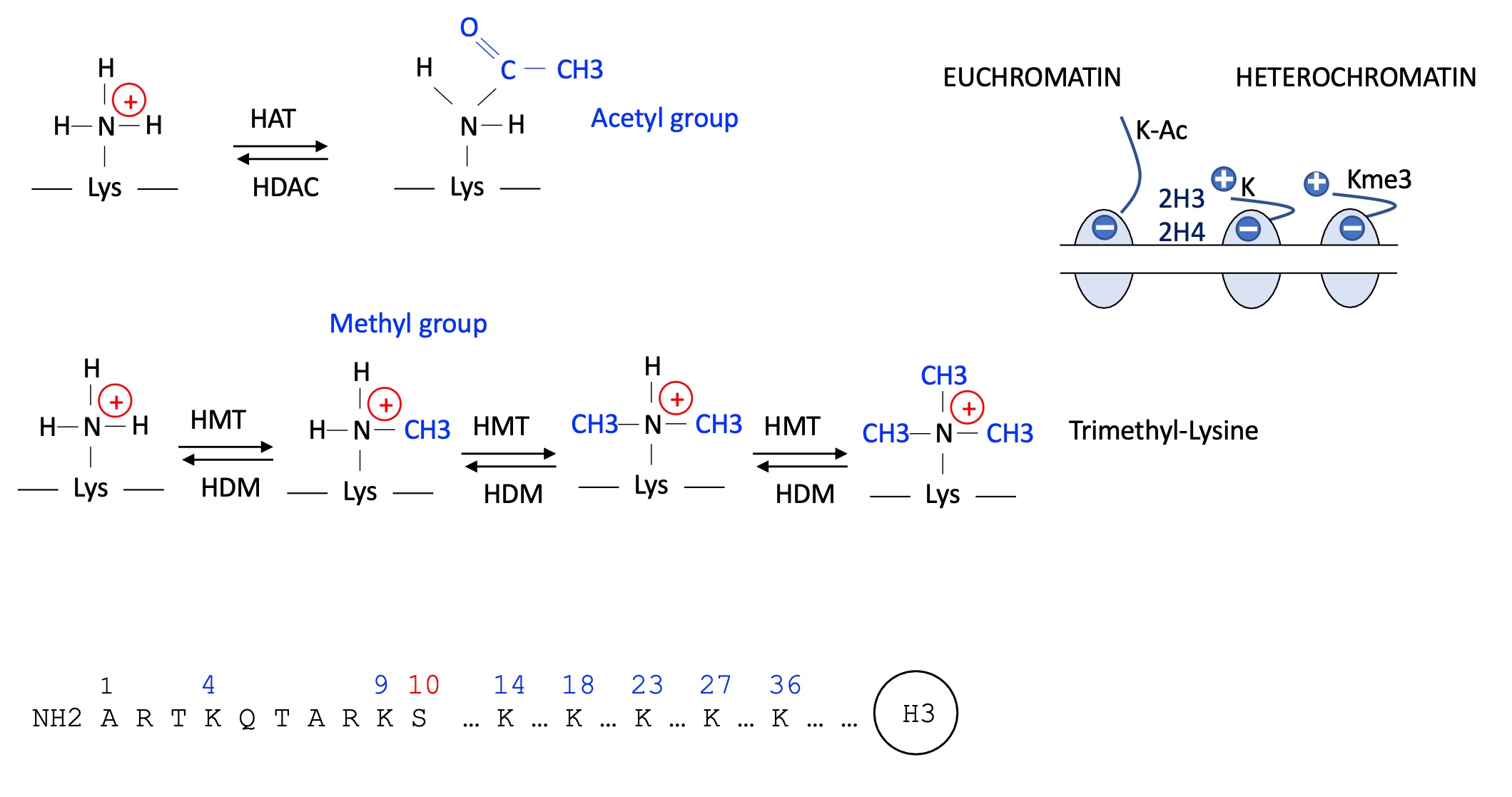
1.4.2 how does acetylation and methylation on Lysine of stone tail affect transcriptional activity
when acetylation of K occurs it looses its positive charge —> liberates tail of the histone —> loosens chromatin
= Euchromatin (genes are accessible and so activation of transcription)
when K methylated it is done so multiple times and is tri-methylated —> positive charge remains —> cremation remains compact
= Heterochromatin (genes are inaccessible so inhibition of transcription)
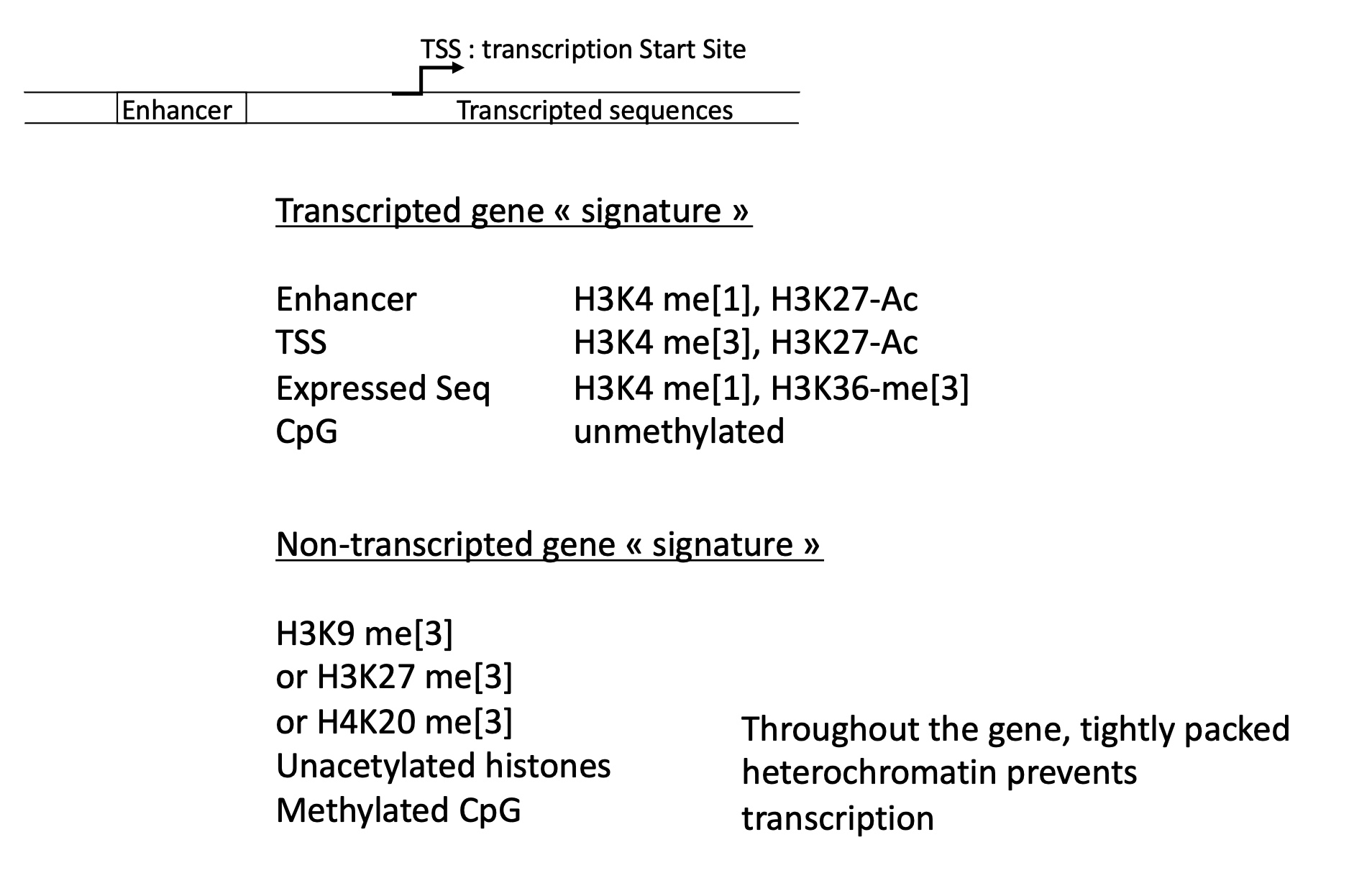
1.4.3 what histone marks are correlated to activation/repression of transcription
activation of gene transcription :
H3K4-me1,H3K27-ac
CpG un-methylated
repression of gene transcription :
H3K9-me3
un-acetylated histones
methylated CpG
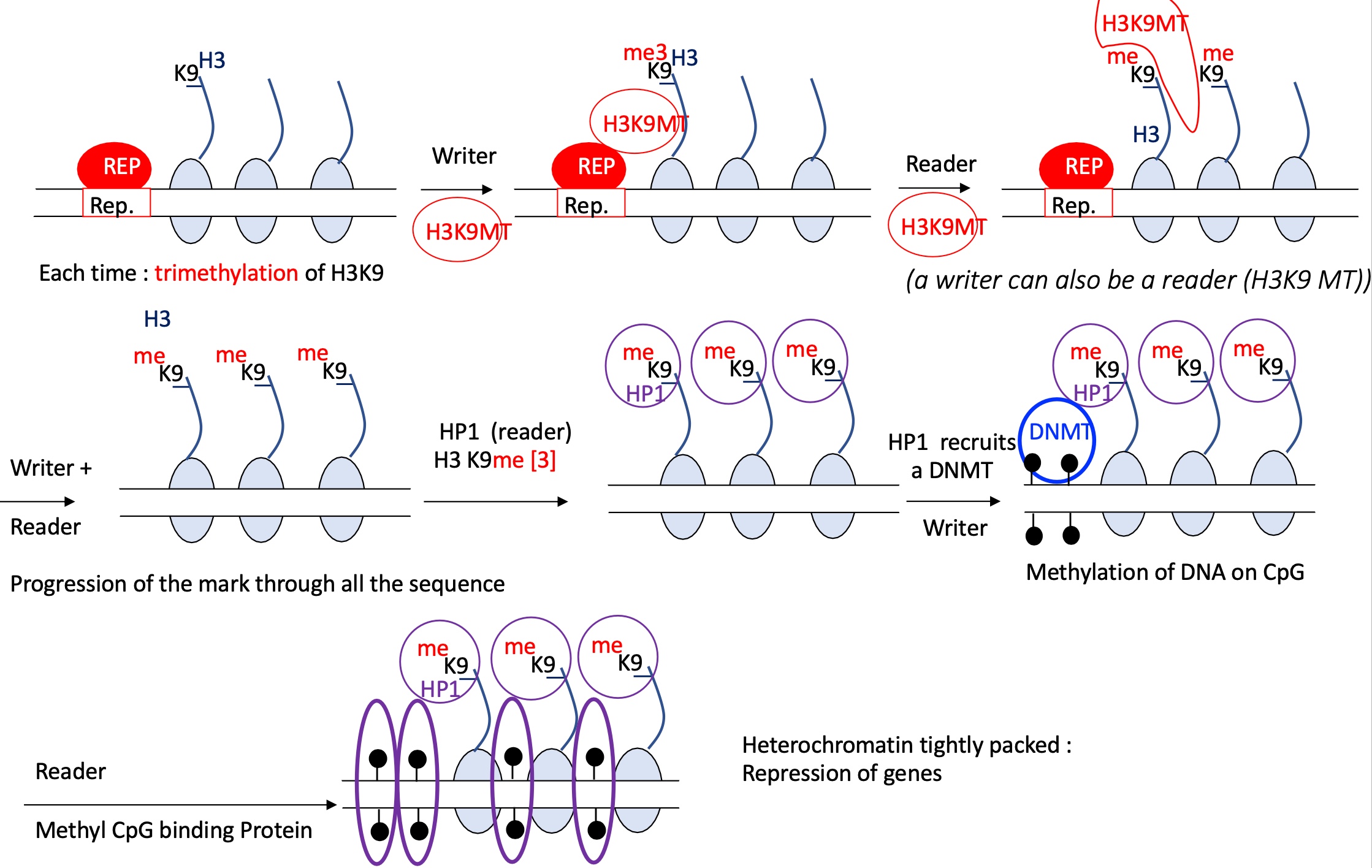
1.4.4 how is heterochromatin formed and gene expression inhibited
REP binds to REp binding site
a writer = H3K9MT methylates H3K9-me3
a reader = H3K9MT reads the repressor methylation signal and continues to ti-methylate H3K9 on each nucleosome
progression of the mark along the sequence
a reader = HP1 recognises the H3K9-me3
this recognition recruits DNMT which methylates CpG on DNA —> heterochromatin
inaccessible genes and therefore of transcription
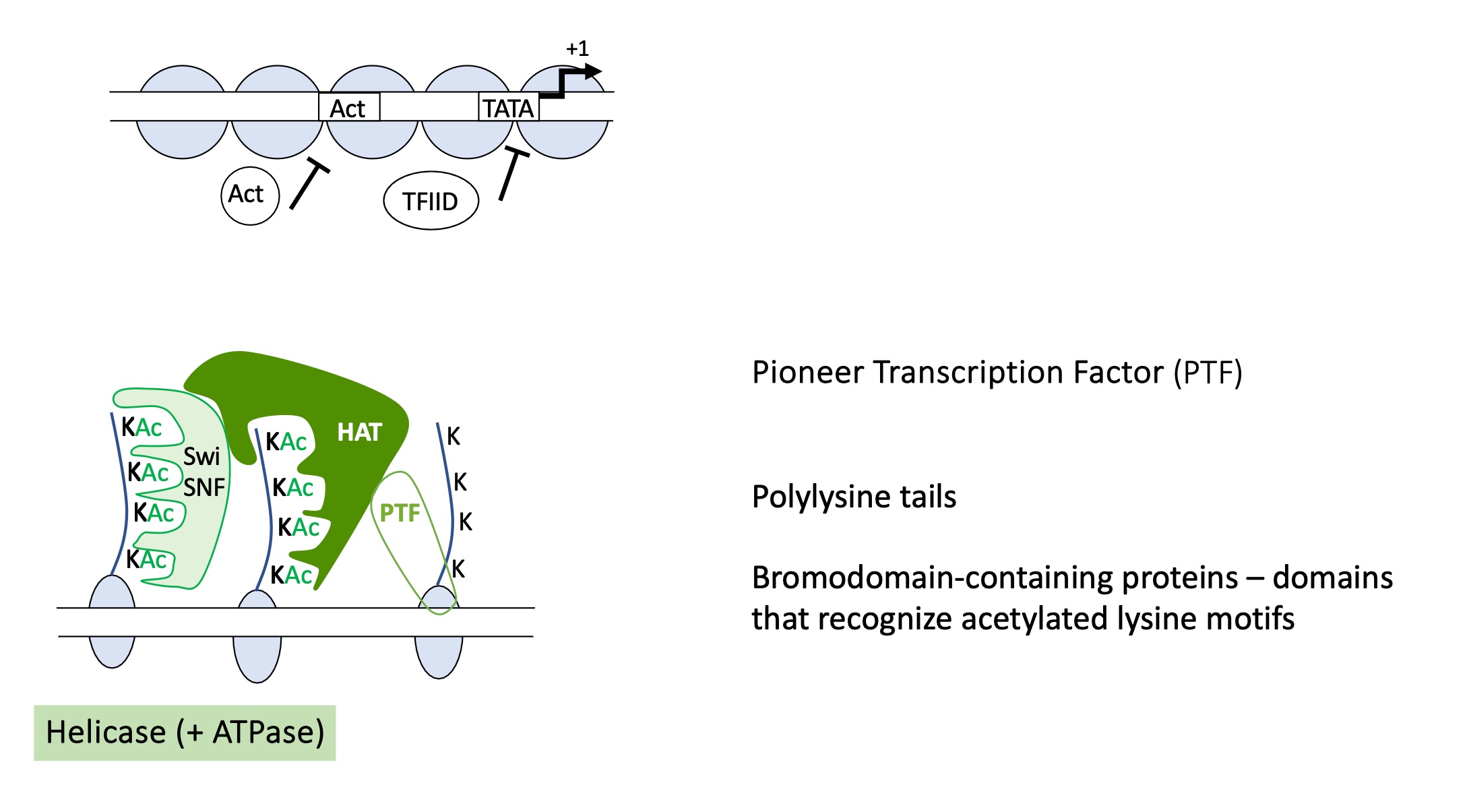
1.4.5 how is euchromatin formed
A pioneer transcription factor binds to its specific DNA sequence making the DNA more accessible.
The pioneer factor recruits a writer enzyme ( a histone acetyltransferase (HAT) ) which acetylates lysine residues on histone tails (e.g., H3K9ac, H3K27ac).
A reader protein with a bromodomain recognises and binds to the acetylated lysines on histones.
Chromatin remodeling complexes (such as SWI/SNF) are recruited — these have ATPase activity that uses ATP hydrolysis to slide or evict nucleosomes, exposing promoter and enhancer DNA.
euchromatin formed, genes accessible
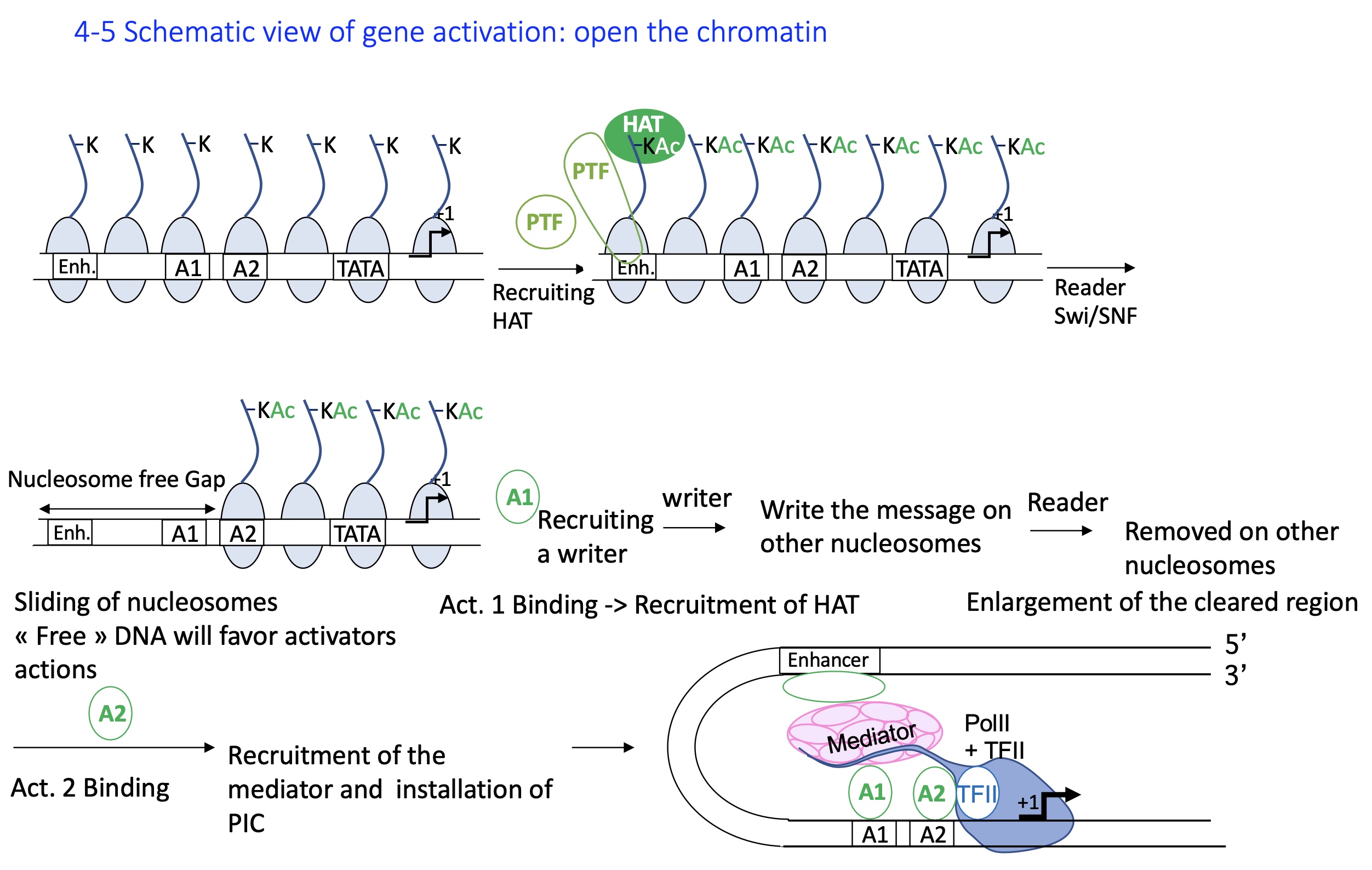
1.4.5 how does the formation of euchromatin activate gene expression
Closed chromatin (heterochromatin): Histones are deacetylated, DNA is tightly packed, and transcription is inactive.
A PTF pioneer transcription factor binds its enhancer site even in condensed chromatin and recruits a HAT (histone acetyltransferase).
HAT acetylates histone tails, loosening DNA–histone interaction and marking chromatin for activation.
Reader complexes (e.g., Swi/SNF) recognize the acetyl marks and slide or remove nucleosomes, creating nucleosome-free regions.
Activator binding: The open DNA allows activator transcription factors to bind promoters/enhancers and recruit more HATs, extending euchromatin formation.
Mediator and PIC recruitment: Activators recruit the Mediator complex, which helps assemble the Pre-Initiation Complex (PIC) with RNA polymerase II.
=> Transcription initiation : The open euchromatin and assembled PIC enable RNA Pol II to begin transcription.
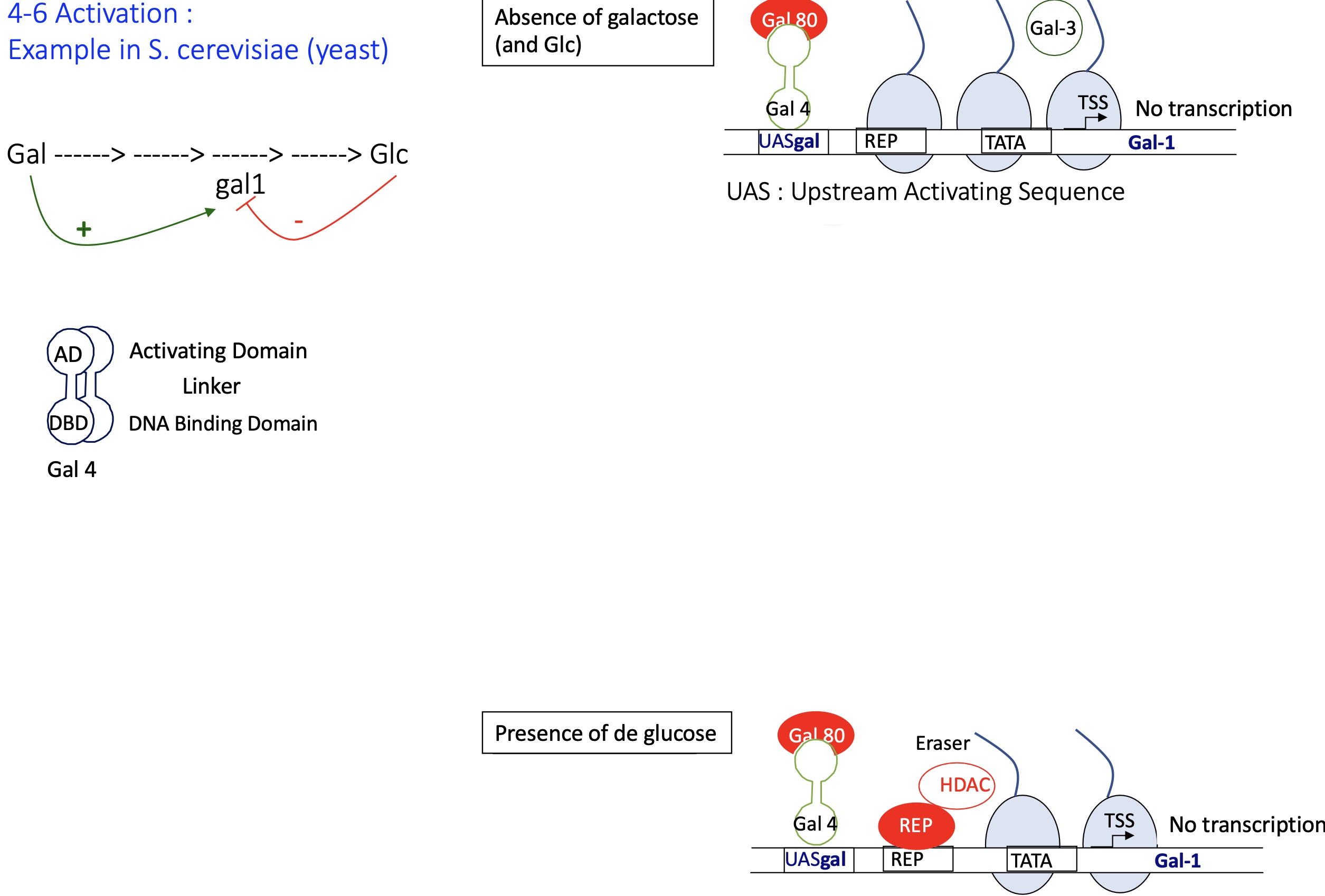
1.4.6 How does the repression of gene expression in S.cerevisiae (yeast) by Gal4
absence of galactose (and Glc) :
the DBD of Gal4 (Transcription factor) binds to its binding site UAS ( upstream activation sequence)
Gal 3 remains free and inactive in the cytoplasm - as its not bound to Gal it doesn’t bind to Gal 80
Gal 80 binds to AD of Gal4 —> inhibiting its activity and unable to recruit transcription machinery (Mediator and PIC)
chromatin stays condensed
—> no transcription of GAL genes
presence of Glc :
Gal 80 binds to AD of Gal4 —> inhibiting recruitment of transcription machinery
activation of a repressor that’s binds to its binding sequence
recruitment of HDAC histone de-acetylase —> removes Ac histone marks and chromatin becomes condense (heterochromatin)
—> no transcription
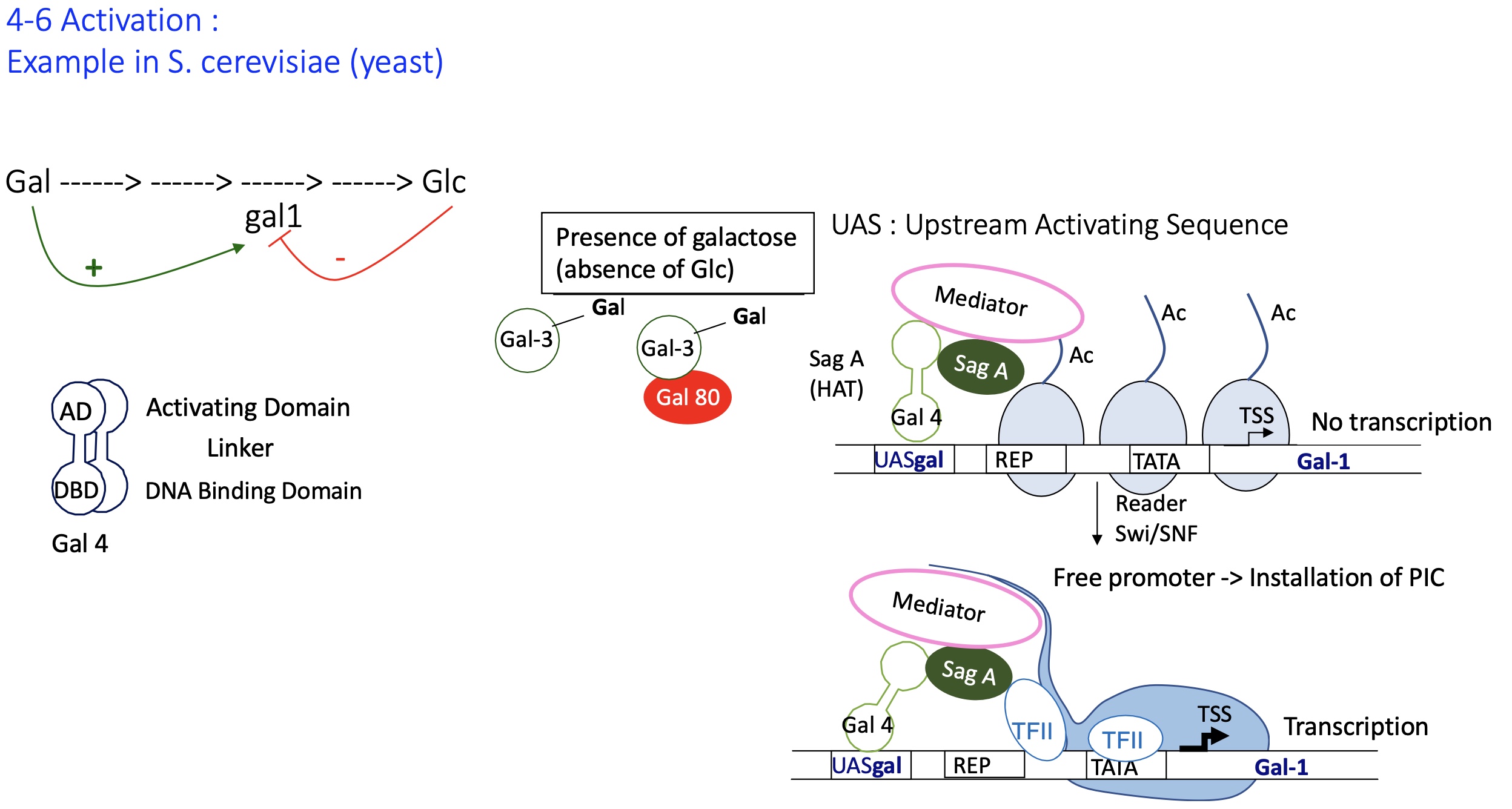
1.4.6 How does the activation of gene expression in S.cerevisiae (yeast) by Gal4
presence of galactose (absence of Glc) :
Gal 3 binds w/ Gal 80 —> repressing the activity of Gal 80 and can no longer bind to Gal4
Gal4 no longer inhibited by Gal80
Gal4 bound to UAS recruits writer SagA that acylated histone tails
installation of mediator but no transcription
reader Swi/SNF recognises Ac histones and allows the sliding of nucleosomes
promoter is free and TFII can now bind and permit installation of PIC takes place
transcription of GAL genes takes place e.g genes coding for enzymes that metabolise Gal
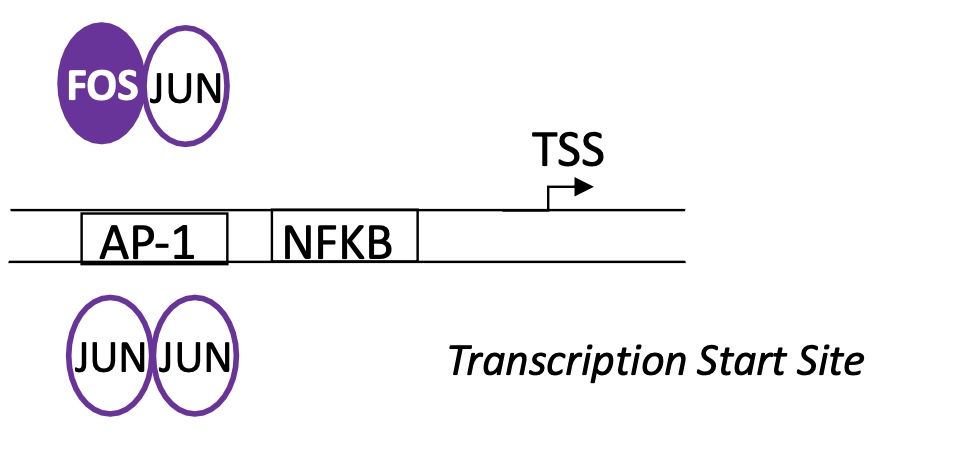
1.4.7 binding sites of AP1 and NFKB in macrophages to regulate activity of induced genes by an infection by a pathogen
AP1 = DNA binding for FOS-JUN and JUN-JUN dimers
NFKB binding site is located after AP1 BS —> NFKB binds here
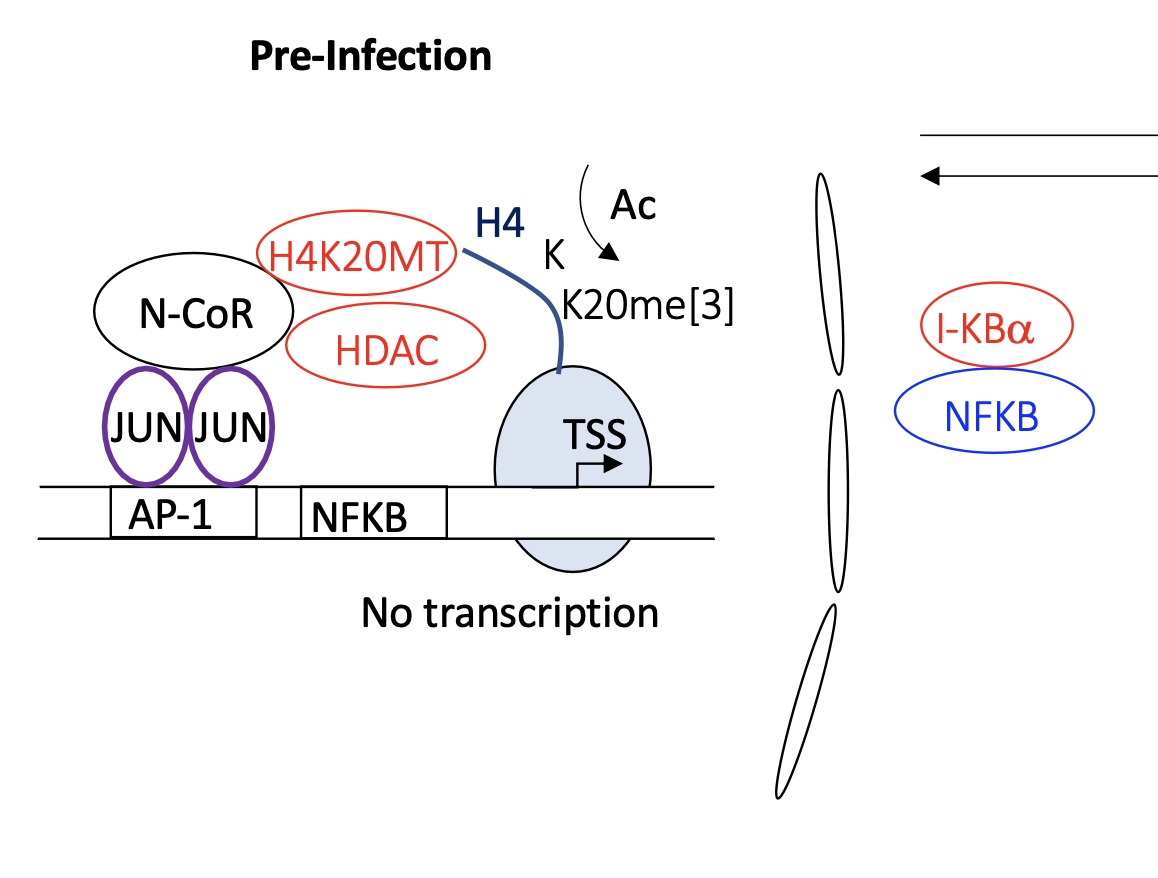
1.4.7 pre-infected macrophage how is the transcription of genes regulated
pre-infected cell :
JUN-JUN binds to AP1
this allows recruitment of several proteins
H4K20MT —> adding me to histone tails
HDAC —> remove Ac from histone tails
chromatin remains condensed and genes are unavailable for transcription (nucleus)
NFKB remains bound to IKBa (inhibitor) in the cytoplasm —> NFKB is inactive
no transcription
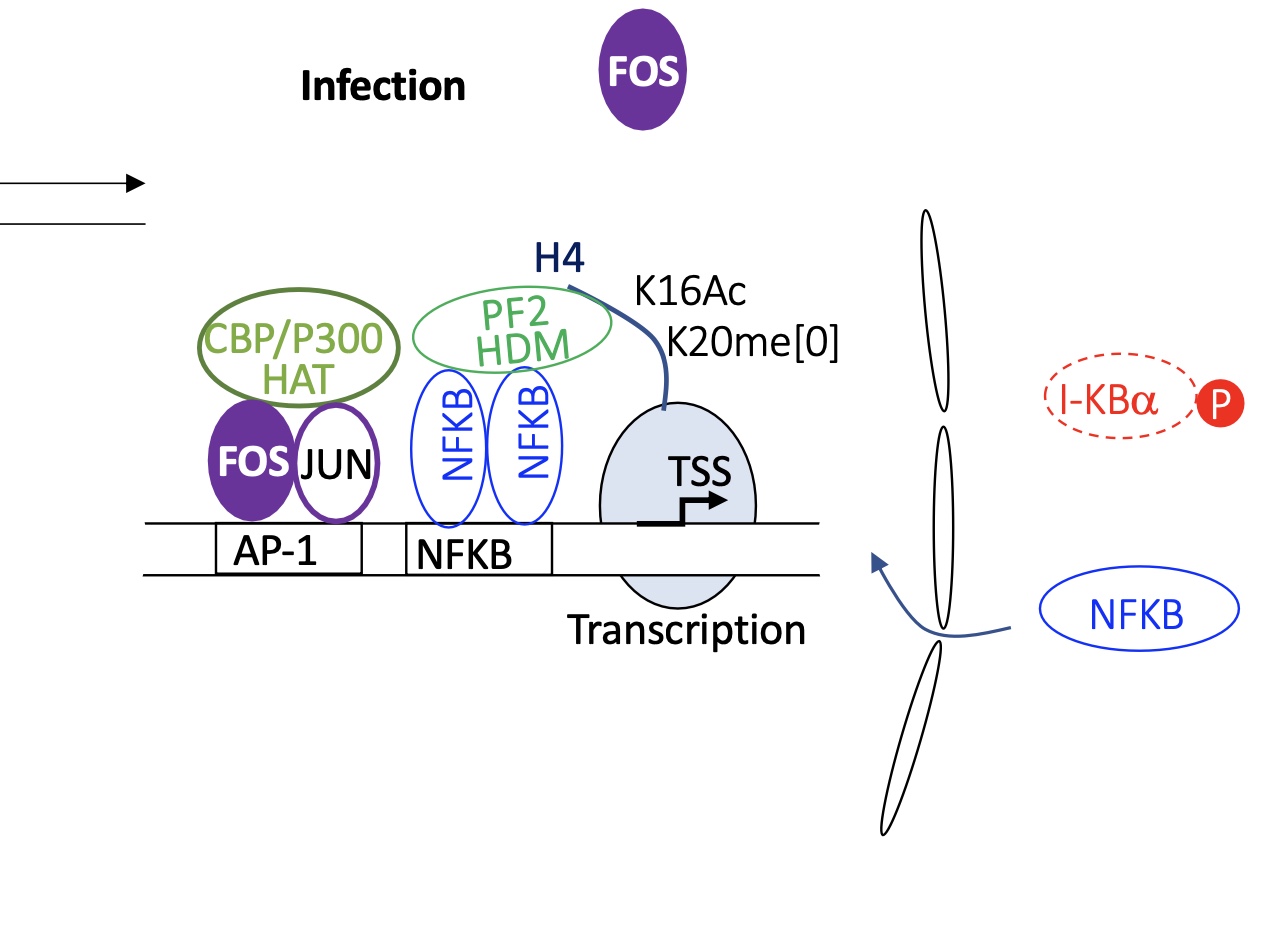
1.4.7 infected macrophage how is the transcription of genes regulated
infected :
FOS-JUN dimer binds to AP1
recruitment of HAT Histone Acetylase transferase proteins
IKBa is phosphorylated chi liberates it from interaction w/ NFKB
NFKB is free In the cytoplasm and translocates into nucleus to bind to its BS
recruits histone de-methylase
euchromatin —> chromatin open
transcription of immune response genes takes pace
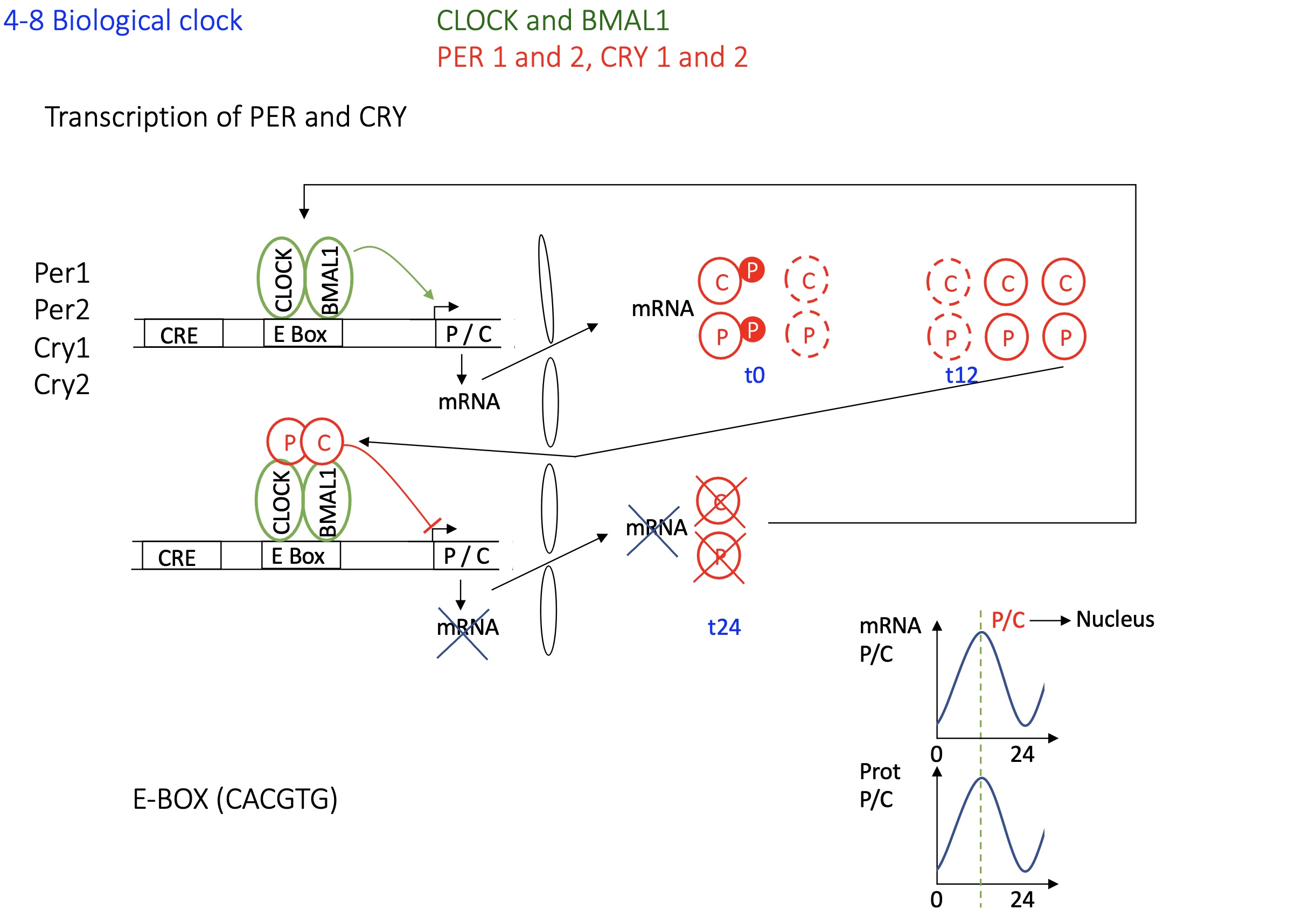
1.4.8 transcription of PER and CRY control biological clock
Activation Phase:
CLOCK and BMAL1 (Act) form a heterodimer and bind to E-box elements in the promoters of Per and Cry genes —> activation of transcription of Per and Cry mRNAs.
Translation and Post-translational Modification:
Per and Cry proteins are translated in the cytoplasm
They undergo phosphorylation
Formation of Repressor Complex:
Per and Cry form dimers
The P-C dimer translocates into the nucleus when there’s an accumulation of P-C dimer in cytoplasm
Negative Feedback:
In the nucleus, the P-C dimer binds to CLOCK-BMAL1 on E-box, inhibiting further transcription of Per and Cry
Degradation Phase and Cycle Reset:
Over time, P-C dimer detaches from Act proteins when too little P or C protein in the cytoplasm
new transcription of Per and Cry begin
Cycle Continuation
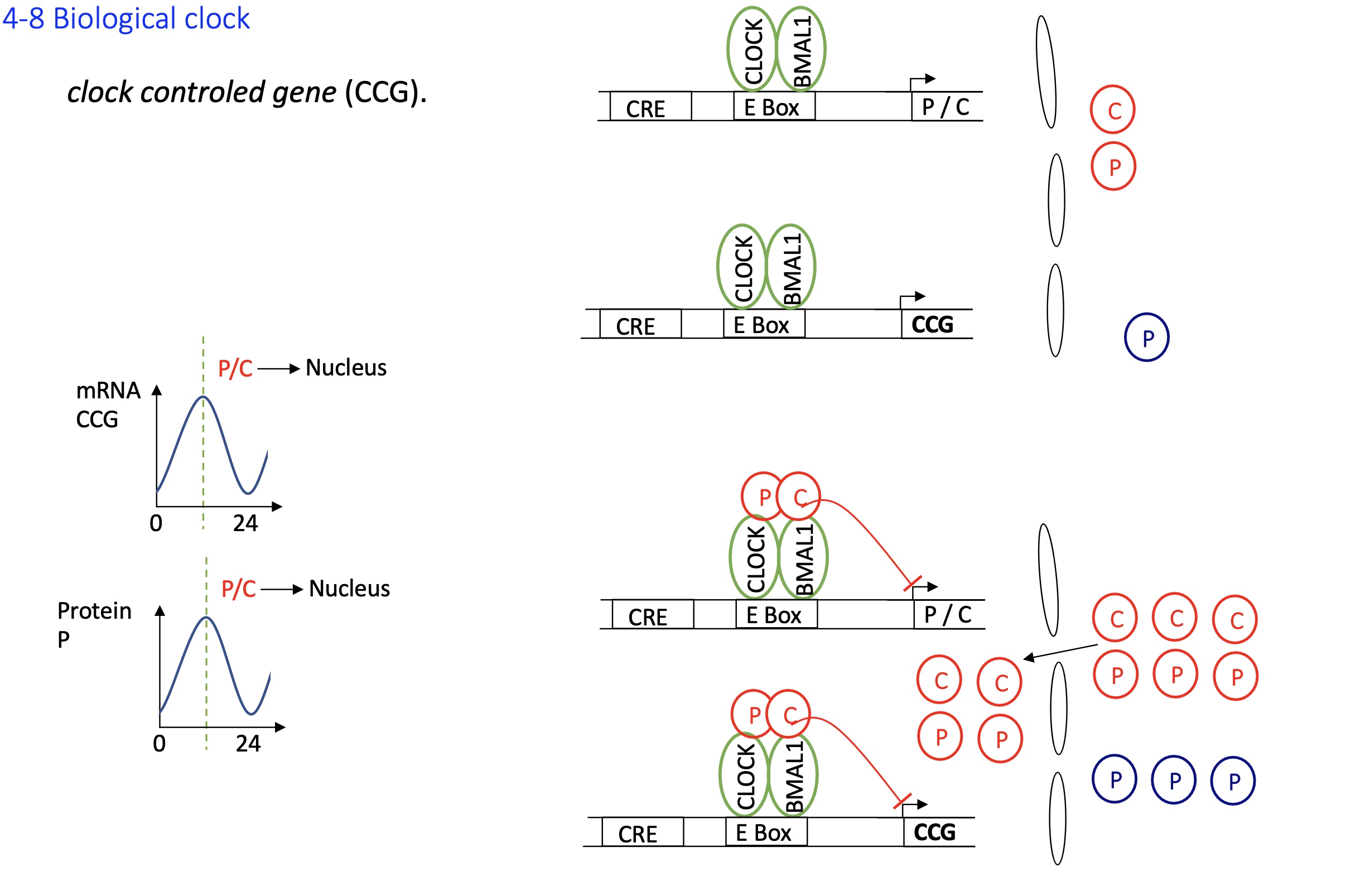
1.4.8 how is the clock control gene CCG regulate gene expression
CCGs have E-boxes elements in their promoters
When CLOCK-BMAL1 is active (i.e., not inhibited by PER-CRY), it binds to these sites and activates transcription of CCGs
when C and P accumulates the C-P dimer translocates into nucleus and binds to Act proteins on e-box the transcription of CCGs are inhibited
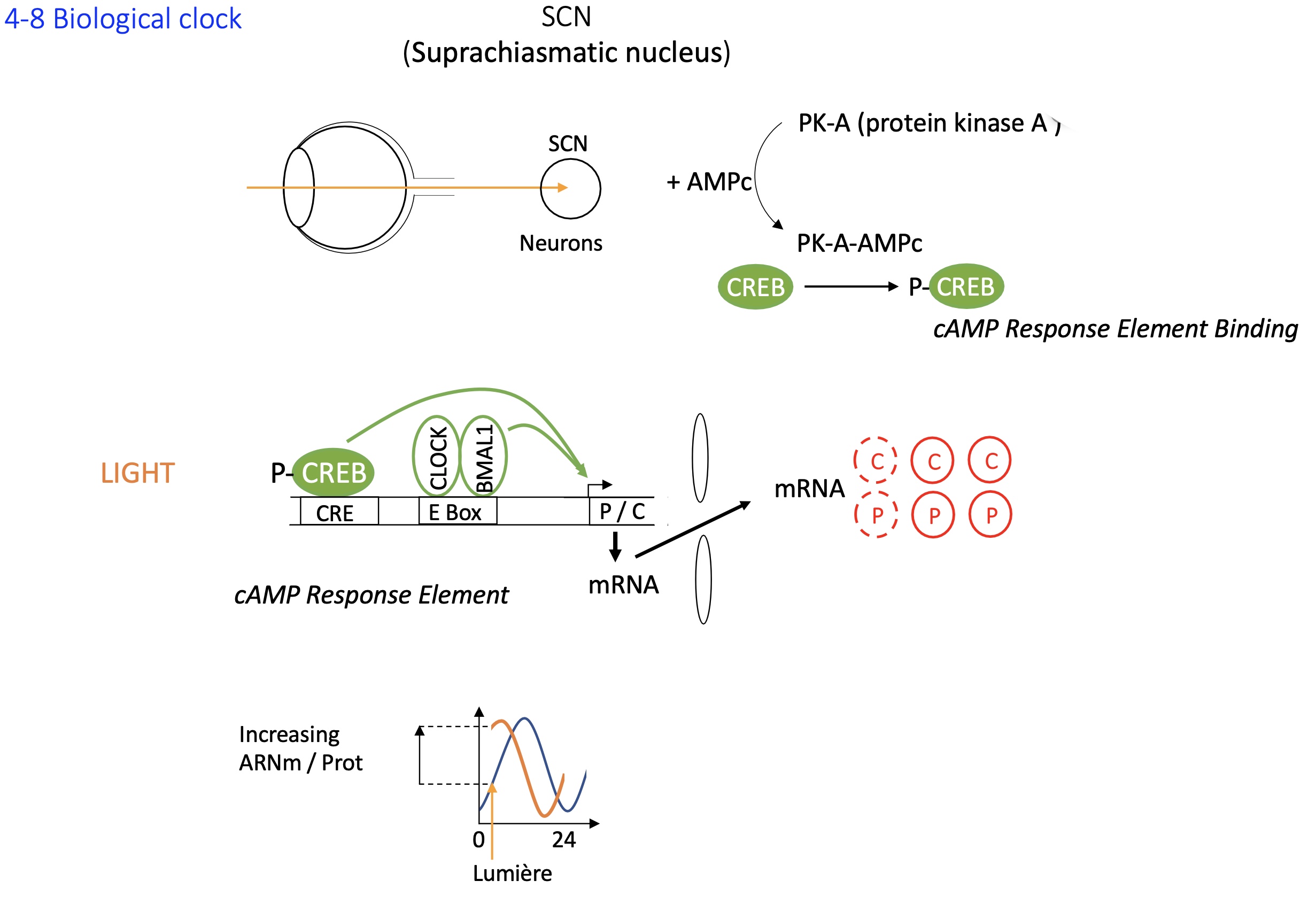
1.4.8 biological clock : SCN suprachiasmatic nucleus
sigh stimulates SCN neurons
PKA + AMPc —> phosphorylates CREB
P-CREB binds distal binding site (CRE BS)
this along w/ the binding of CLOCK & BMAL1 to e-box activates the transcription of Per and Cry resetting the clock
Separately, the CLOCK:BMAL1 complex regulates circadian transcription independently of light input:
CLOCK and BMAL1 are TF that form a heterodimer and bind to E-box elements in promoters of Per and Cry genes.
PER and CRY proteins accumulate, form complexes, and then inhibit CLOCK:BMAL1 activity — this creates the ~24-hour oscillation.
1.4.9 how is the transcriptional memory established
Activation of gene transcription
A signal activates an activator (TF₁) that binds to its binding site/enhancer in the promoter region of gene A
Gene A is transcribed and translated into protein A (TF₂).
TF₂ binds to an activator binding site in its own promoter, reinforcing its own transcription
This self-sustaining transcriptional loop establishes transcriptional memory, allowing the gene to stay active even after the original signal is gone
Epigenetic marks/histone modifications :
Upon activation due to a signal, the TF recruits a writer (e.g., histone acetyltransferases)
These “writer” enzymes add activating histone marks, at the promoter/enhancer
These epigenetic marks keep chromatin in an open, transcriptionally active state = euchromatin
The marks persist, maintaining continued transcription — even in the absence of the original stimulus.
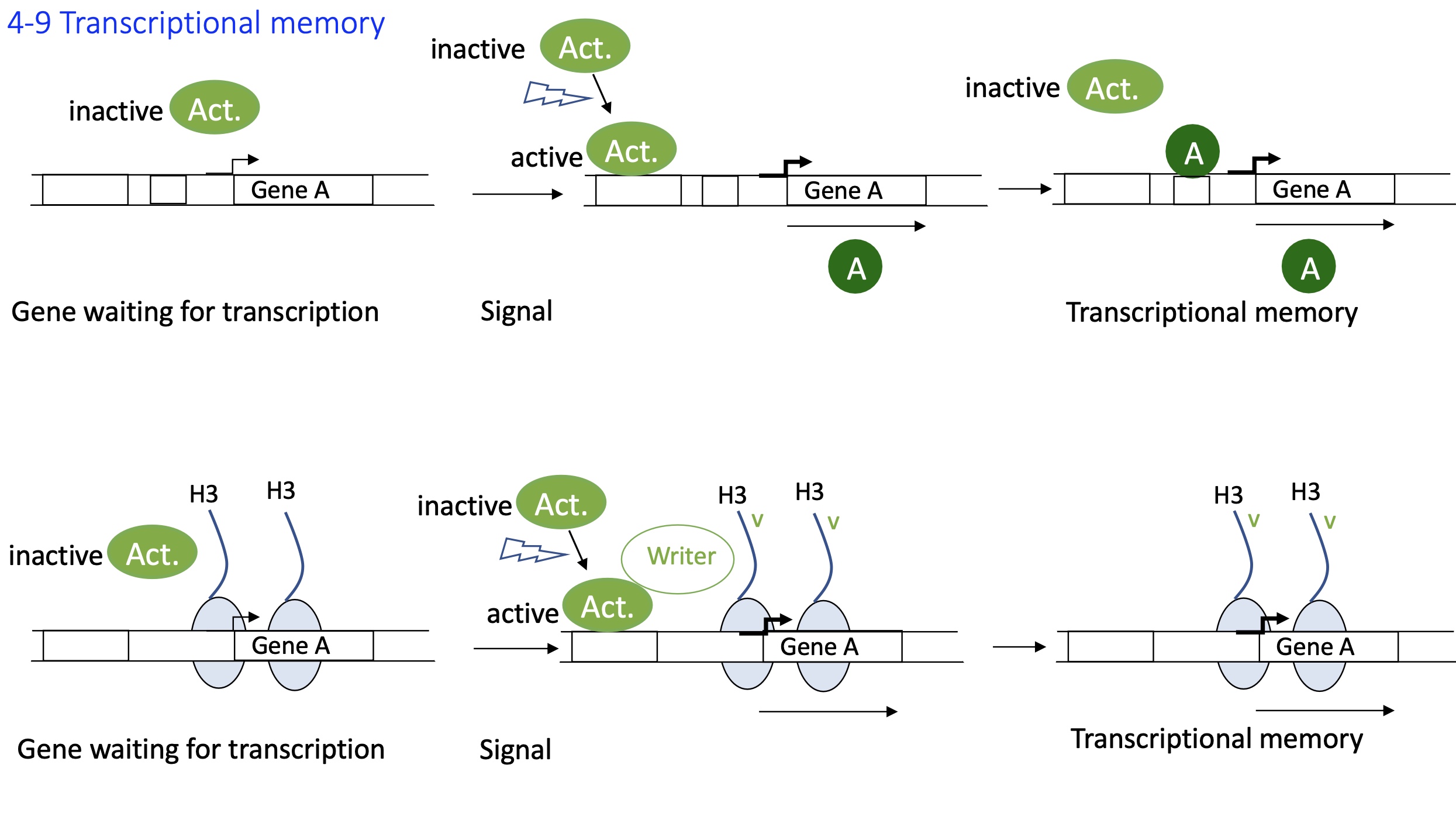
1.4.9 how is transcriptional memory used in cell division
Epigenetic marks/Histone modification :
TF/act protein activated due to a signal
TF binds to enhancer and recruits writer
Histone modification takes place and remains on the newly synthesised cells
writer can recognise histone tag on these newly synthesised cells
gene A will have the same signal in new cells
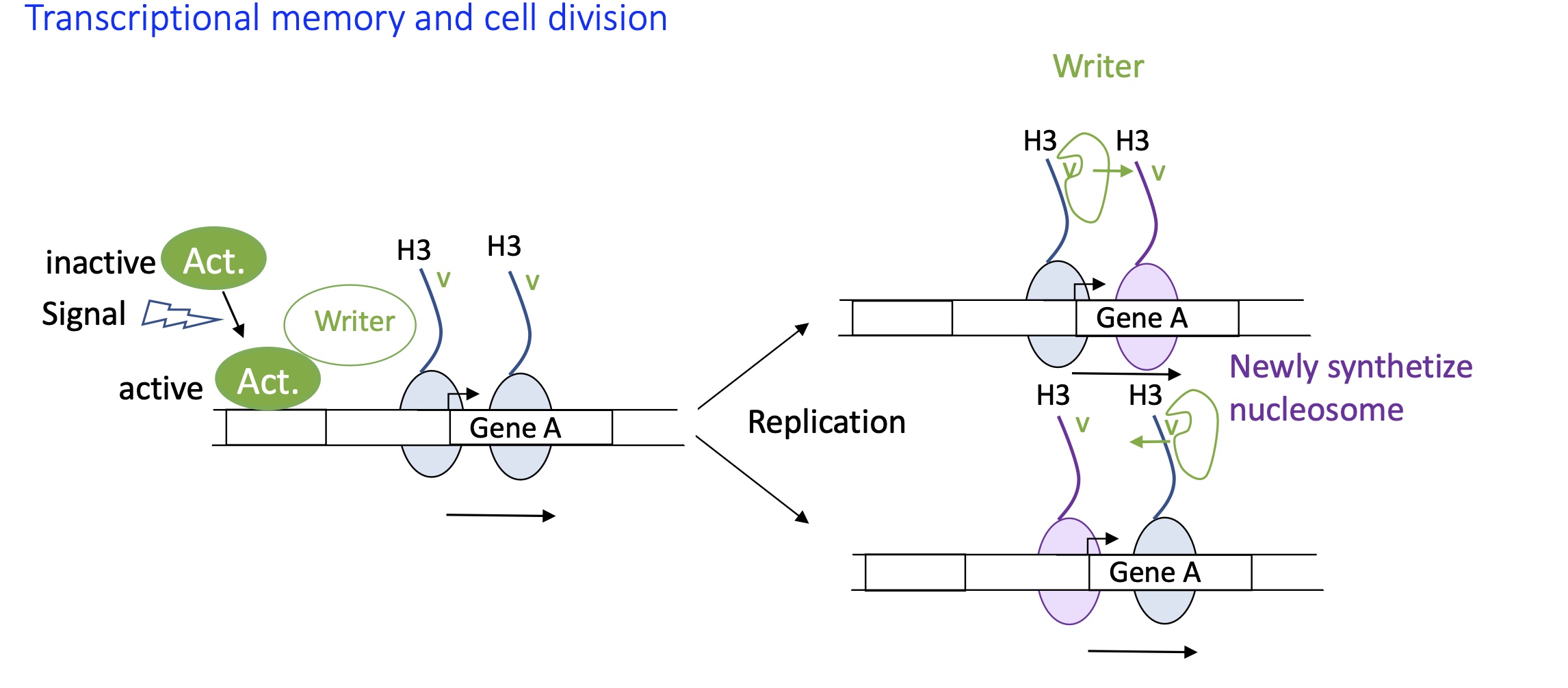
1.4.10 Control of known transcription initiation by a lncRNA (2 known roles)
known role :
lncRNA HOT AIR repression of HOX D gene : recognises specific sequences that will recruit silencing writers methyl transferase and de-methylase —> gene silencing
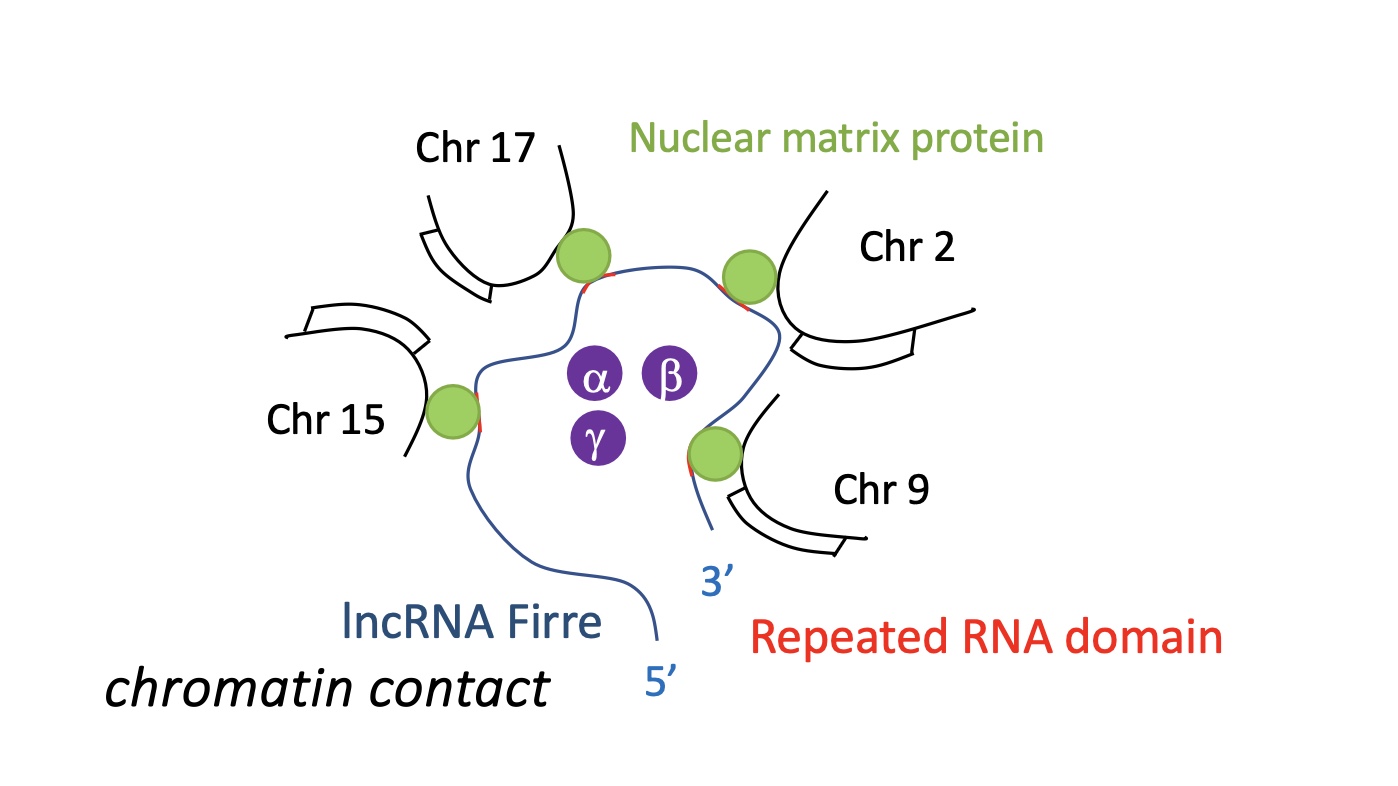
lncRNA Firre : nuclear organiser, 8 different mRNA sequences on lncRNA Firre recognises a specific nuclear matrix proteins. this protein recognises specific sequence on chromosome and brings the chromosomes together to create chromatin contact
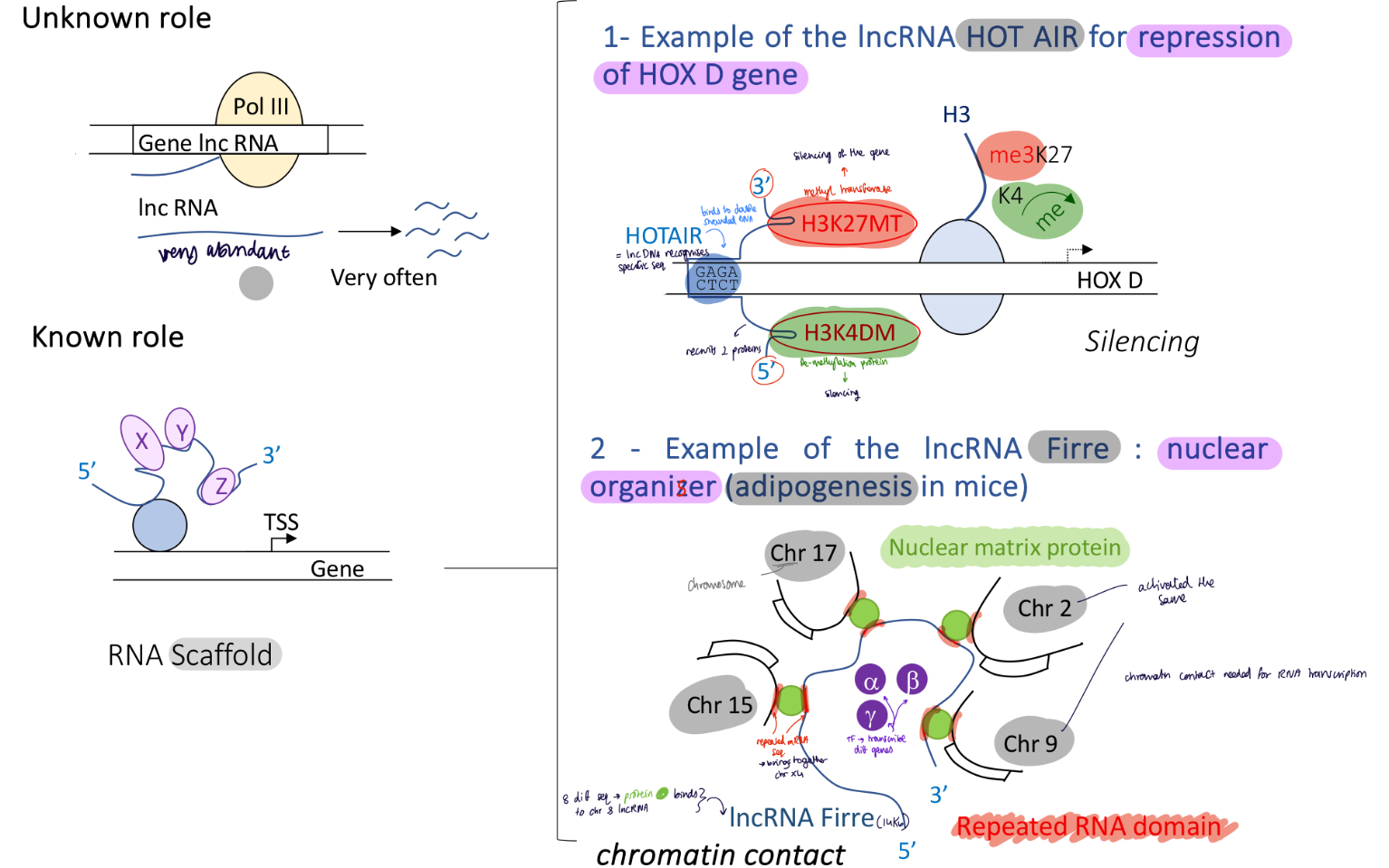
1.4.11 3D gene organisation and topology associated domains thanks to activator and mediator
If two genes are too far away on different chromosomes :
Act will bind to enhancer in chromosome 1
mediator recruited which will bring the two chromosomes together and induce transcription go both genes
If two genes are too far away on the same chromosomes :
DNA bending protein will bind to the chromosomes and bed the the DNA sequence to bring two genes tother. transcription activated by Act + mediator
1.4.11 how does 3D genome organisation take place
inside chromosomes territories there are TAD domains (topologically associated domain) that are isolated by CTCF insulator proteins so that only the genes present on TAD are transcribed
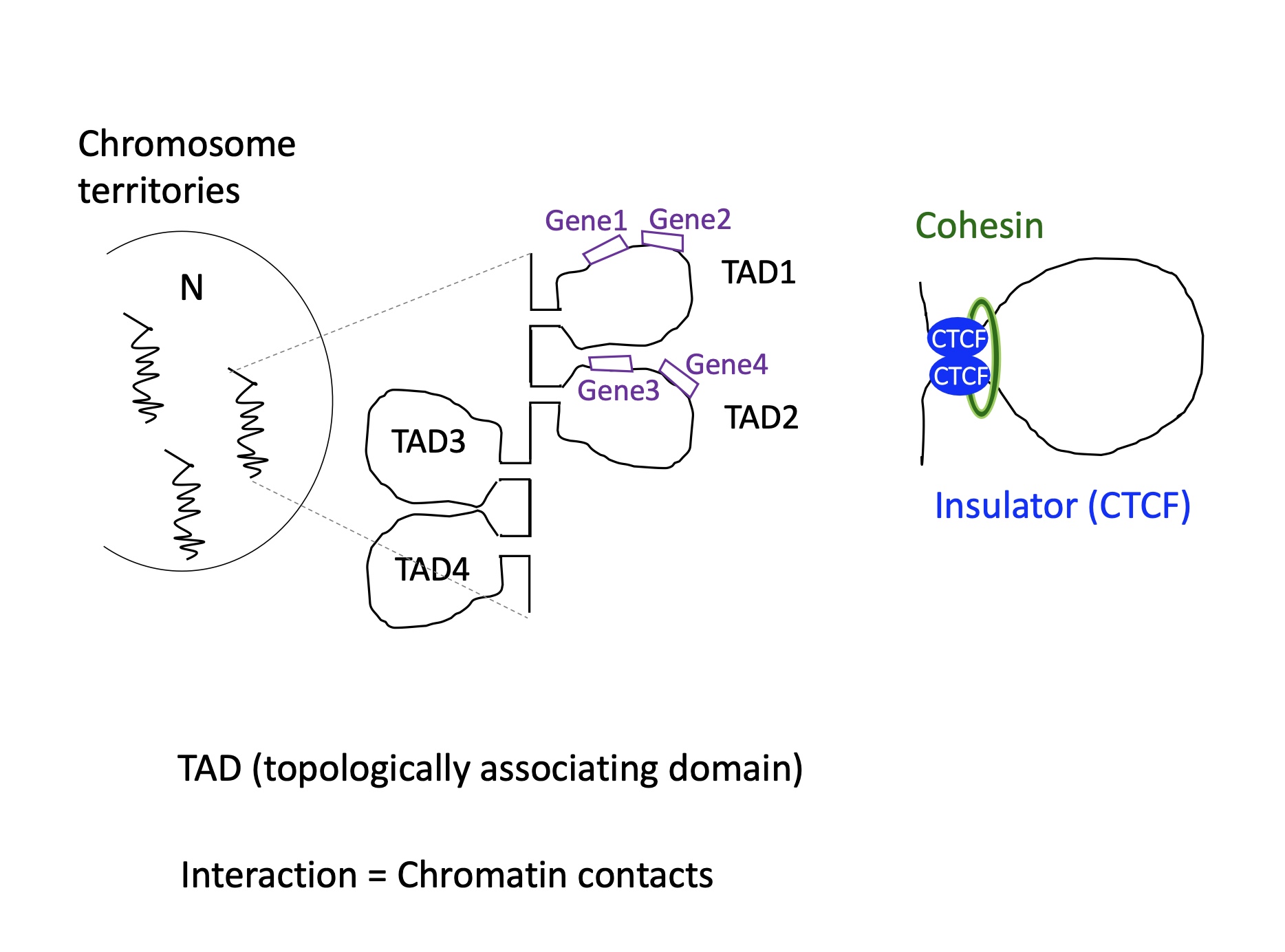
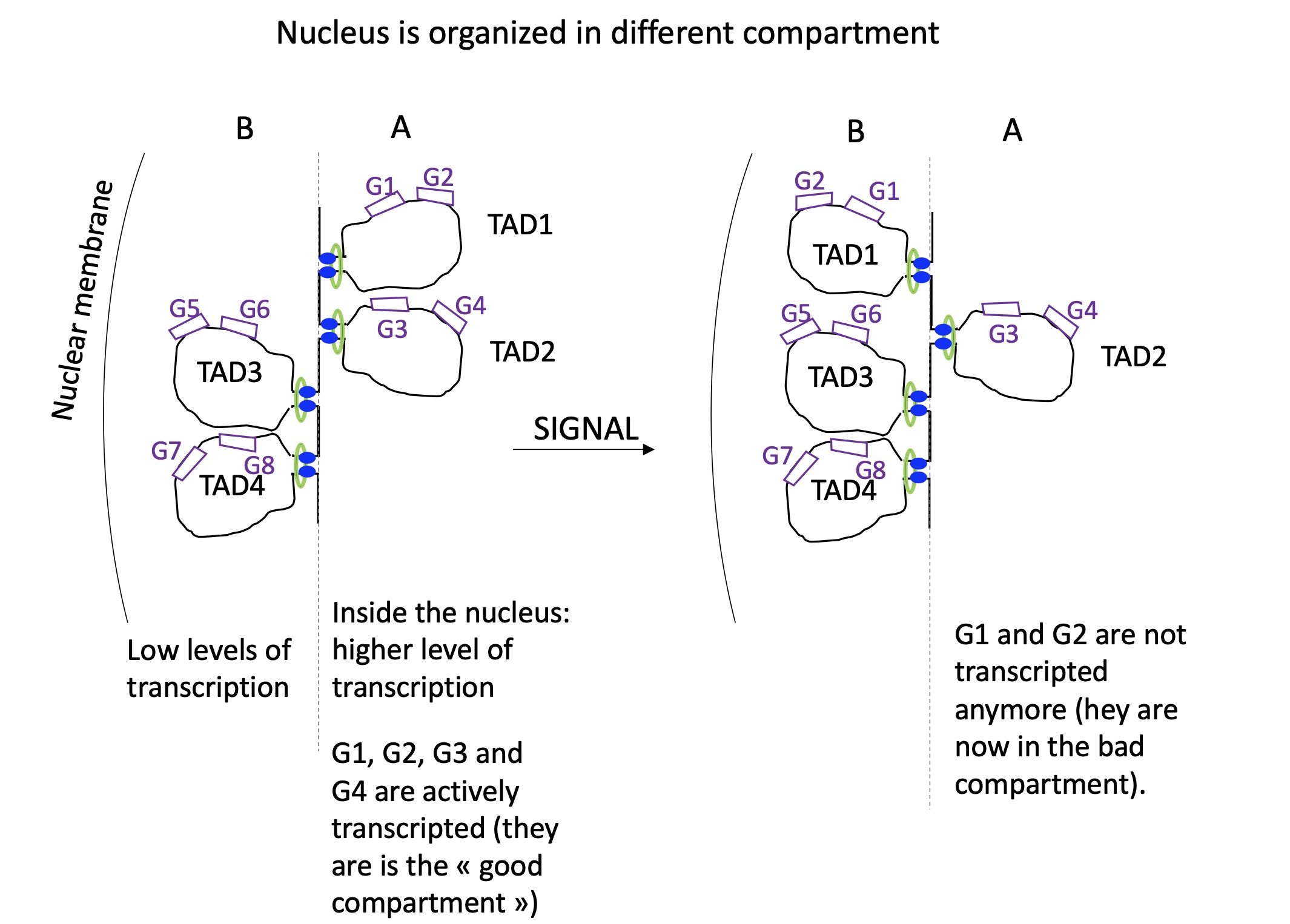
1.4.11 following a signal what happens the nucleus organisation and TAD reorientation
in the nucleus we have two compartments : A = compact (heterochromatin) and B = less compact and central (euchromatin)
following a signal TAD domain can change department which will change the expression of genes on TAD
1.4.11 what lncRNAs are involved in 3D genome organisation
lncRNA Firre
lncRNA Xist
1.4.11 how does lncRNA Firre rearrange the genome
lncRNA Firre which assembles multiple chromosomes together by recruiting nuclear matrix proteins that recognises sequences on chromosomes along with specific sequences repeated RNA domains on lncRNA Firre
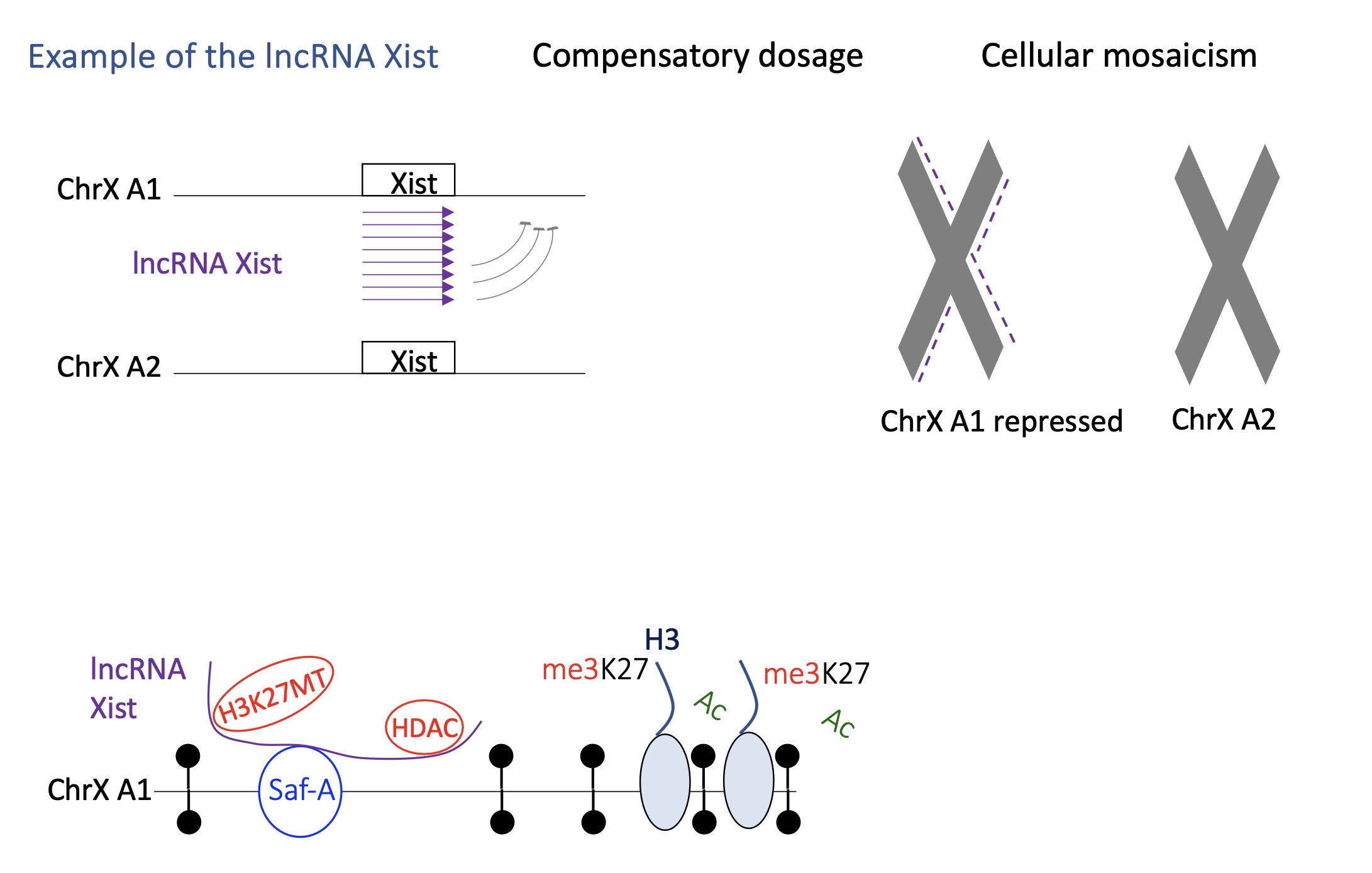
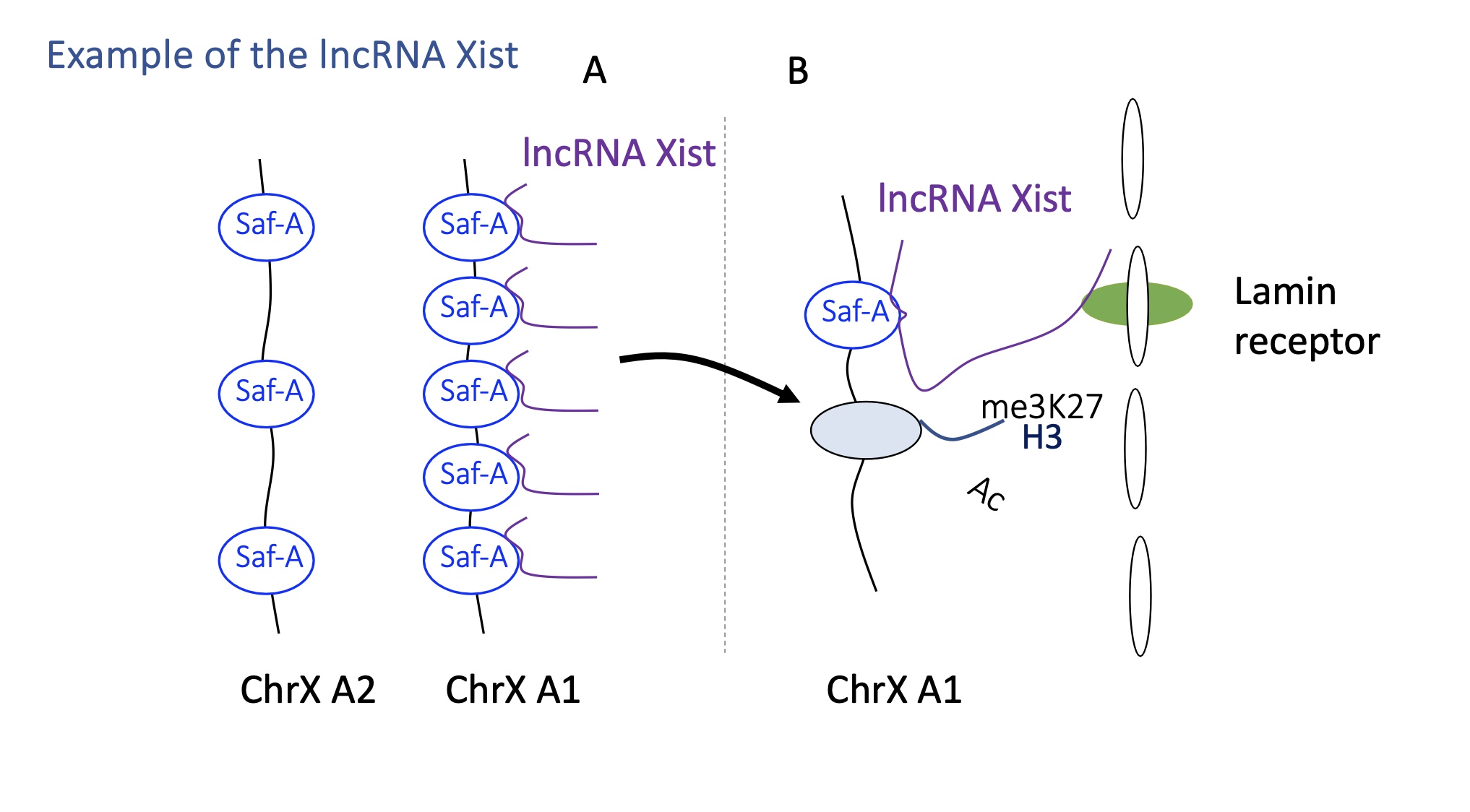
1.4.11 how does lncRNA Xist switch between active/inactive transcription
lncRNA Xist acts on one of the chromosomes X in females where only one chromosome X remains active during the embryonic development
Xist condenses the chromosome to inactivate it :
Xist is only expressed but the chromosome that will be inactivated
Xist will recognise a protein SafA present on chromosome X which helps it attach to chromosome
once bound to SafA —> recruitment of writers that silence gene expression e.g H3K27MT and HDAC
the inactivated chromosome X is brought o compartment B (Heterochromatin)
Xist interacts w/ lamin receptor on nuclear membrane which allows the inactive chromosome to remain in the heterochromatin/B heterochromatin
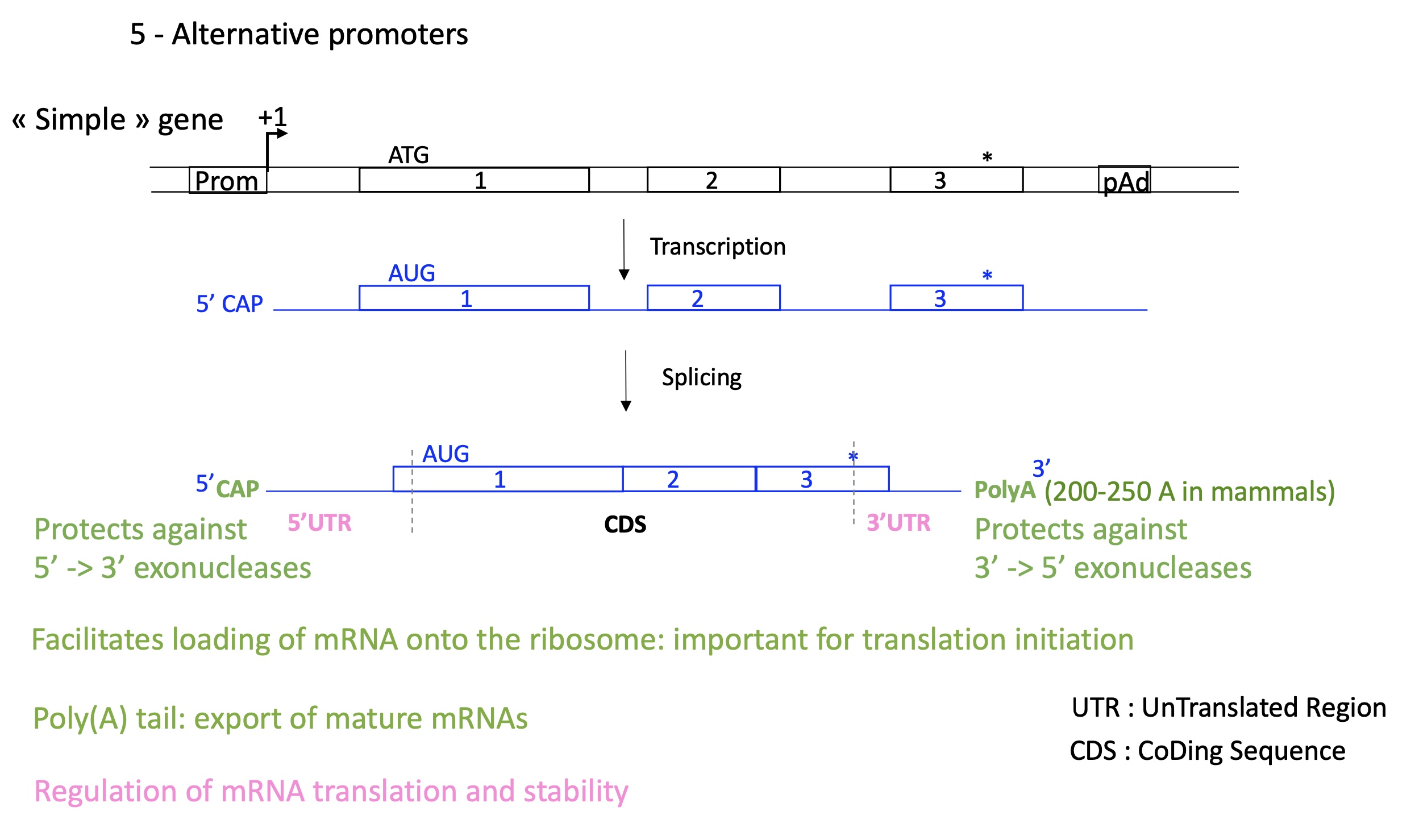
1.5 how does alternative promoters control transcription initiation in simple genes
5’ CAP & 3’ Poly A : protects against exonuclease
5’ CAP facilitates loading of mRNA on ribosome (translation)
3’ polA export of mature mRNAas
regulate mRNA translation and stability
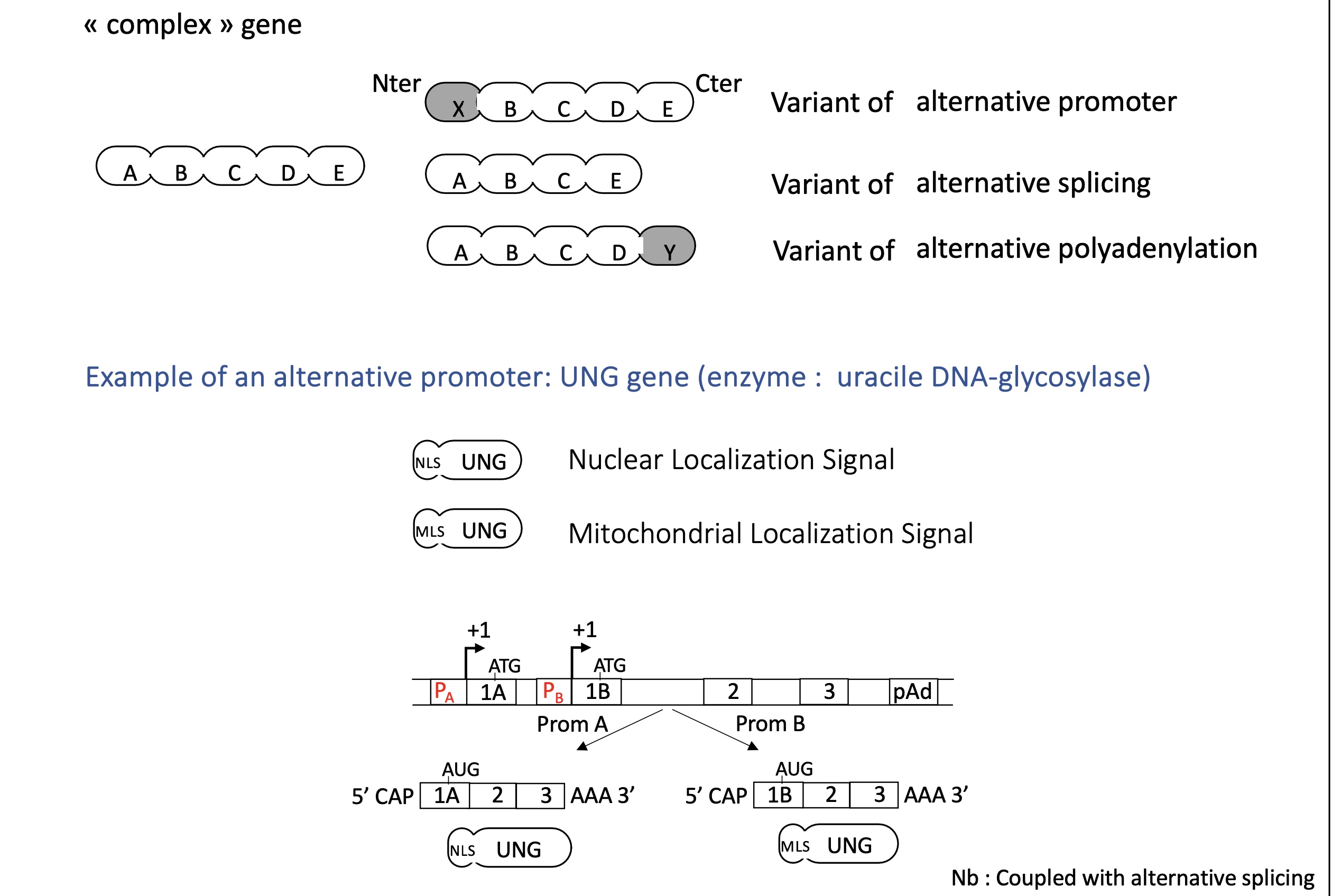
1.5 how does alternative promoters control transcription initiation in complex genes
Alternative promoters allow complex genes to initiate transcription from different start sites under different regulatory conditions.
alternative promoter can change protein addressing e.g nuclear/mitochondrial localisation signal
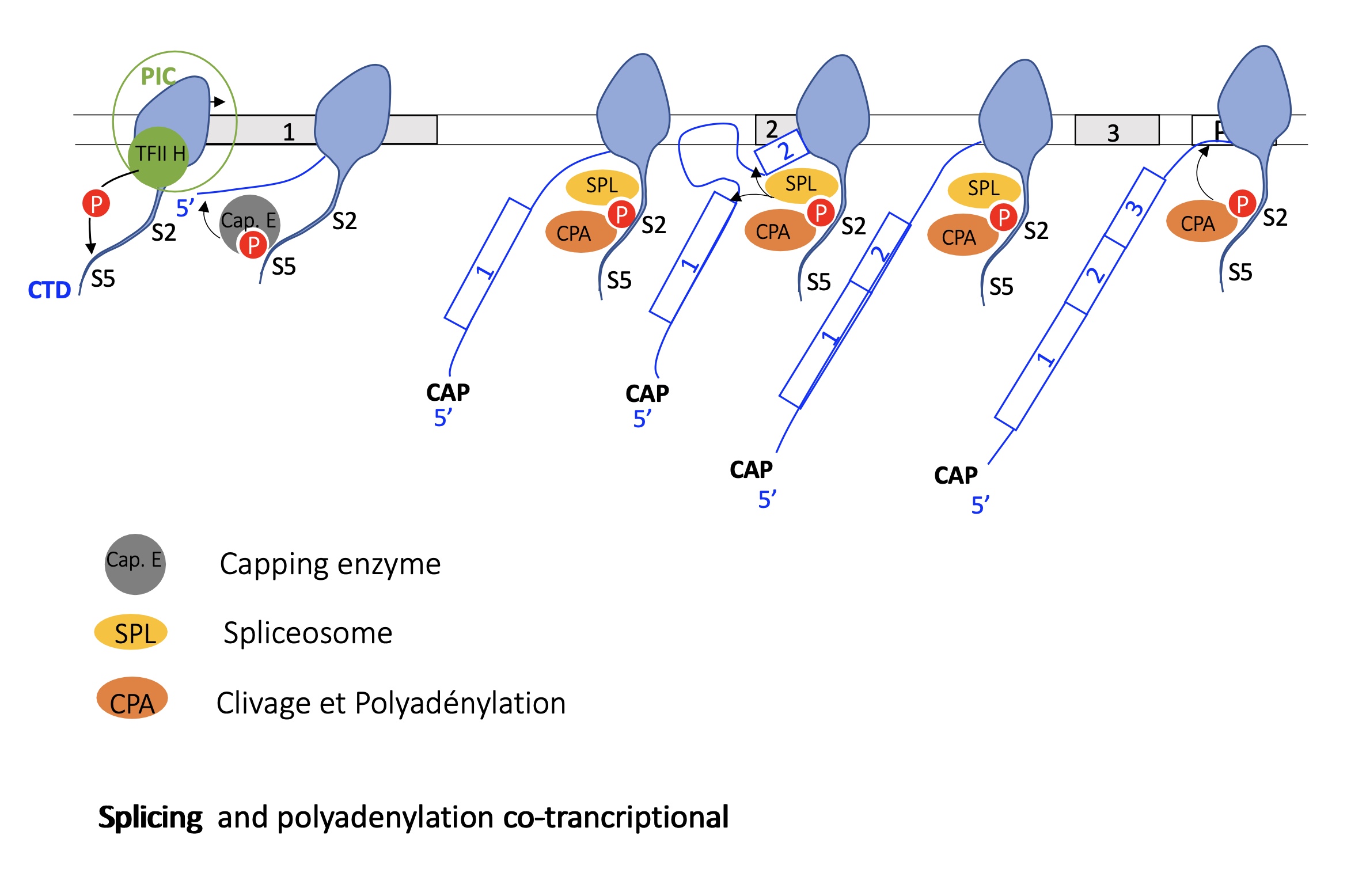
the role of RNA Pol II CTD after transcription initiation
hybridisation of PIC to promoter = initiation of transcription
phosphorylation of S5 by TFIIH on RNA pol II CTD recruits Capping enzyme —> capping of mRNA during translation
once capping is done the phospho-SER5 is dephosphorylated
phosphorylation of ser2 : phospho-S2 recruits splicing complex (SPL + CPA) —> co-transcriptional splicing takes place
SPL dissociates at the end of transcription and CPA remains and stops transcription at polyA site
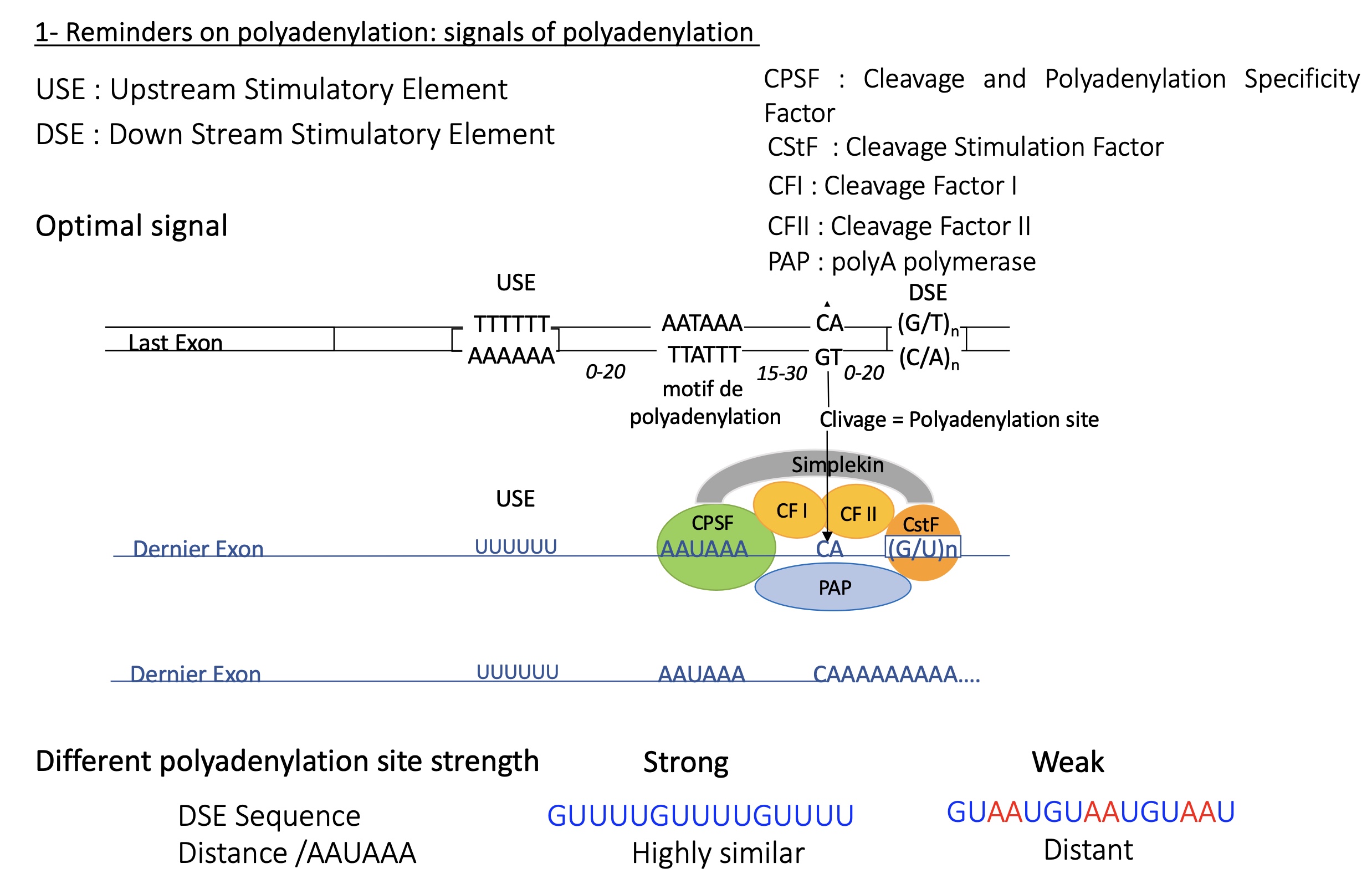
3.1 how does the cleavage cleavage at polyA on mRNA take place
DNA sequence :
USE = upstream stimulatory element (T/A)
polyadenylation motif (T/A)
polyadenlyation/cleavage site
DSE =down stream stimulatory element
mRNA transcribed : factors recruited due to specific sequence on mRNA from DNA
CPSF (cleavage and polyadenylation specific factor) binds to polyadenylation motif
CF I/II (cleavage factors) and PAP (polyA polymerase) binds to polyadenylation site
CstF (cleavage stimulation factor) binds to DSE
the further the DSE sequence resembles the consensus sequences the healer it is and therefore polyA is less
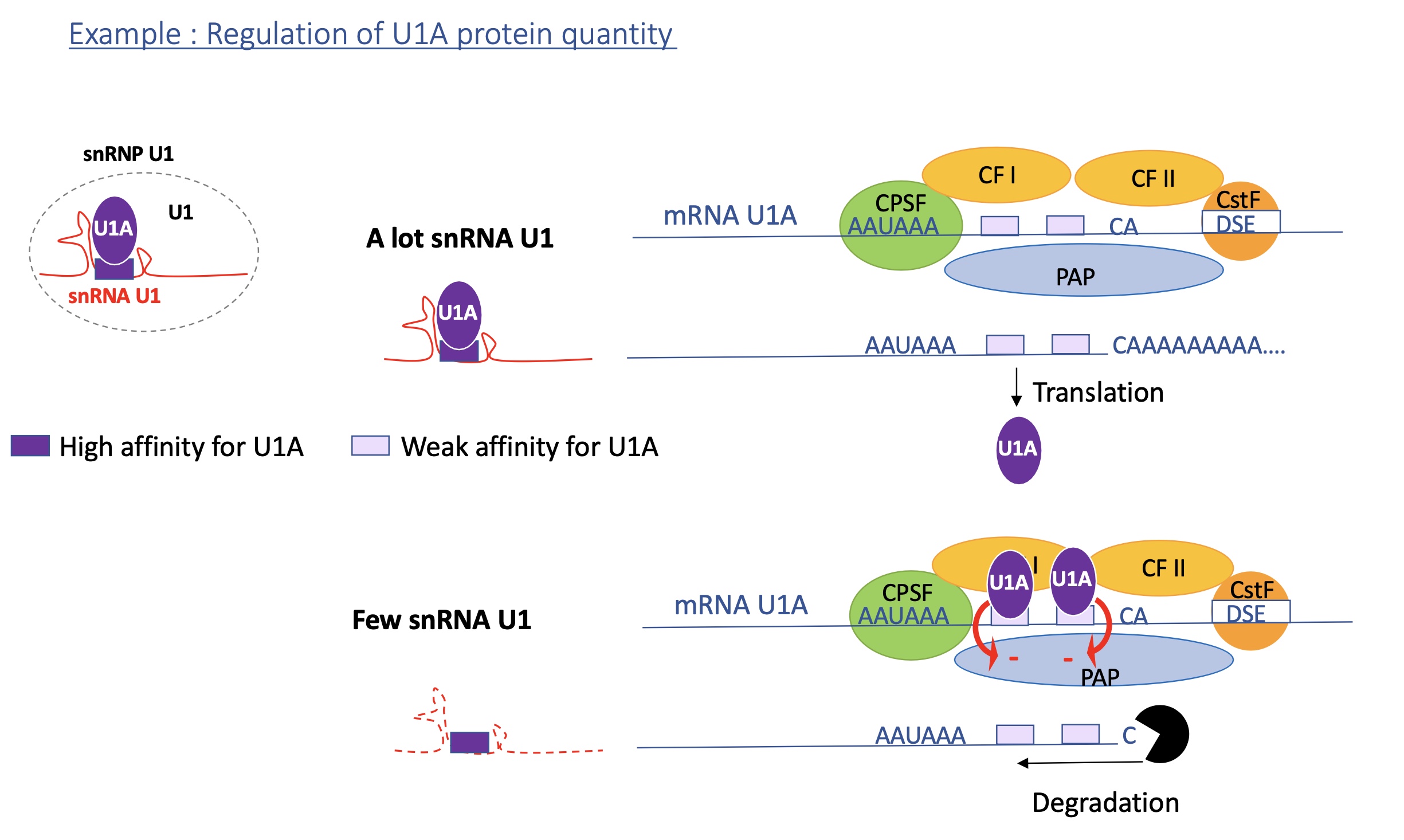
3.2 polyadenylation efficiency can be controlled by protein such as U1A protein, how?
When U1A protein levels are high (few snRNA U1 complex):
Excess U1A now binds to both U1A binding sites in its own mRNA’s 3′ UTR.
When two U1A molecules bind, they interact with poly(A) polymerase (PAP) and inhibit its activity.
Result:
❌ No (or very short) poly(A) tail added →
❌ mRNA is unstable and rapidly degraded by exonucleases.This reduces production of more U1A protein.
When U1A protein levels are low (lots of snRNA U1 complex):
There is little U1A available in the cell as lots bound to snRNA
The U1A mRNA has two weak U1A binding sites in its 3′ UTR, near the polyadenylation signal (AAUAAA).
Because U1A levels are low, these sites remain mostly unbound.
This allows poly(A) polymerase (PAP) to bind normally →
✅ Efficient polyadenylation → stable U1A mRNA → translation of more U1A protein.
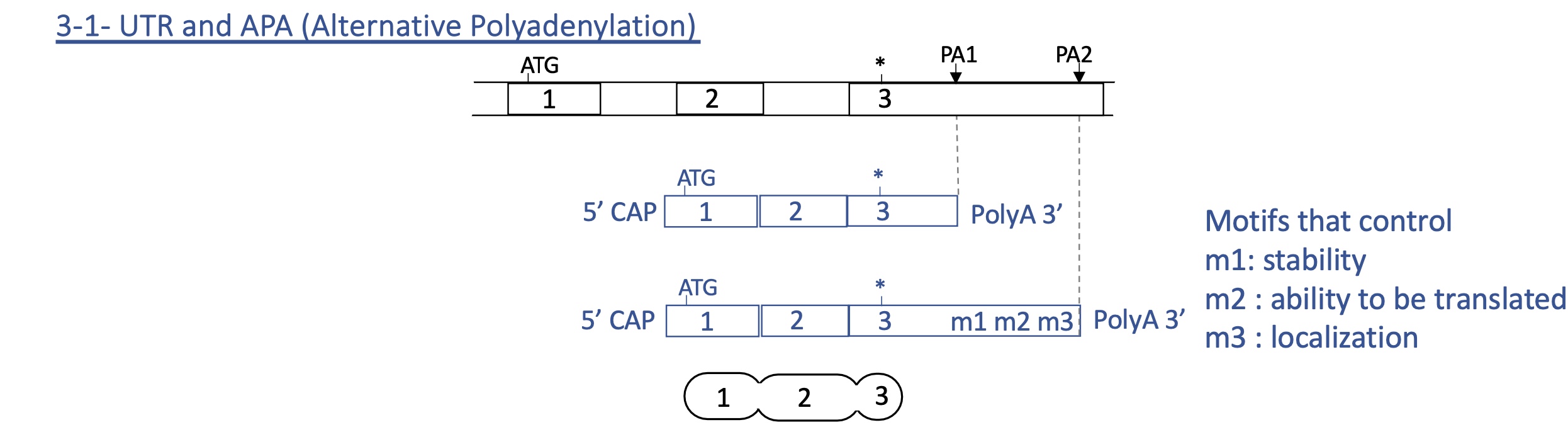
3.3. strength of alternative polyadenylation sites
proximal sites PA1 = weak
distal sites PA2 = strong
3.3. 3’ UTR (untranslated region) and alternative PA
if PA1 used : little to no effect on stability, localisation or ability to be translated
if PA2 used : can effect stability, localisation and ability to be translated
=> final protein remains unchanged but its quantity and localisation can vary depending on PA used
3.3. CD coding region and alternative PA
if PA are places in different coding regions
PA1 used —> shorter coding sequence translated
PA2 used —> longer coding sequence translated
=> depending on PA used proteins may differ
3.3. how does the choice of alternative PA effect transcription
APA (alternative polyadenylation) modulates :
level of cstF (cleavage stimulation factor)
elongation rate of RNA pol II
splicing
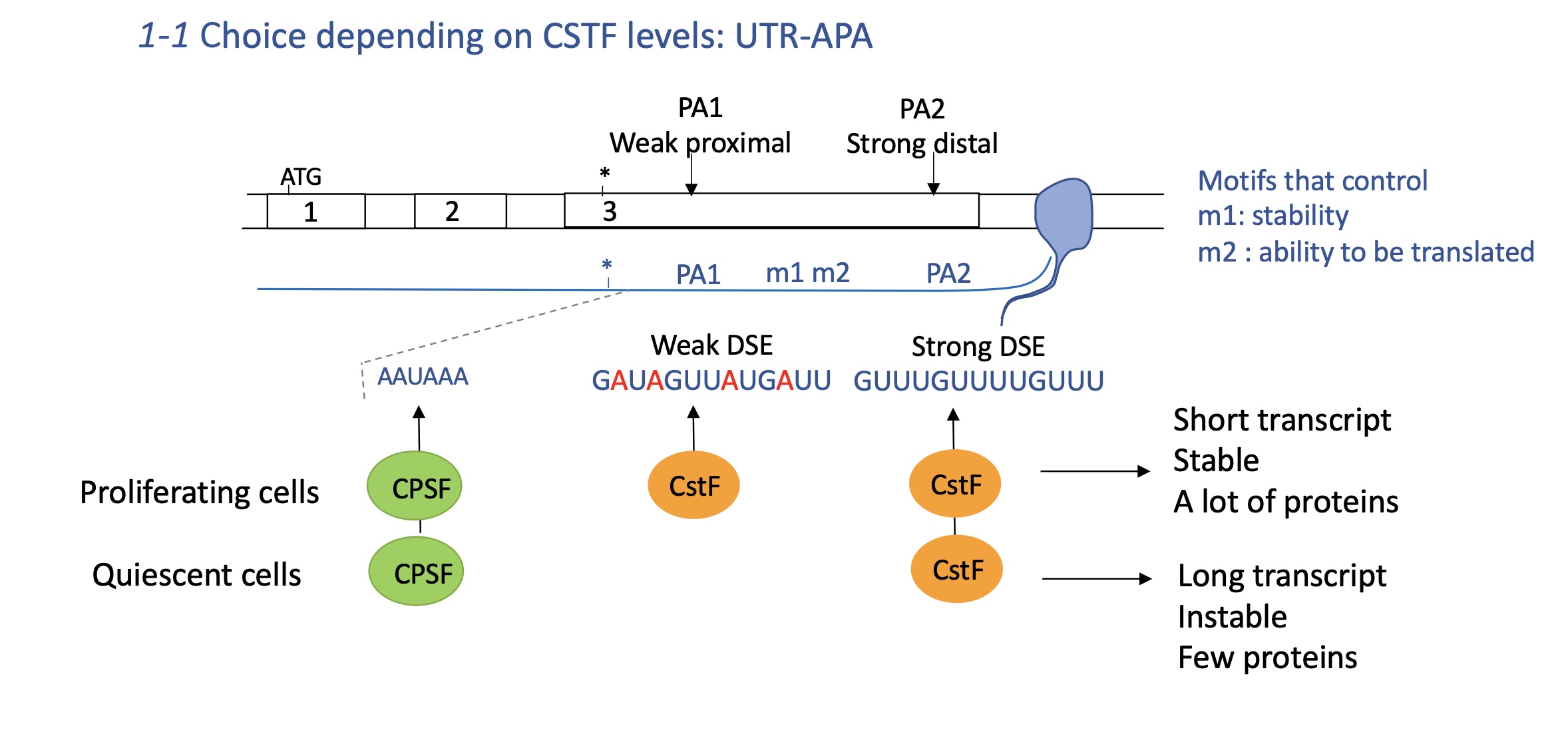
3.3.1 how does the choice of APA in UTR effect CstF levels
CPSF (cleavage and polyA specificity) and CstF (cleavage stimulation factor)
USE Upstream stimulator element
DSE downstream stimulator element
Quiescent cells : non proliferating cells so need less CPSF and CstF
CPSF binds to USE and little CstF binds to strong distal DSE PA2
mRNA is longer and has motifs that control stability and ability to be translated
longer transcript —> instable and few proteins translated
proliferating cells : needs more CPSF and cstF
CPSF binds to USE and CstF binds to both DSE of PA1 and PA2
increased polyA at PA1 —> short transcript w/out motifs that control stability and ability to be translated
proteins translated are stable and many are produced (necessary for proliferation )
3.3.1 how does the choice of APA in CR coding region effect CstF levels
CstF cleavage stimulator factor
DSE downstream stimulator element
few CstF :
CstF binds to DSE to strong distal PA2
coding sequence between PA1 and PA2 excluded
—> Ig in the membrane of Bcell = BCR
lots of CstF :
CstF binds to DSE of PA1 and PA2 —> PA1 favoured
coding sequence between PA1 and PA2 spliced/included
—> secreted Ig from pasta cell
this can effect whether Ig will be transmembrane or cytoplasmic
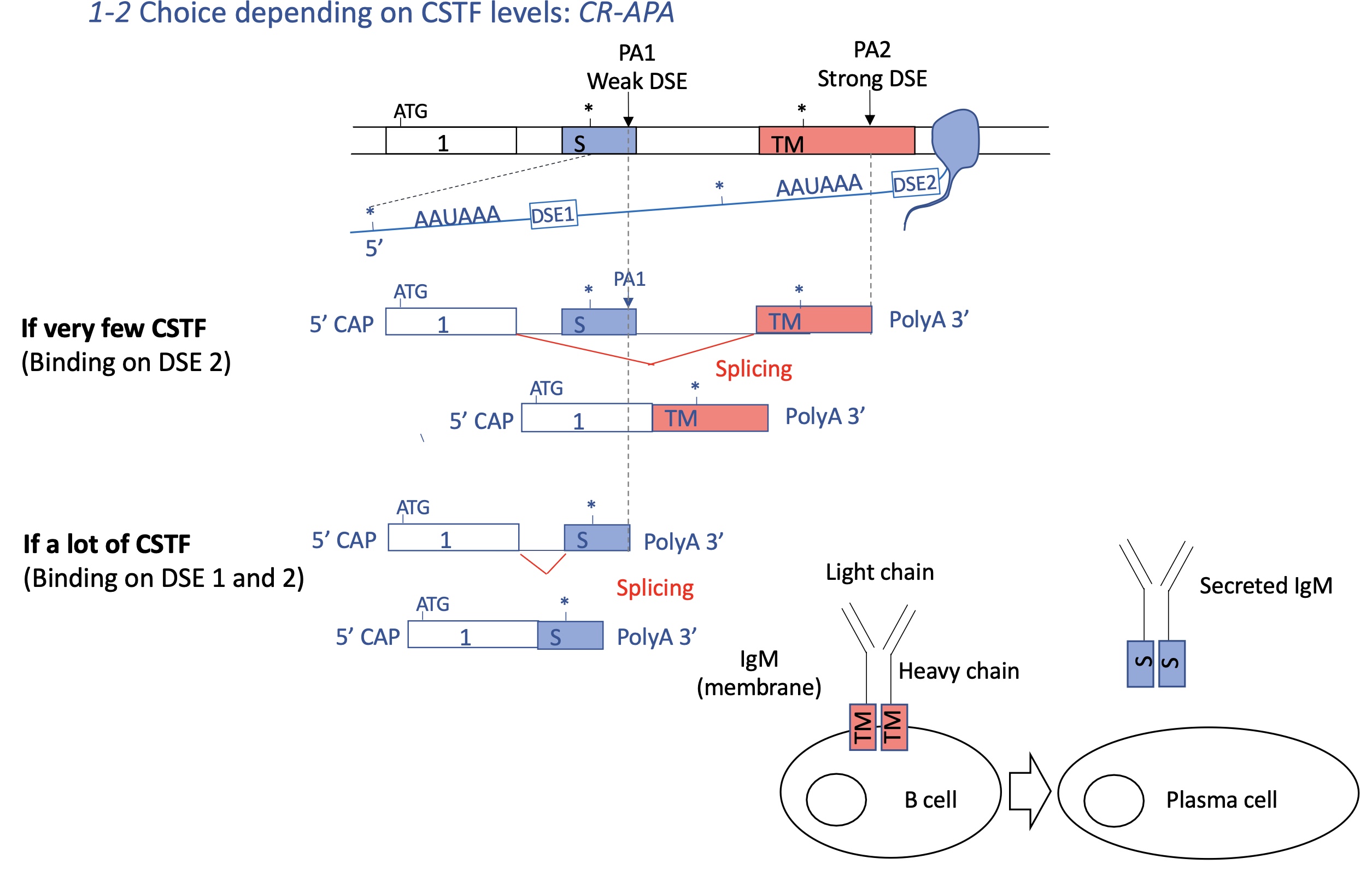
3.3.2 how does the choice of PA effect elongation rate of RNA pol II
protein attached to RNA pol II can modify its conformation which changes its speed of transcription
A slower Pol II gives RNA processing complexes more time to act, promoting use of proximal poly(A) sites/PA1
faster Pol II often skips these and uses distal sites/PA2, producing longer mRNA
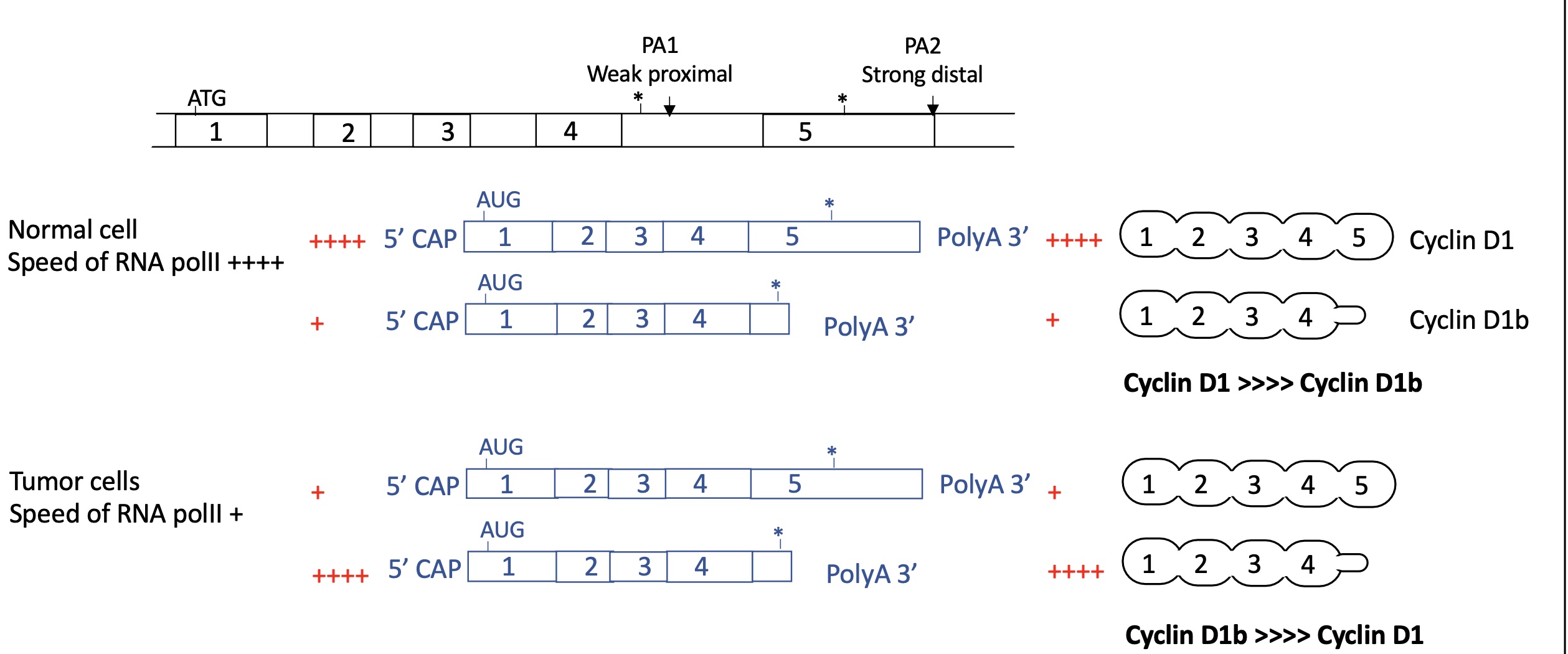
3.3.2 how does the speed of RNA pol II effect the use of APA and so change the expression of cyclin D1
normal cell :
ARN pol II moves rapidly
so proximal PA1 is skipped and distal strong PA2 is used
long mRNA w/ 5 exons corresponding to cyclin D1
Cyclin D1 >> cyclin D1b
tumour cell :
RNA pol II moves slow
proximal PA 1 used as factors have more time to bind and induce polyA
short mRNA w/ 4 exons expressed corresponding to cyclin D1b
Cyclin D1 << cyclin D1b
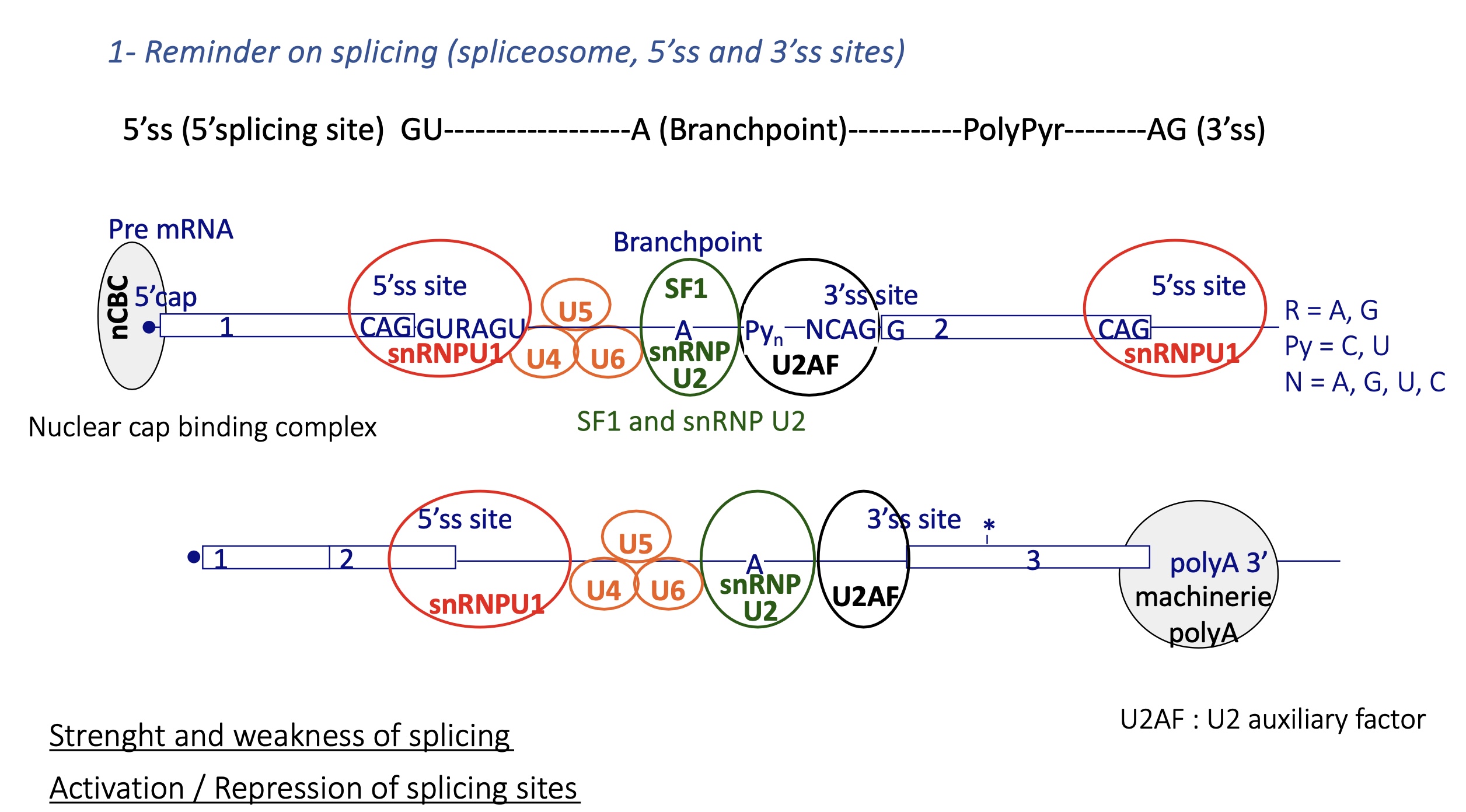
3.3.3 how does splicing take place
premRNA :
nuclear Cap binding complex binds to 5’ CAP —> define beginning of exon and prevents degradation
snRNPU1 at the 5’Splicing Site SS of the intron
U2AF binds to 3’SS = auxiliary factor
SF1 and snRNP U2 is recruited at branch point thanks to U2AF and binds to branch point
snRNP U2 recruites other snRNP U4/5/6 and SF1 is removed
= spliceosome
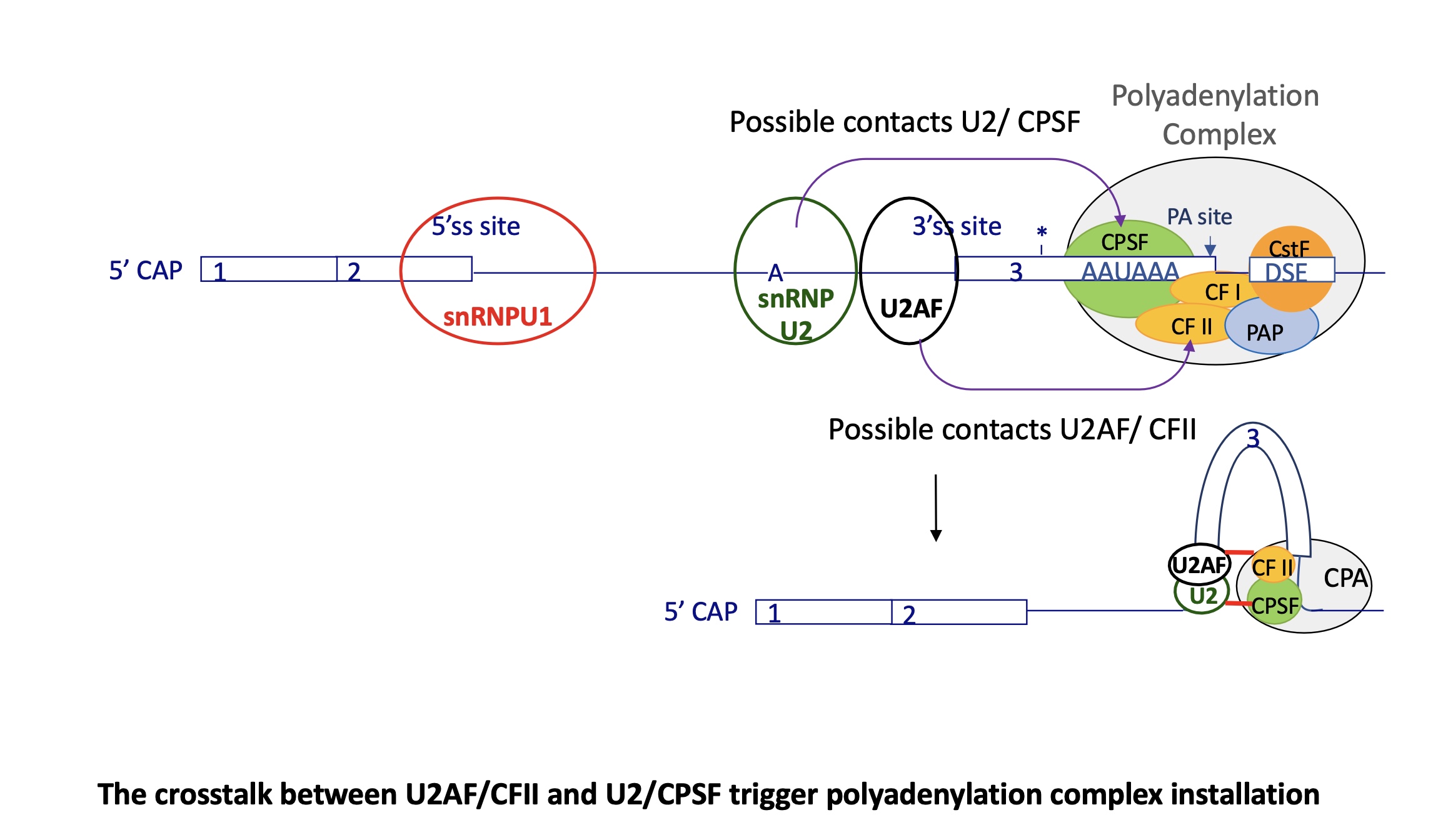
3.3.3 how does splicing change the choice of polyA (APA)
the interaction of U2Af at 3’SS w/ CFII (cleavage factor)
+ the interaction of snRNP U2 at branch point w/ CPSF (cleavage and polyA specific factor) triggers polyA complex installation
this causes the exon to be caught in this crosslink
is the control of splicing another way of controlling transcription ?
YES
splicing sites can have different strengths depending on how close they are to the consensus sequence : pyr (C/U)
weak sequence : pur (A/G)

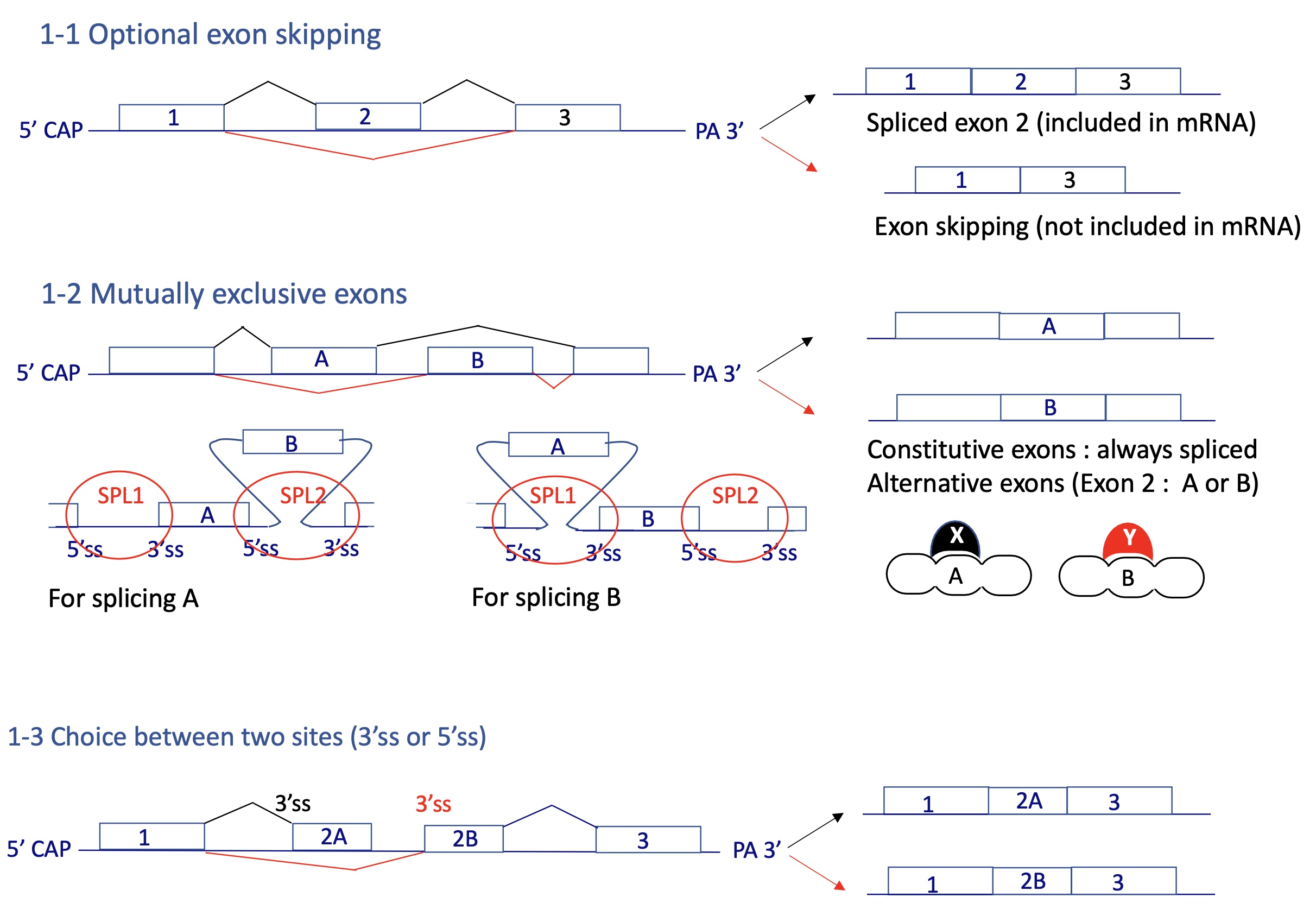
what are the different types of alternative splicing
optional exon skipping
mutually exclusive exons
choice between two SS
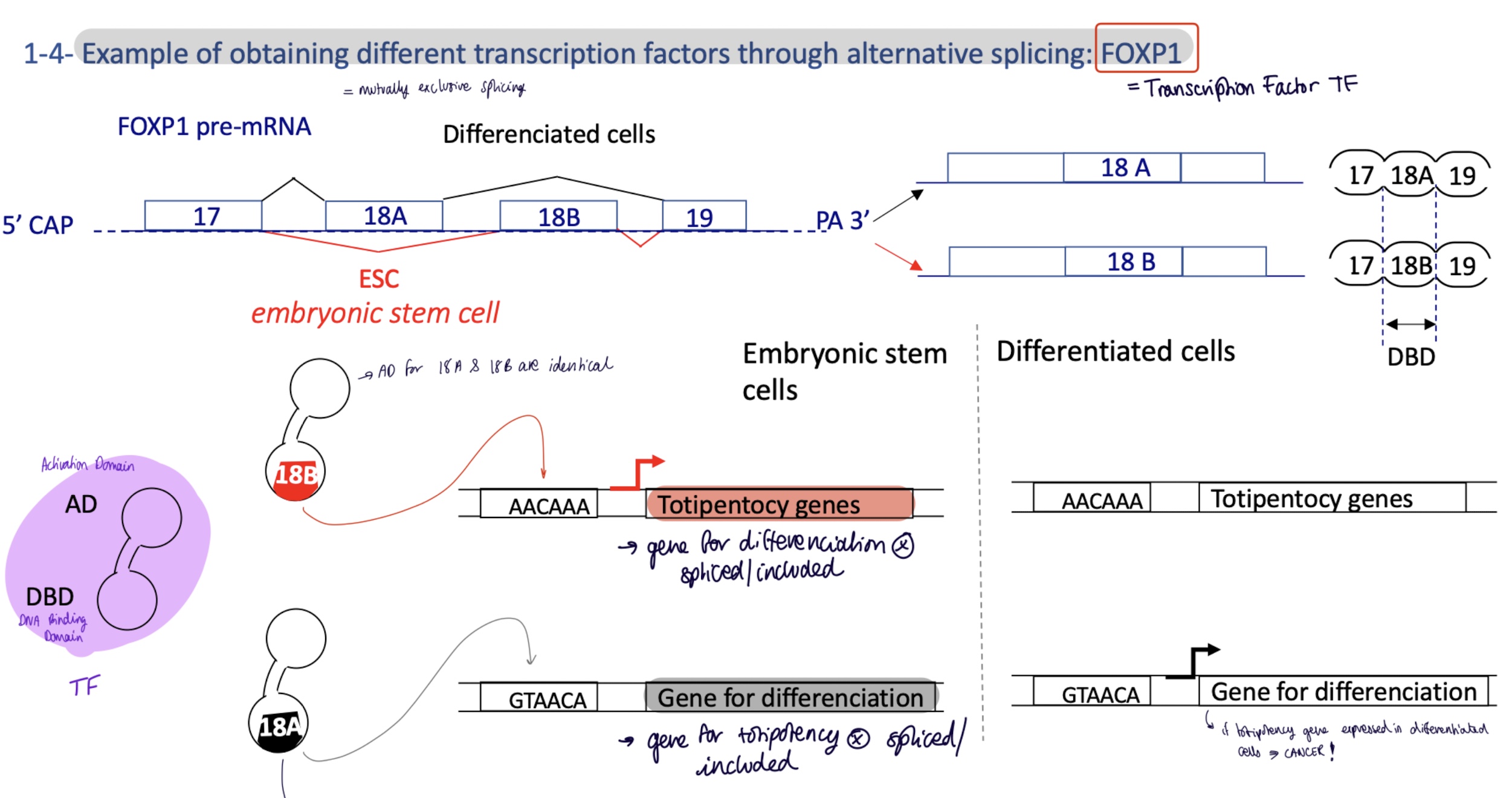
4.1 how does alternative splicing change the expression of genes for totipotent and differentiating cells
mutually exclusive splicing of TF FOXP1 where gene 18A/B codes for DBD DNA Binding Site
differentiated cells :
gene 18A is spliced/included
FOXP1 w/ 18A DBD will bind to promoter for genes for differentiation
embryonic totipotent stem cells :
gene 18B is spliced/included
FOXP1 w/ 18B DBD binds to promoter for totipotency genes
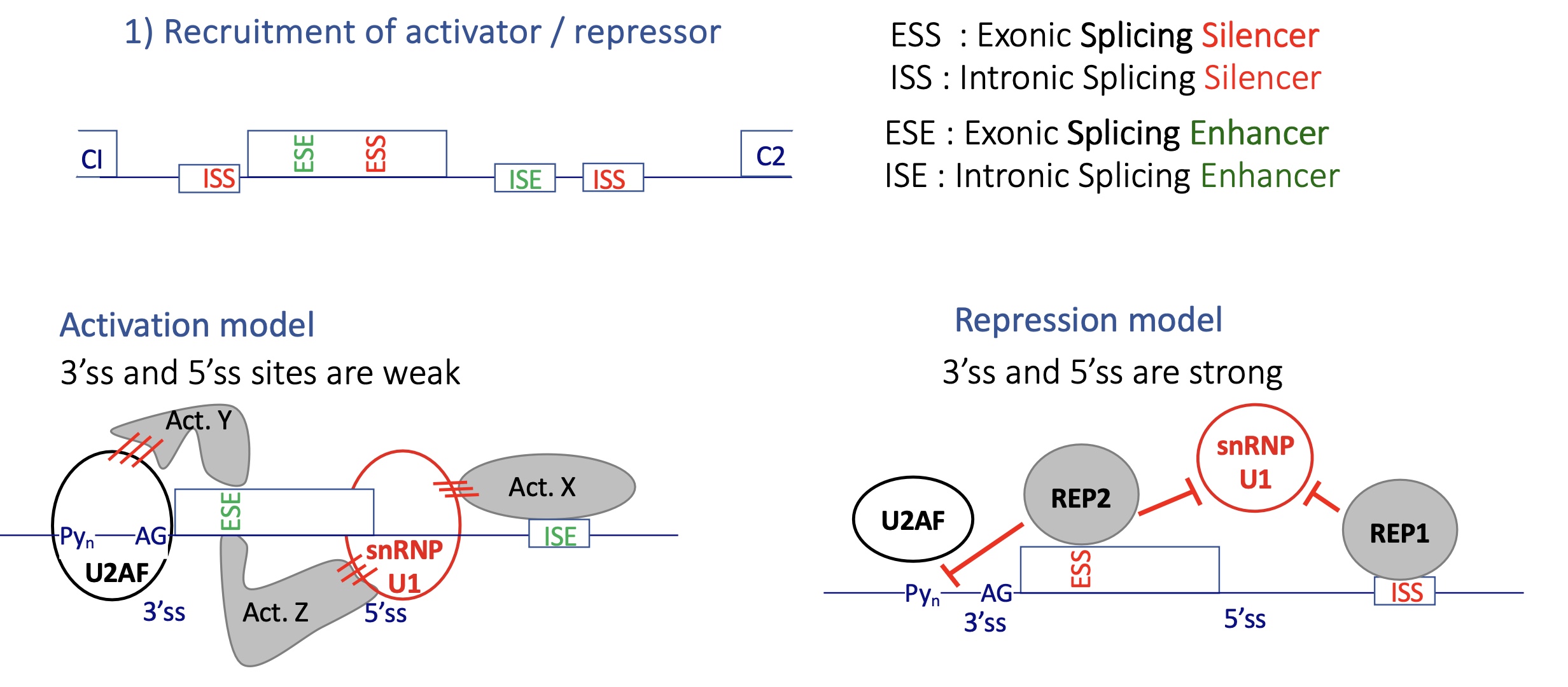
4.2 how is alternative splicing controlled by activator/repressor
activation model : 3’ and 5’ SS are weak
act proteins bind to E/ISE exonic/intronic splicing enhancer
insuring the binding of splicing factors (U2Af and snRNP U1) to 3’SS and 5’SS
repression model : 3’ and 5’ SS are too strong
Rep binds to E/ISS exonic/intronix splicing silencer
inhibits the installation of splicing factors (U2AF and snRNP) —. no spliceosome
4.2 what are examples of activators and repressors
Activator :
SR protein w/ SR domain that’s contains Arg-phosphoSer
RBD ad RNA binding domain
Repressors :
hnRNP —> ESS
PTB —> ISS
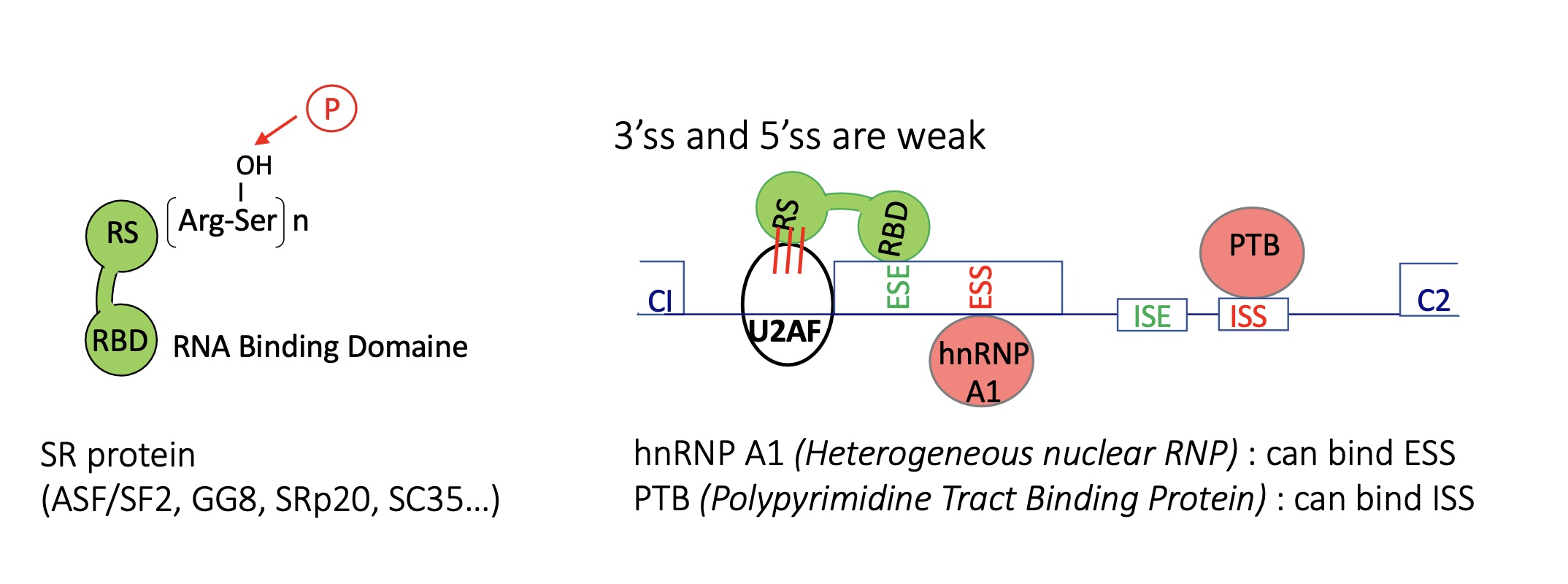
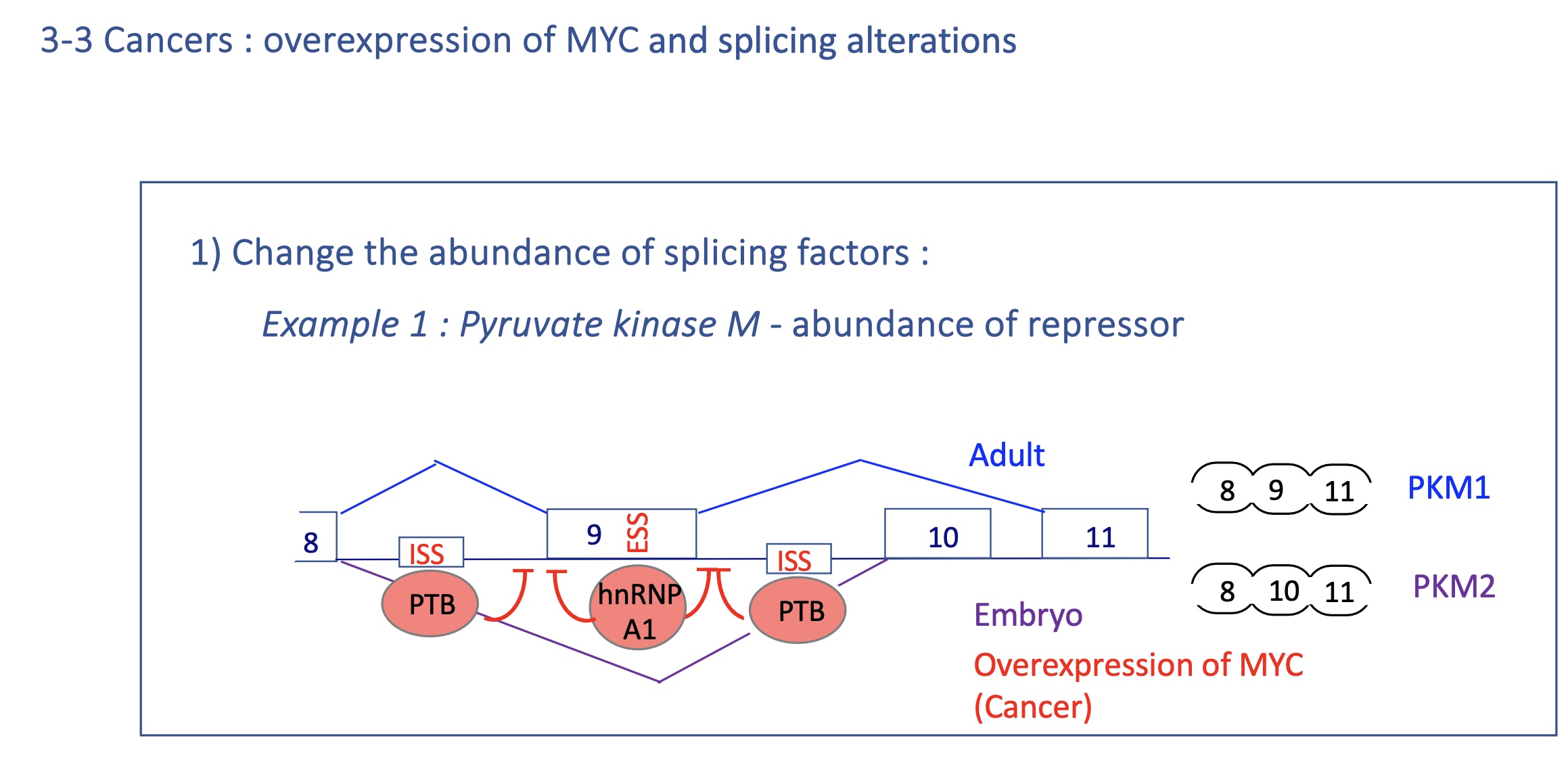
4.2 using the example of pyruvate kinase M how does the abundance of repressors effect transcription
Mutually exclusive splicing
Adult : PKM1 expression
exon 9 spliced and exon 10 excluded
embryo and Cancer : PKM2
exon 10 spliced and exon 9 excluded
due tosplicijg silencers PTB and hnRNP A1 that bind to ESS in exon 9 (silencing of exon 9 splicing) and ISS
in cancer MYC = TF over expresses PTB and hnRNP A1 (splicing silencers)
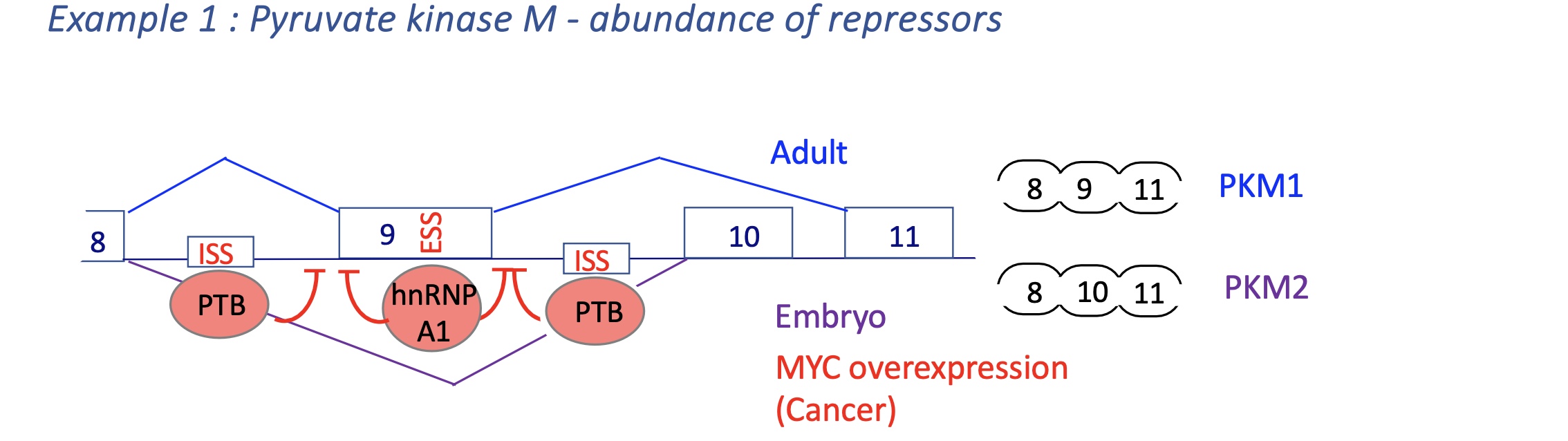
4.2 using an example of MCL1 (involved in apoptosis) how does the abundance of activators the transcription
3’ and 5’ SS of exon 2 are weak
MCL1-L (Long): anti-apoptotic.
MCL1-S (Short): pro-apoptotic (lacks transmembrane domain).
High SR protein ASF/SF2 favors MCL1-L → cell survival.
Stress or pro-apoptotic signaling ↓ ASF/SF2 → exon skipping → MCL1-S → apoptosis.
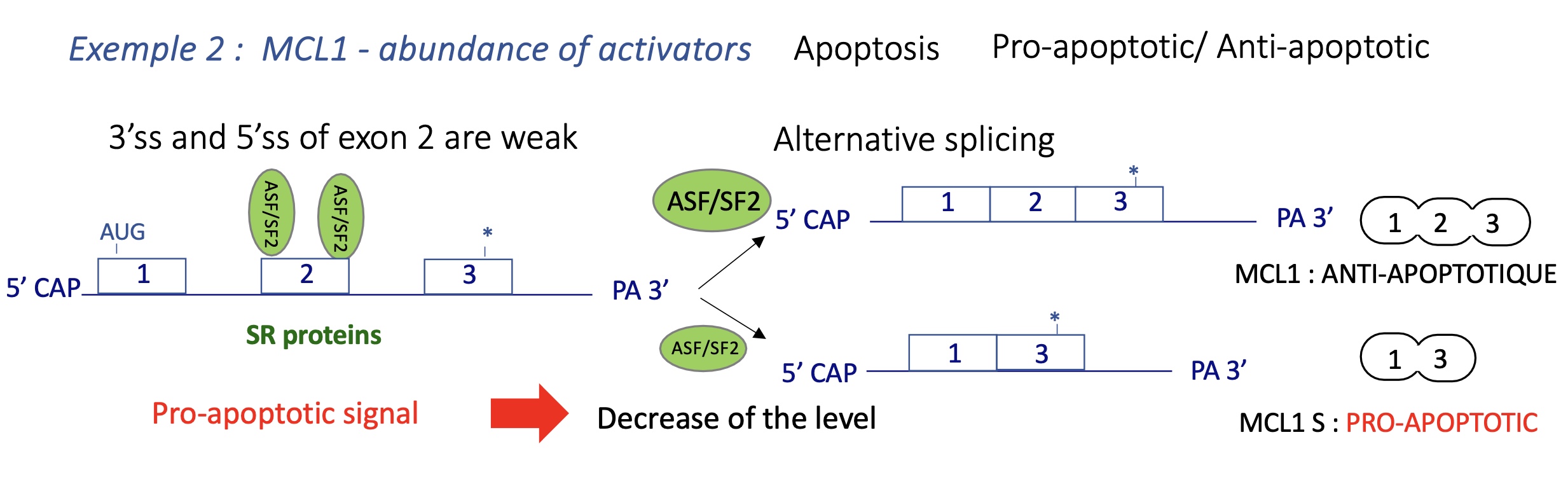
4.2 auto-regularion of SR proteins - change of splicing factor activity
SR proteins promote inclusion of a “poison exon” with a premature STOP codon.
Produces a transcript degraded by Nonsense-Mediated Decay (NMD).
→ Negative feedback keeps SR levels stable.
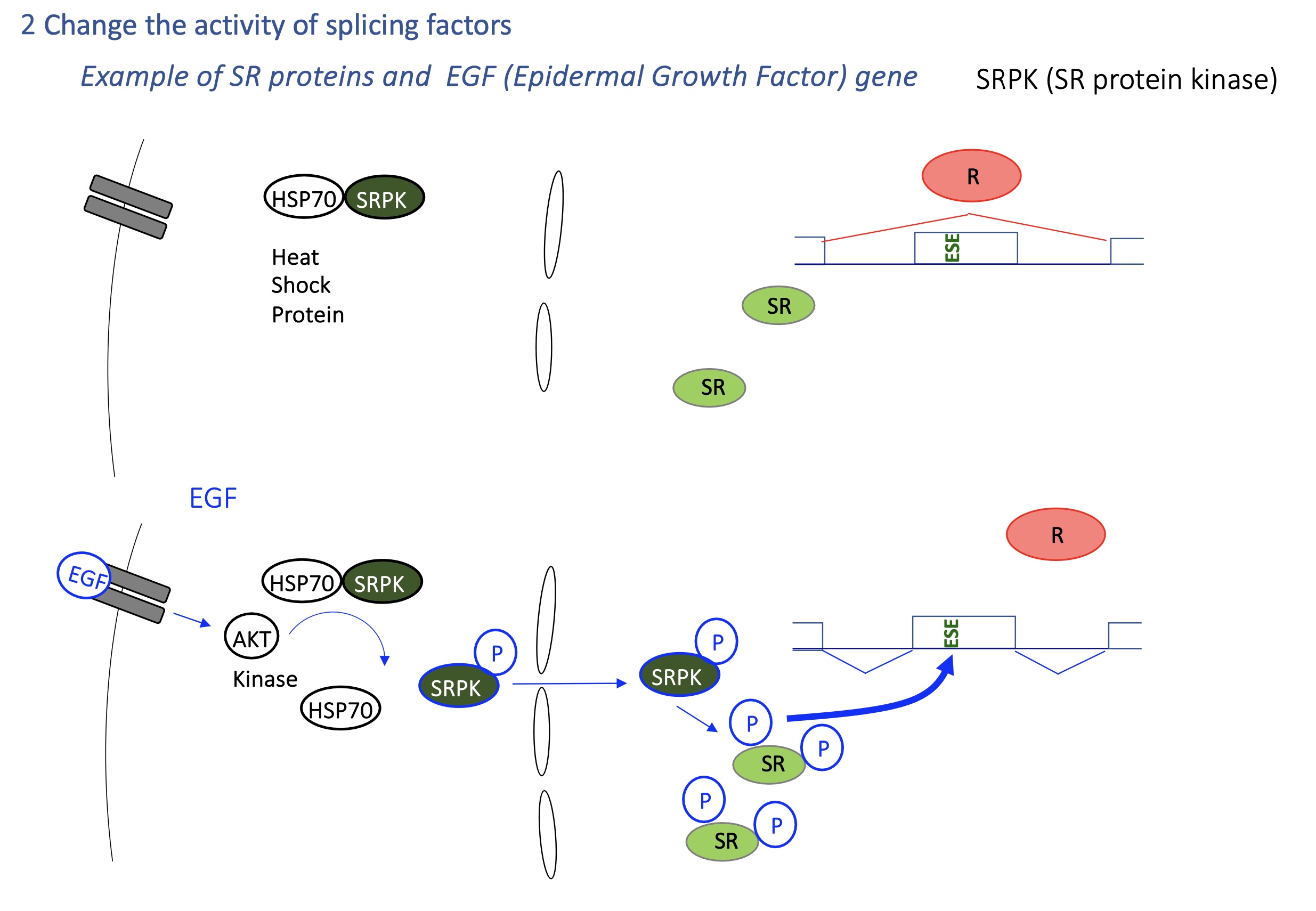
4.2 how is the activity of SR changed by SRPK
SRPK kinases (activated by EGF signaling) phosphorylate SR proteins, affecting localization to translocate inside nucleus and activate splicing activity
Heat shock → activates HSP70 → changes SR protein phosphorylation → global splicing reprogramming.
EGF/AKT pathway → activates SRPK kinases → SR proteins phosphorylated → new exon inclusion pattern.
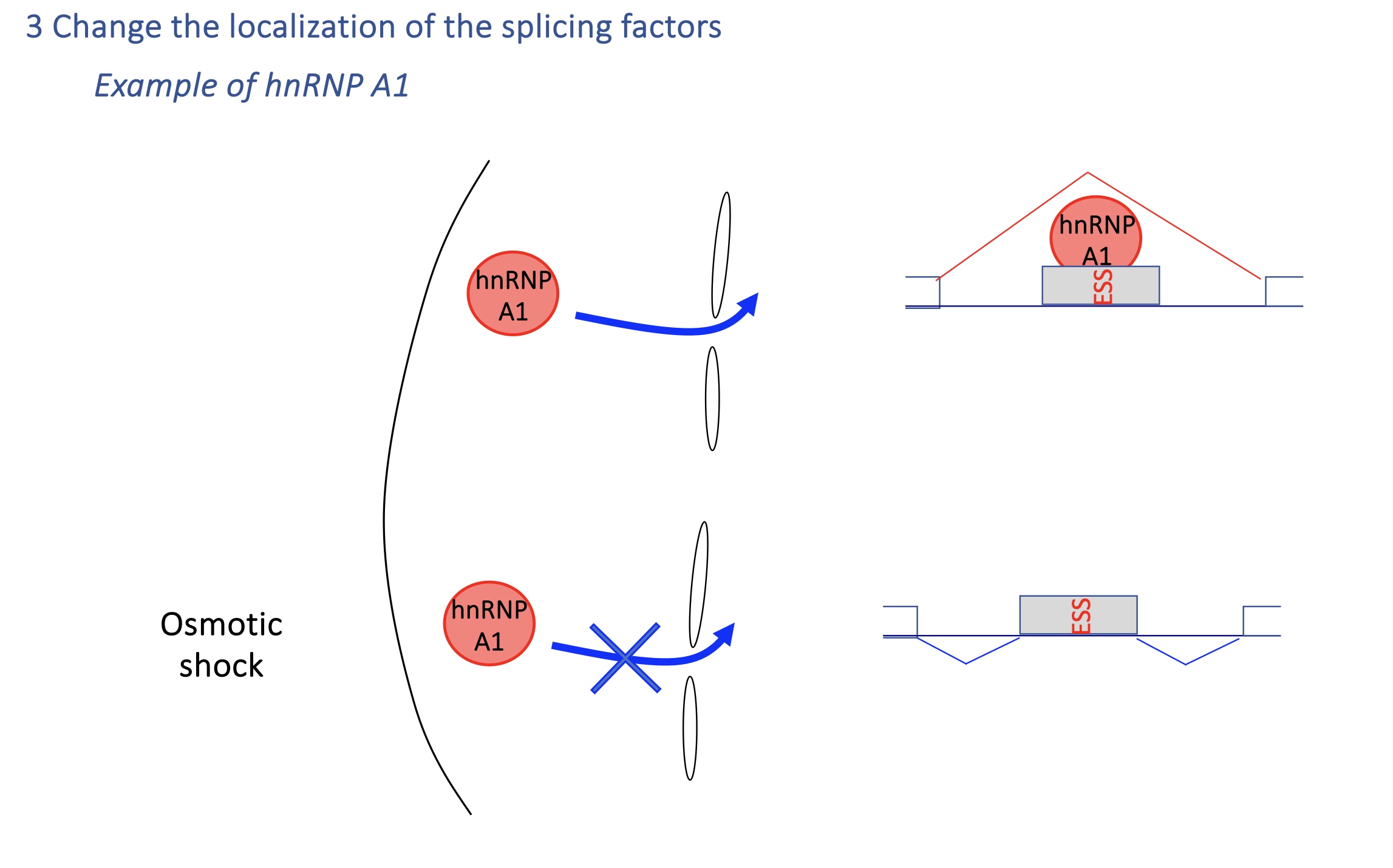
4.2 change of localisation of splicing factor : hnSNP A1
Under osmotic stress, hnRNP A1 shuttles to cytoplasm → less nuclear repression → splicing patterns change.
→ Rapid environmental adaptation.
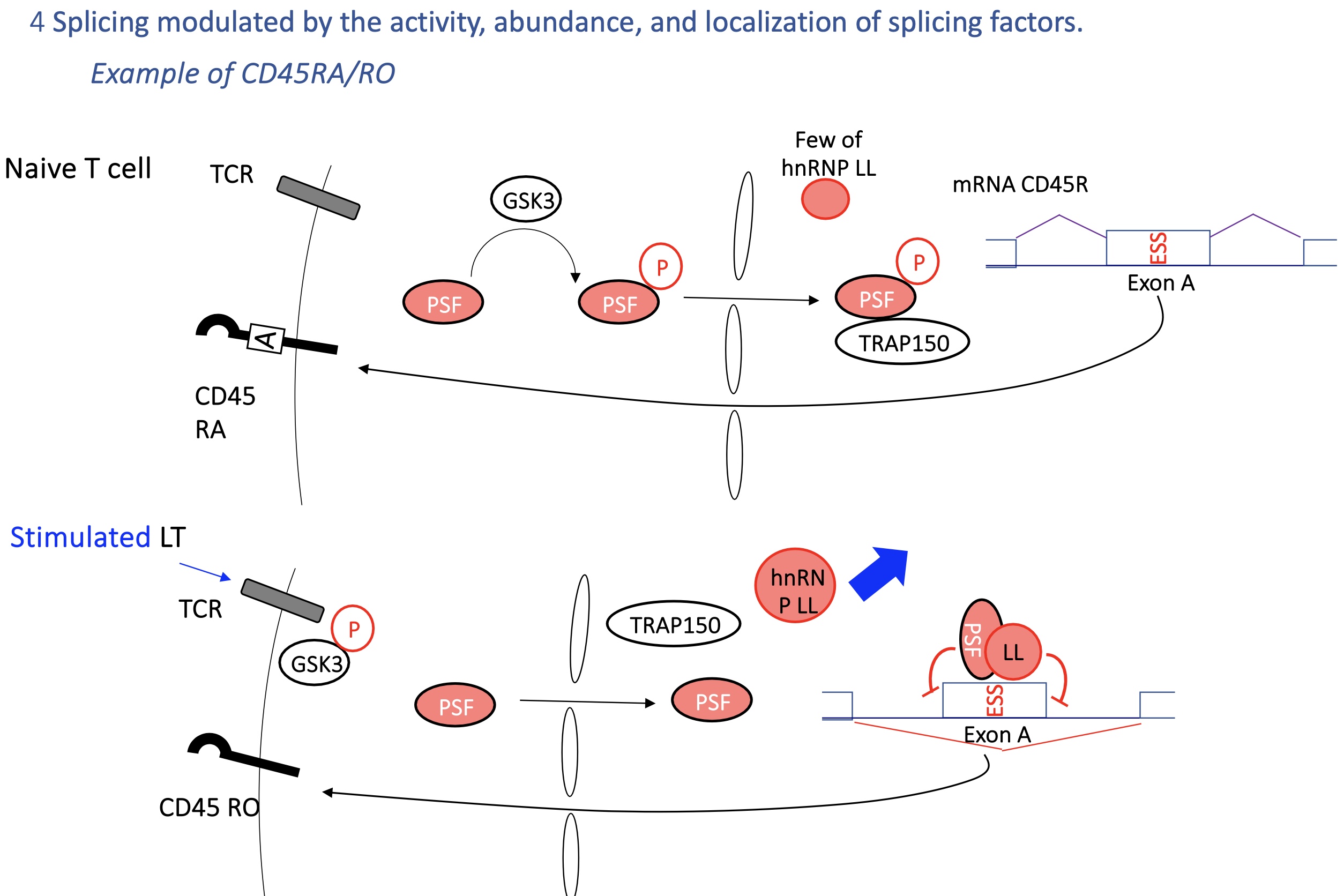
4.2 Immune example: CD45 gene (RA/RO isoforms)
T-cell receptor (TCR) signaling affects hnRNP LL and PSF activity → regulates inclusion of exon A.
Naive T cells (CD45RA) vs activated T cells (CD45RO) produce different CD45 isoforms.
SO
CD45RA (naive T cells): exon A included.
CD45RO (activated T cells): exon A skipped.
Controlled by hnRNP LL and PSF, regulated by TCR-GSK3 signaling.
→ Allows T cells to adapt splicing during activation
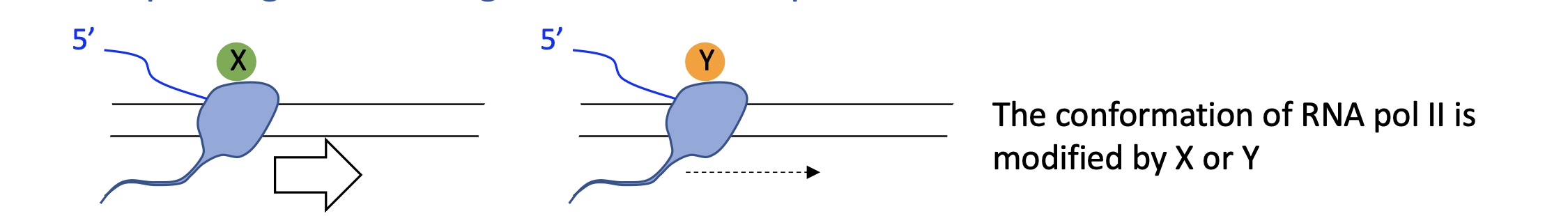
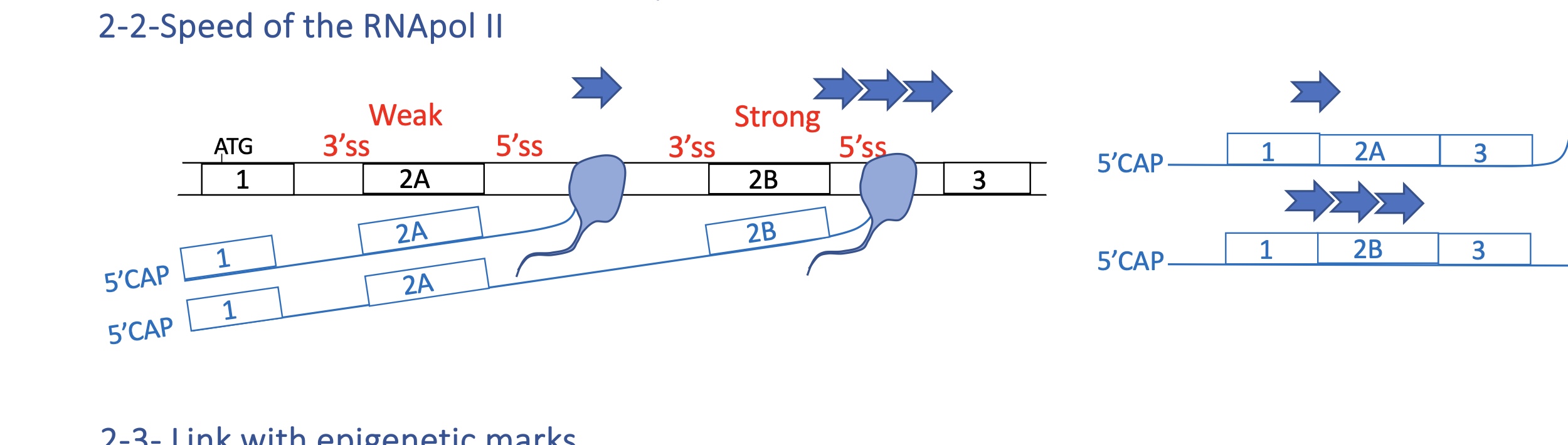
4.2 how does speed of RNA pol II
Slow elongation: spliceosome has more time → inclusion of weak exons.
Fast elongation: spliceosome skips weak sites → exon exclusion.
→ Splicing is co-transcriptional; transcription kinetics directly influence it.
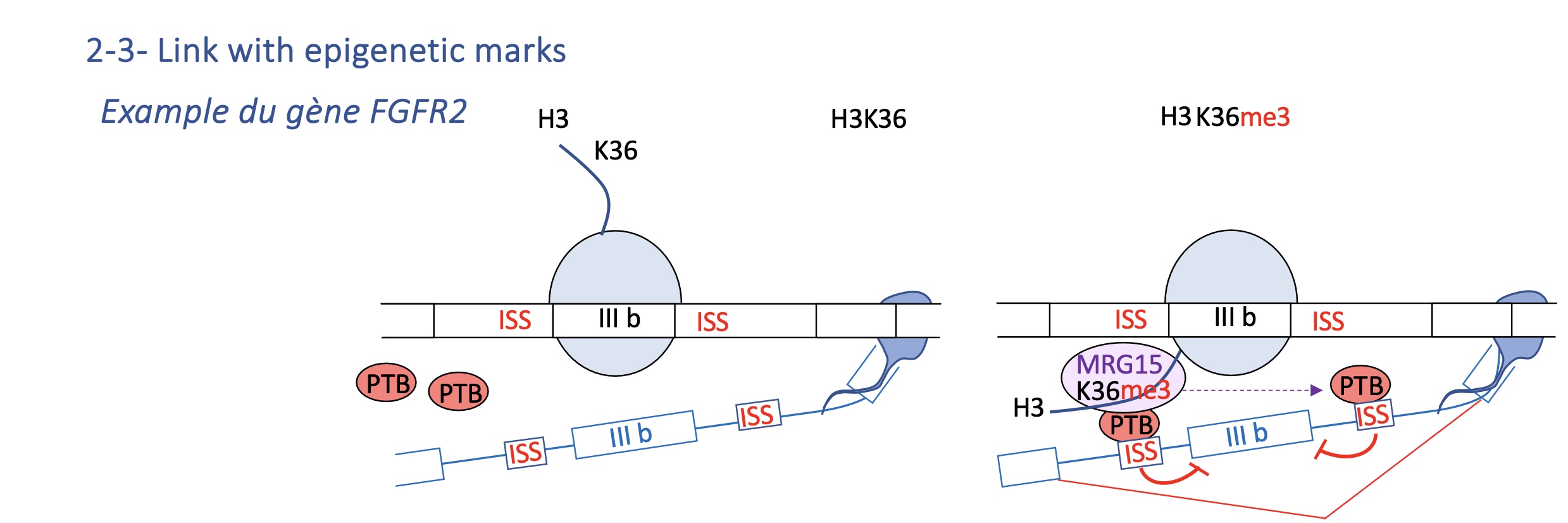
4.2 Epigenetic marks: FGFR2 example
H3K36me3 recruits MRG15, which attracts PTB to silence exon IIIb.
Tissue-specific histone modification determines exon inclusion (e.g. IIIb vs IIIc in FGFR2).
Two mutually exclusive exons (IIIb, IIIc).
H3K36me3 histone mark recruits MRG15, which binds PTB, repressing IIIb in mesenchymal cells.
→ Epithelial vs mesenchymal tissues express different FGFR2 isoforms due to chromatin context.
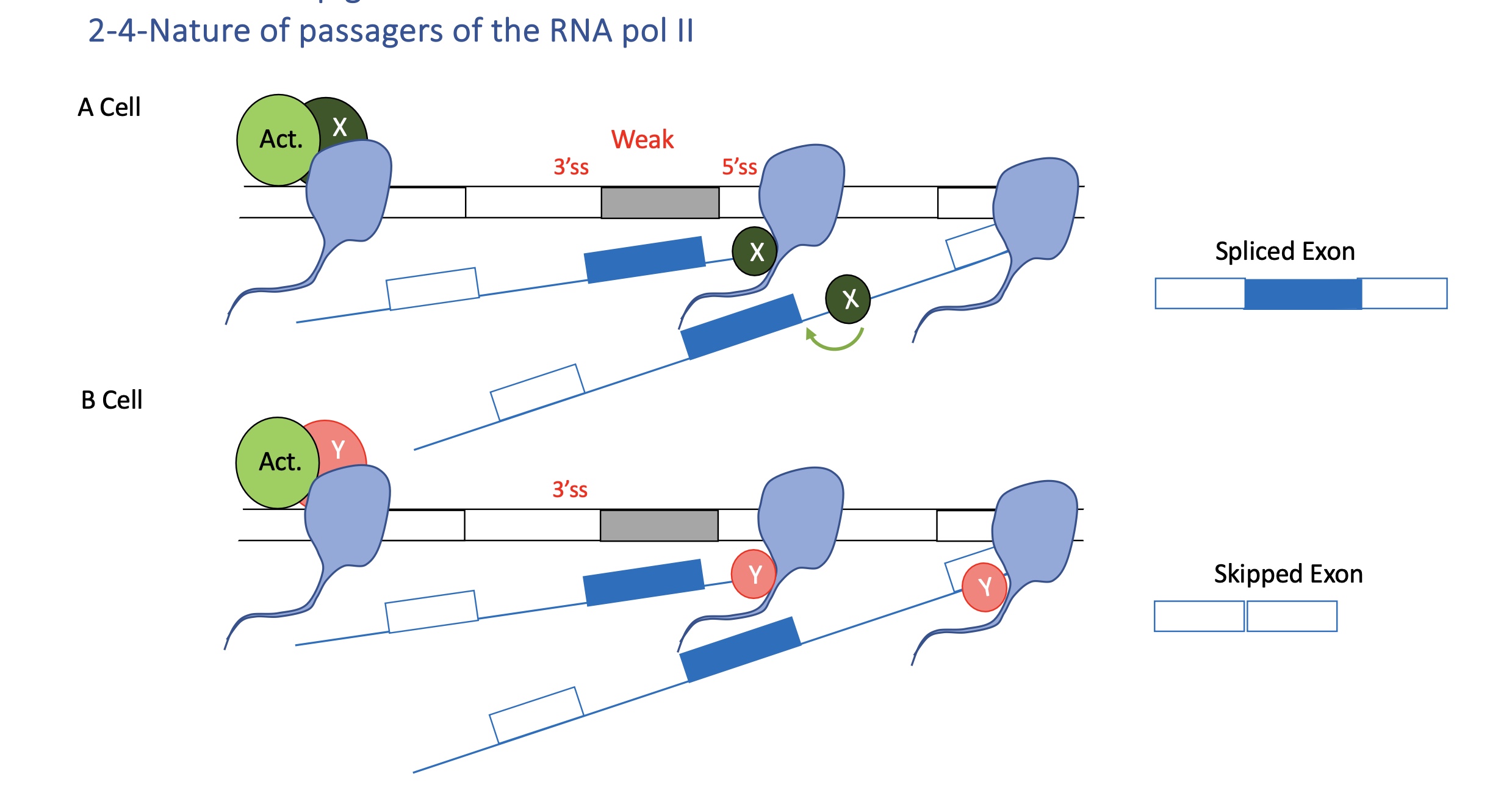
4.2 RNA pol II partners
Different co-factors (X or Y) associated with Pol II determine whether a weak exon is spliced in or skipped.
Pol II’s C-terminal domain (CTD) recruits RNA-processing factors (capping enzymes, splicing factors).
Depending on its phosphorylation state (Ser2/Ser5), it interacts with different splicing regulators → cell-specific outcomes
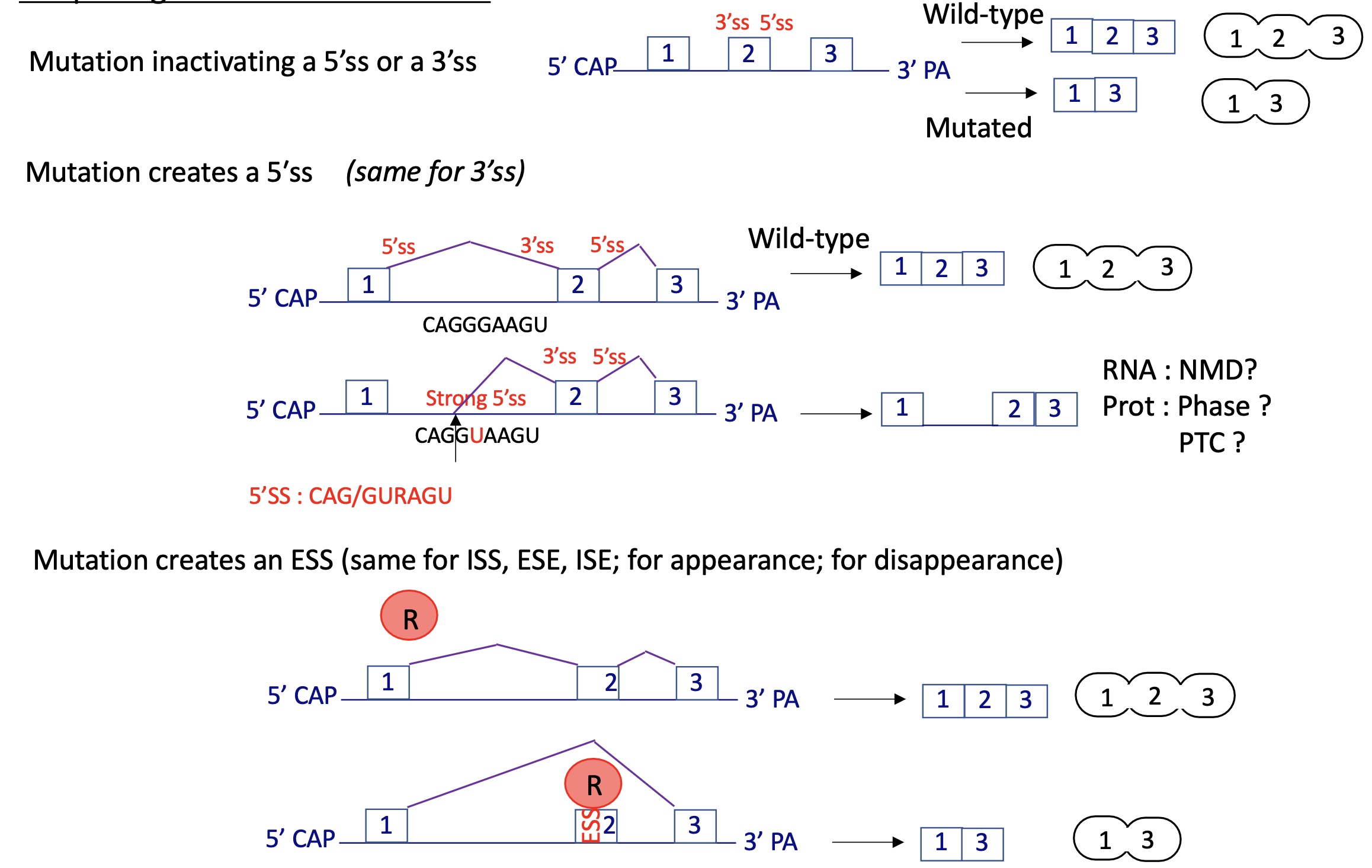
4.3 splicing defects and genetic disease general mechanism
mutation that can :
inactivate 3’SS or 5’SS
create a 5’SS or 3’SS
creating ESS/ISS/ISE/ESE
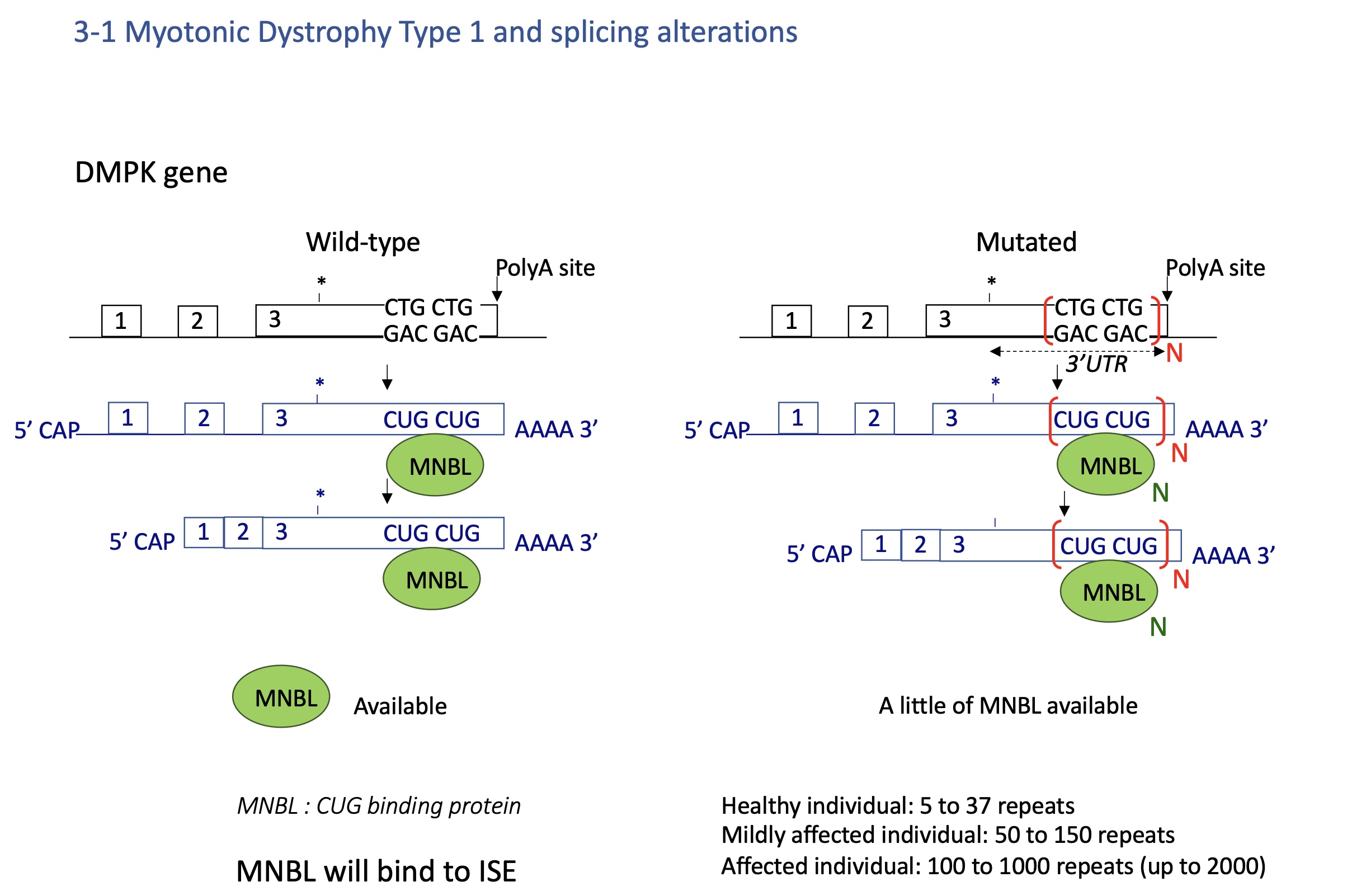
4.3 myotonic dystrophy Type 1 - splicing defect
Expansion of CTG repeats in DMPK 3′UTR.
RNA with long CUG repeats sequesters MBNL1 protein (a splicing regulator).
Loss of MBNL1 → mis-splicing of many pre-mRNAs (e.g., ClC-1, INSR).
→ Muscle weakness, myotonia, insulin resistance.
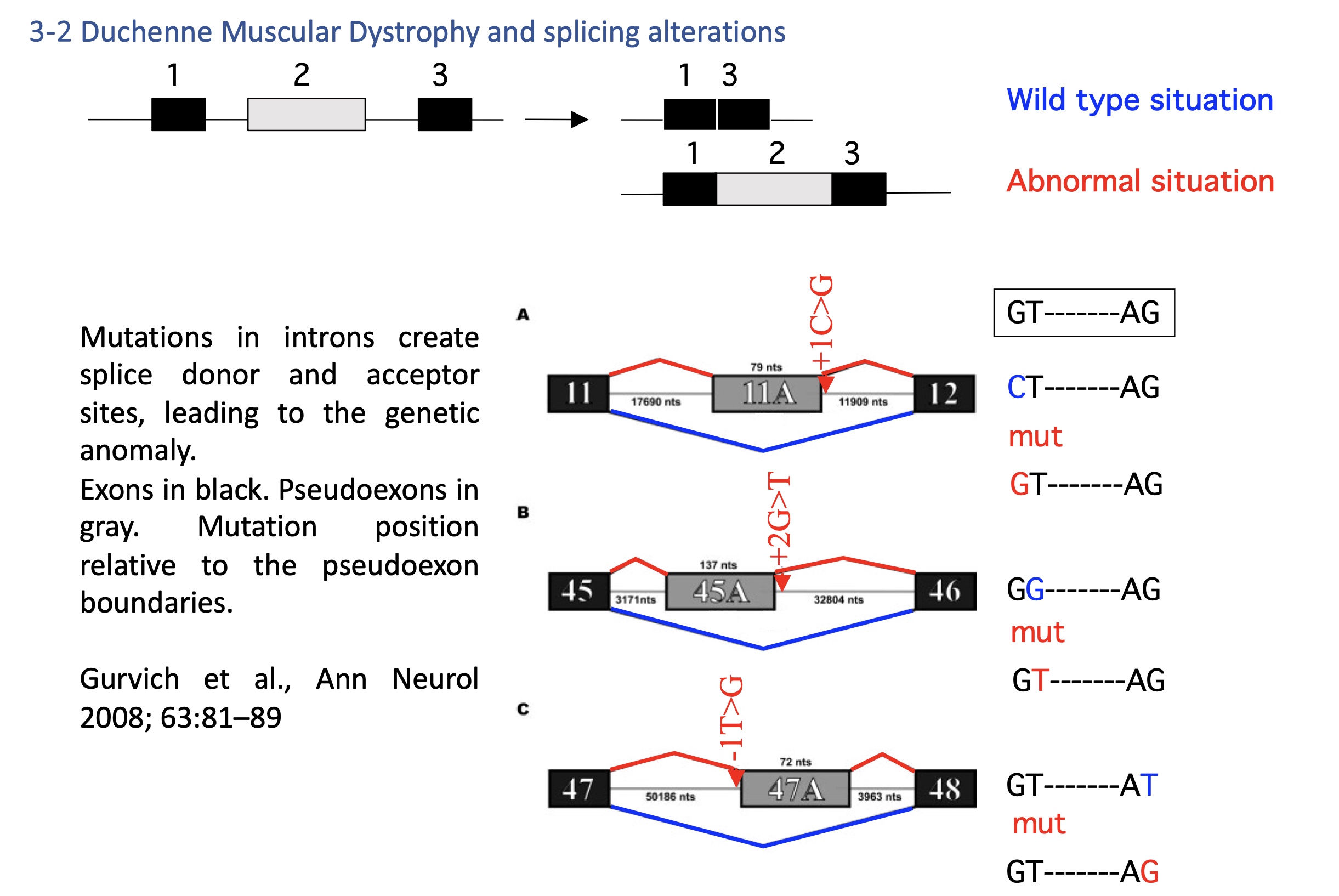
4.2 Duchenne muscular dystrophy DMD - splicing defect
Mutations create cryptic splice sites in introns → inclusion of pseudoexons → nonfunctional dystrophin.
Therapeutic approach: antisense oligonucleotides to block aberrant sites and restore the reading frame
4.2 cancers - splicing defect
MYC overexpression alters splicing factor levels → favors pro-tumor isoforms (e.g., PKM2).
→ Aberrant splicing = hallmark of cancer metabolism and signaling
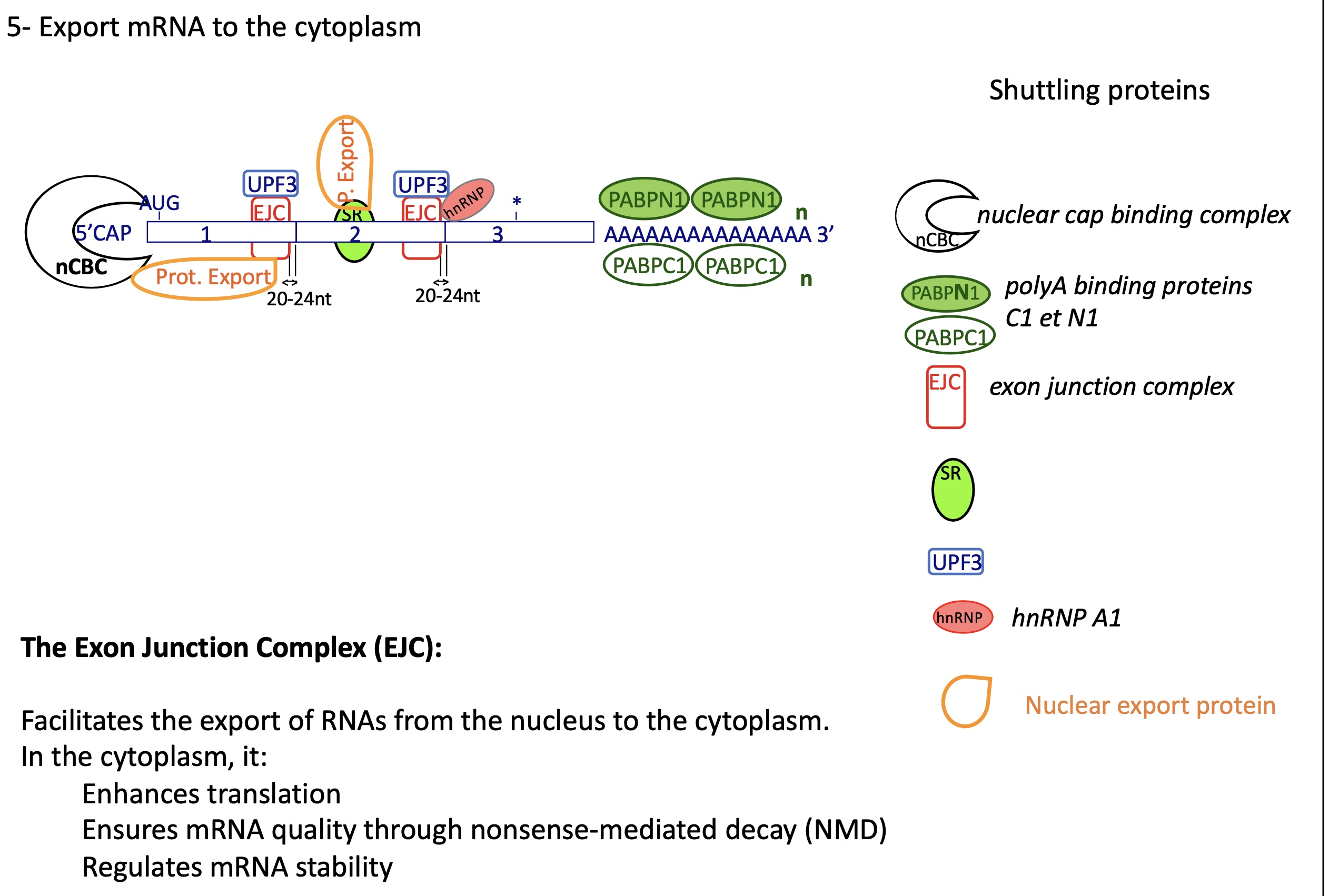
how does a mRNA exported to cytoplasm look like ?
At the 5′ end — the “Cap Structure”
The 5′ cap (7-methylguanosine) is bound by the nuclear Cap-Binding Complex (nCBC) made of:
CBP20
CBP80
Within the coding region — Exon Junction Complexes (EJCs)
After splicing, an EJC (Exon Junction Complex) is deposited ~20–24 nucleotides upstream of each exon–exon junction.
Core components:
eIF4AIII
MAGOH
Y14 (RBM8A)
MLN51 (CASC3)
Associated with UPF3 (and later UPF2, UPF1) for Nonsense-Mediated Decay (NMD) control.
Throughout the mRNA — hnRNPs and SR proteins
hnRNPs (heterogeneous nuclear ribonucleoproteins) coat the mRNA to protect it and help in packaging.
e.g., hnRNP A1, hnRNP C
SR proteins (splicing activators) remain bound and help recruit export adaptors.
= shuttling proteins
At the 3′ end — Poly(A) tail and Poly(A)-binding proteins
The poly(A) tail (≈200 adenines) has two major binding proteins:
PABPN1 (nuclear) — binds the tail in the nucleus during processing.
PABPC1 (cytoplasmic) — replaces PABPN1 after export and interacts with eIF4G to stimulate translation.