MCAT Biochemistry - DNA and Biotechnology
1/99
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
100 Terms
deoxyribonucleic acid (DNA)
stores the information in our cells and selectively shares that information when appropriate; passed from generation to generation; replicated in a carefully regulated process designed to keep the genome safe from degradation and free from errors; generally double-stranded (dsDNA)
ribonucleic acid (RNA)
biological macromolecule that plays several essential roles in the coding, decoding, regulation, and expression of genes; generally single-stranded (ssRNA)
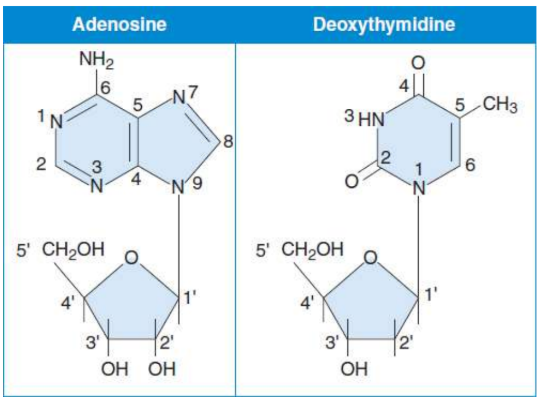
Nucleosides
composed of a five-carbon sugar (pentose) bonded to a nitrogenous base; formed by covalently linking the base to C-1′ of the sugar
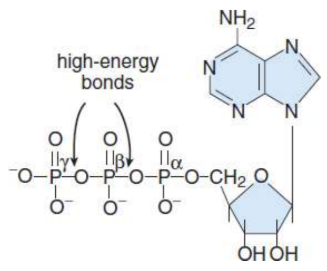
Nucleotides
formed when one or more phosphate groups are attached to C-5′ of a nucleoside; named according to the number of phosphates present; high-energy compounds because of the energy associated with the repulsion between closely associated negative charges on the phosphate groups
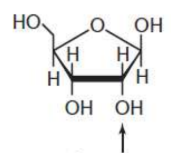
ribose
pentose in RNA
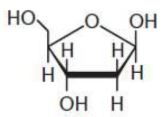
deoxyribose
pentose in DNA
Bases, Nucelosides, Nucleotides
Adenine - Adenosine (Deoxyadenosine) - AMP (dAMP) ADP (dADP) ATP (dATP)
Guanine - Guanosine (Deoxyguanosine) - GMP (dGMP) GDP (dGDP) GTP (dGTP)
Cytosine - Cytidine (Deoxycytidine) - CMP (dCMP) CDP (dCDP) CTP (dCTP)
Uracil - Uridine (Deoxyuridine) - UMP (dUMP) UDP (dUDP) UTP (dUTP)
Thymine - (Deoxythymidine) - (dTMP) (dTDP) (dTTP)
Sugar–Phosphate Backbone
alternating sugar and phosphate groups; determines the directionality of the DNA; read from 5′ to 3′; joined by 3′–5′ phosphodiester bonds
5′ end of DNA
−OH or phosphate group bonded to C-5′ of the sugar
3′ end of DNA
has a free −OH on C-3′ of the sugar
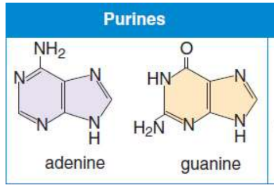
Purines
contain two rings in their structure
Adenine & Guanine (Pure As Gold)
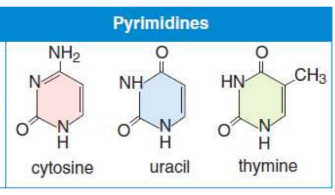
Pyrimidines
contain only one ring in their structure
Cytosine, Thymine & Uracil (CUT the Pie)
aromatic
any unusually stable ring system that adheres to the following four specific rules:
cyclic
planar
conjugated
Huckel’s rule (4n+2)
ex. Benzene, purines, pyrimidines
Watson–Crick model of DNA structure
three-dimensional structure of DNA; double-helical nature; specific base-pairing that would be the basis of a copying mechanism
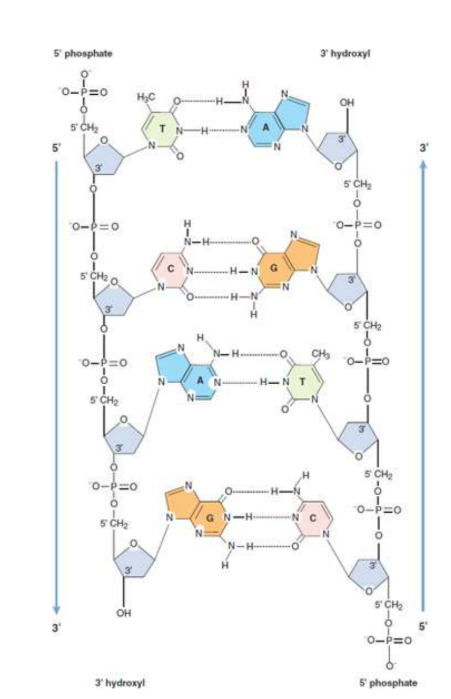
double helix
two linear polynucleotide chains of DNA are wound together in a spiral orientation along a common axis
antiparallel: strands are oriented in opposite directions, i.e one strand has polarity 5′ to 3′ down the page, the other strand has 5′ to 3′ polarity up the page
sugar–phosphate backbone is on the outside of the helix with the nitrogenous bases on the inside
specific base-pairing rules: hydrogen bonds and the hydrophobic interactions between bases provide stability to the double helix structure
complementary base-pairing
An adenine (A) is always base-paired with a thymine (T) via two hydrogen bonds.
A guanine (G) always pairs with cytosine (C) via three hydrogen
bonds.
Chargaffs rules.
the amount of A equals the amount of T, and the amount of G equals the amount of C.
total purines will be equal to total pyrimidines overall.
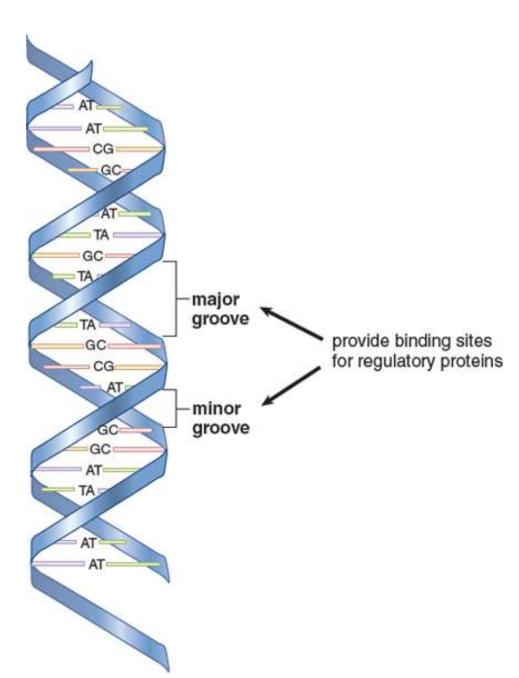
B-DNA
right-handed double helix; makes a turn every 3.4 nm and contains about 10 bases within that span; Major and minor grooves can be identified between the interlocking strands and are often the site of protein binding
Z-DNA
zigzag appearance; left-handed helix that has a turn every 4.6 nm and contains 12 bases within each turn; high GC-content or a high salt concentration may contribute to the formation; unstable and difficult to research
denaturation
“melting” of the double helix into two single strands that have separated from each other by conditions (heat, alkaline pH, formaldehyde, urea) that disrupt hydrogen bonding and base-pairing; none of the covalent links between the nucleotides in the backbone of the DNA break during this process.
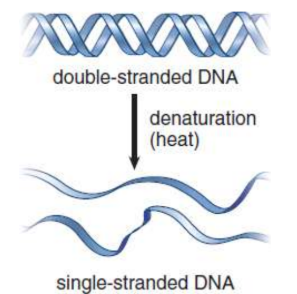
reannealing
single strand brought back together
probe DNA
DNA with known sequence used in PCR or to identify a gene of interest
chromosomes
a package of DNA containing part or all of the genetic material of an organism
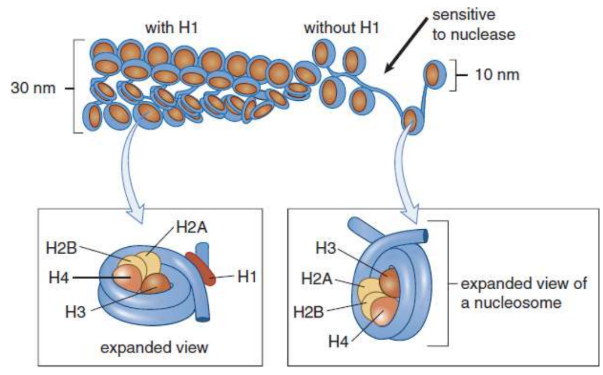
histones
small basic proteins found around DNA as coiling tool
five found in eukaryotic cells
chromatin
DNA wounds around histones
nuclesome
Two copies each of the histone proteins H2A, H2B, H3, and H4 form a histone core and about 200 base pairs of DNA are wrapped around this protein complex
nucleoproteins
proteins that associate with DNA; typically are acid-soluble and tend to stimulate processes such as transcription
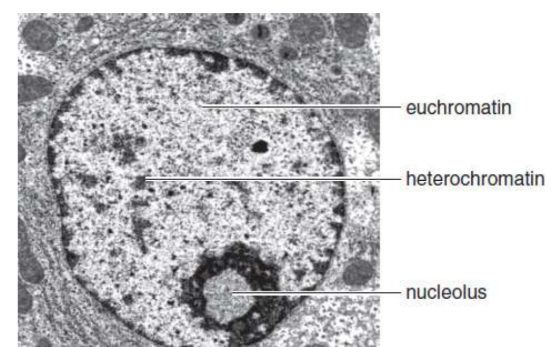
heterochromatin
small percentage of the chromatin remains compacted during interphase; dark under light microscopy and transcriptionally silent; DNA with highly repetitive sequences and high GC content
During cell division, the two sister chromatids can therefore remain connected until microtubules separate the chromatids during anaphase
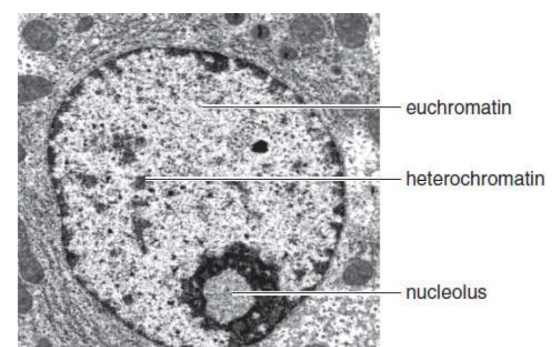
euchromatin
dispersed chromatin; light under light microscopy; genetically active DNA
telomere
simple repeating unit (TTAGGG) at the end of the DNA that accounts for DNA replication not extending to the end
telomerase
can replace lost telomere sequences; more highly expressed in rapidly dividing cells
centromeres
region of DNA found in the center of chromosomes; ‘sites of constriction’ because they form noticeable indentations; heterochromatin
replisome/replication complex
a set of specialized proteins that assist the DNA polymerases
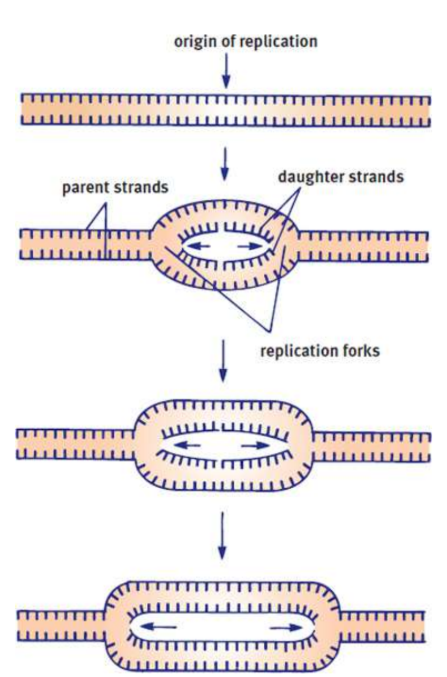
origins of replication
DNA unwinds at points to begin replication
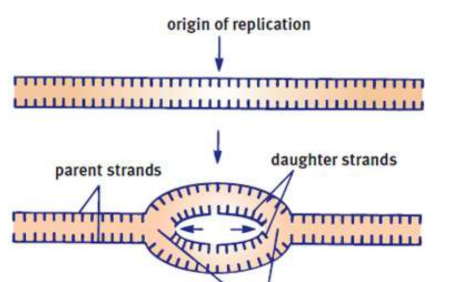
replication forks
DNA replication extends out to either side of an origin on each strand
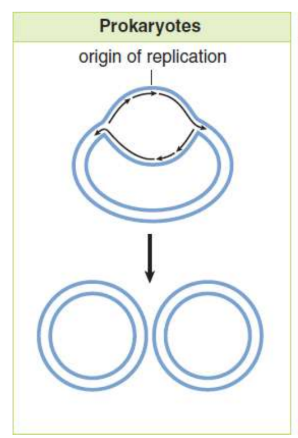
bacterial chromosome
closed, double-stranded circular DNA molecule with a single origin of replication; two replication forks eventually meet, resulting in the production of two identical circular molecules of DNA
Eukaryotic replication
must copy many more bases compared to prokaryotic; slower process
each eukaryotic chromosome contains one linear molecule of double-stranded DNA having multiple origins of replication; As the replication forks move toward each other and sister chromatids are created, the chromatids will remain connected at the centromere
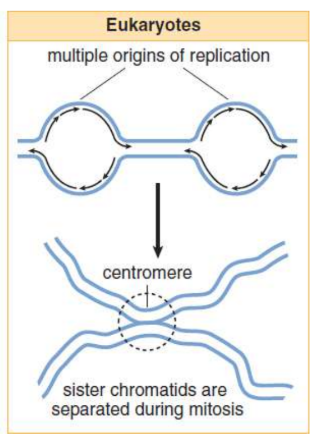
sister chromatids
identical copies (chromatids) formed by the DNA replication of a chromosome, with both copies joined together by a common centromere
fluoroquinolone
antibiotics that target the prokaryotic topoisomerase DNA gyrase; stops bacterial replication and slows infection
used for pneumonia and infections of the genitourinary system
end with –floxacin (ciprofloxacin, moxifloxacin, and so on)
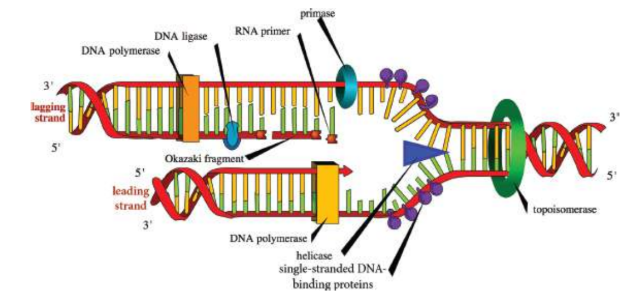
Helicase
enzyme responsible for unwinding the DNA, generating two single-stranded template strands ahead of the polymerase
single-stranded DNA-binding proteins
bind to the unraveled strand, preventing both the reassociation of the DNA strands and the degradation of DNA by nucleases
nuclease
an enzyme capable of cleaving the phosphodiester bonds that link nucleotides together to form nucleic acids
Supercoiling
wrapping of DNA on itself as its helical structure is pushed ever further toward the telomeres during replication
DNA topoisomerases
introduce negative supercoils to alleviate this torsional stress and reduce the risk of strand breakage; working ahead of helicase, nicking one or both strands, allowing relaxation of the torsional pressure, and then resealing the cut strands
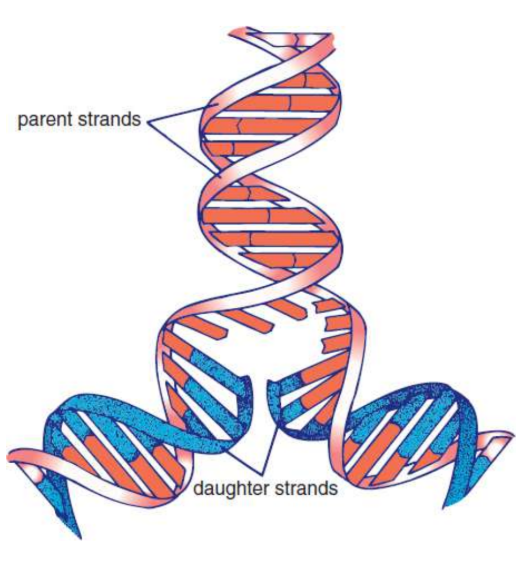
parental strands
original DNA that gets replicated
daughter strands
the result of DNA replication
semiconservative replication
one parental strand is retained in each of the two resulting identical double-stranded DNA molecules
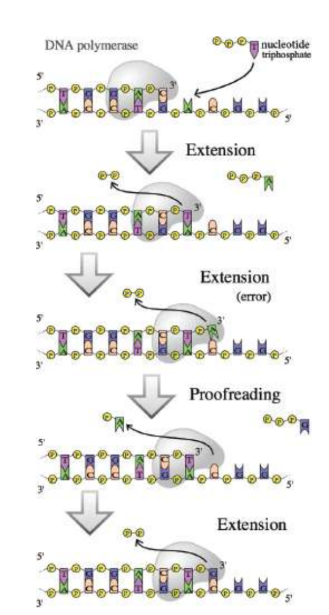
DNA polymerases
are responsible for reading the DNA template, or parental strand, and synthesizing the new daughter strand; read in 3’ to 5’ while synthesising 5’ to 3’
leading strand
copied in a continuous fashion, in the same direction as the advancing replication fork, will be read 3′ to 5′ and its complement will be synthesized in a 5′ to 3′ manner
lagging strand
copied in a direction opposite the direction of the replication fork; parental strand has 5′ to 3′ polarity; Okazaki fragments
Okazaki fragments
small strands created as the replication fork continues to move forward on the lagging strand and DNA polymerase
primase
synthesizes a short RNA primer (roughly 10 nucleotides) in the 5′ to 3′ direction to start replication on each strand for DNA to hook on to
DNA polymerase III
prokaryotes; begin synthesizing the daughter strands of DNA in the 5′ to 3′ manner
DNA polymerases α, δ, and ε
in eukaryotes; begin synthesizing the daughter strands of DNA in the 5′ to 3′ manner
DNA polymerase I
in prokaryotes; remove RNA primer and adds dNA nucleotides where primer was
RNase H
in eukaryotes; removes RNA primer
DNA polymerase δ
in eukaryotes; adds DNA nucleotides where the RNA primer had been
DNA ligase
seals the ends of the DNA molecules together between replication forks and Okazaki fragments creating one continuous strand of DNA
sliding clamp.
DNA polymerases δ and ε are assisted by the PCNA protein, which assembles into a trimer; strengthen the interaction between these DNA polymerases and the template strand
DNA polymerase γ
replicates mitochondrial DNA
DNA polymerases β
DNA repair
Cancer
disease fro m unmitigated cellular growth/division
metastasis
a migration of cancer to distant tissues by the bloodstream or lymphatic system
oncogenes
Mutated genes that cause cancer; dominant
ex. src
proto-oncogenes
oncogenes prior to mutation
antioncogenes
Tumor suppressor genes encode proteins that inhibit the cell cycle or participate in DNA repair processes
ex. p53, Rb
proofreading
two double-stranded DNA molecules will pass through a part of the DNA polymerase enzyme to correct improper matches in base pairs
Methylation
the template strand has existed in the cell for a longer period of time, and therefore is more heavily methylated
mismatch repair
detect and remove errors introduced in replication that were missed during the S phase of the cell cycle
eukaryotes: genes MSH2 and MLH1
prokaryotes: MutS and MutL
nucleotide excision repair (NER)
cut-and-patch process
specific proteins scan the DNA molecule and recognize the lesion because of a bulge in the strand. An excision endonuclease then makes nicks in the phosphodiester backbone of the damaged strand on both sides of the thymine dimer and removes the defective oligonucleotide. DNA polymerase can then fill in the gap by synthesizing DNA in the 5′ to 3′ direction, using the undamaged strand as a template. Finally, the nick in the strand is sealed by DNA ligase.
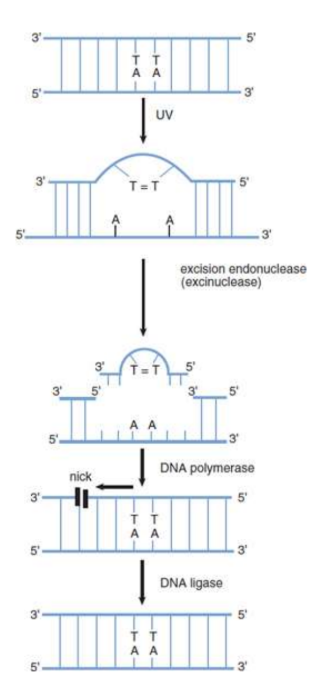
excision endonuclease/encinuclease
makes nicks in the phosphodiester backbone of the damaged strand on both sides of the thymine dimer and removes the defective oligonucleotide
base excision repair
cytiosine→uracil in DNA or other small, non-helix-distorting mutations; the affected base is recognized and removed by a glycosylase enzyme, leaving behind an AP site recognized by an AP endonuclease that removes the damaged sequence from the DNA
apurinic/apyrimidinic (AP)/abasic site.
point on DNA without a base
AP endonuclease
removes the damaged AP site sequence from the DNA
Recombinant DNA
DNA molecules formed by laboratory methods that bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome
DNA cloning
produce large amounts of a desired sequence; ligate the DNA of interest into a piece of nucleic acid
vector
a piece of nucleic acid that will carry the desired foreign gene
recombinant vector
a piece of nucleic acid that carries the desired foreign gene
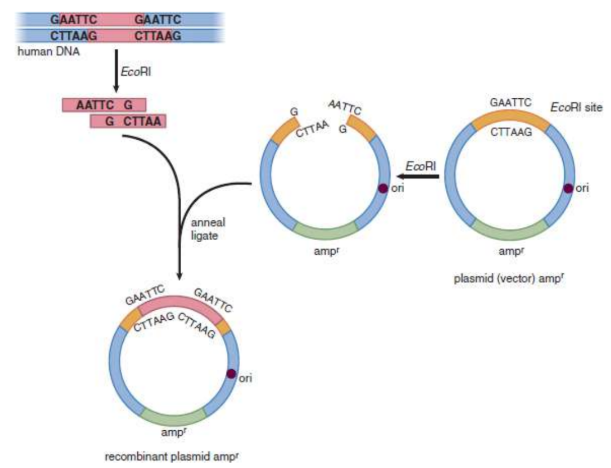
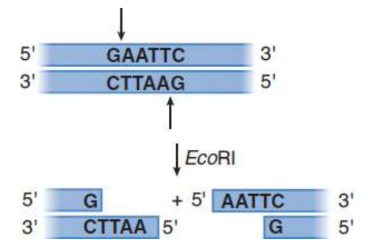
Restriction enzymes/endonucleases
enzymes that recognize specific (typically palindromic) double-stranded DNA sequences and cut through the backbones of the double helix; orginally found in bacteria; some leave offset ‘sticky’ ends, others leave blunt ends
DNA libraries
large collections of known DNA sequences; in sum, these sequences could equate to the genome of an organism
Genomic libraries
contain large fragments of DNA, and include both coding (exon) and noncoding (intron) regions of the genome
complementary DNA (cDNA)
constructed by reverse-transcribing processed mRNA; lacks noncoding regions, such as introns, and only includes the genes that are expressed in the tissue from which the mRNA was isolated
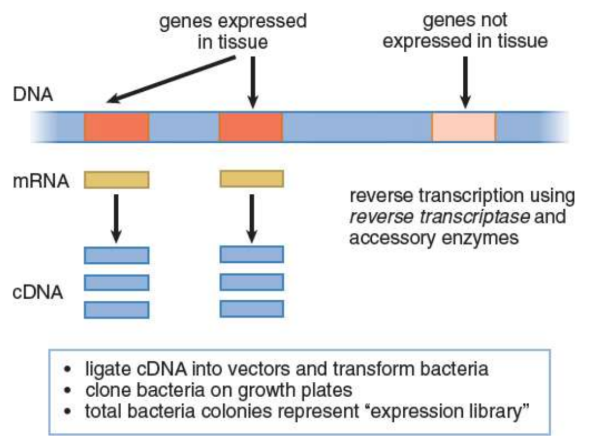
expression libraries
collections of known cDNA
Hybridization
joining of complementary base pair sequences
Polymerase chain reaction (PCR)
automated process that can produce millions of copies of a DNA sequence without amplifying the DNA in bacteria; used to identify criminal suspects, familial relationships, and disease-causing bacteria and viruses; requires primers, nucleotides, DNA polymerase, heat
primers
small bit of DNA complementary to the DNA that flanks the region of interest used in PCR; high GC conetnt
Taq (thermus aquaticus) polymerase
enzyme from a extremophile bacteria, resists denaturing to heating during PCR
Gel electrophoresis
technique used to separate macromolecules, such as DNA and proteins, by size and charge
agarose gel
preferred gel for DNA electrophoresis`
Southern blot
used to detect the presence and quantity of various DNA strands in a sample using gel electrophoresis; All molecules of DNA are negatively charged → anode; gel—the longer the DNA strand, the slower it will migrate in the gel
probe
copies of a single-stranded DNA sequence; labeled with radioisotopes or indicator proteins, both of which can be used to indicate the presence of a desired sequence
dideoxyribonucleotide
contain a hydrogen at C-3′, rather than a hydroxyl group; thus, once one of these modified bases has been incorporated, the polymerase can no longer add to the chain; used in sequencing
DNA sequencing
template DNA, primers, an appropriate DNA polymerase, all four deoxyribonucleotide triphosphates, dideoxyribonucleotides (one at a time); the sample will contain many fragments, as many as the number of nucleotides in the desired sequence, each one of which terminates with one of the modified bases; last base for each fragment can be read, and because gel electrophoresis separates the strands by size, the bases can easily be read in order
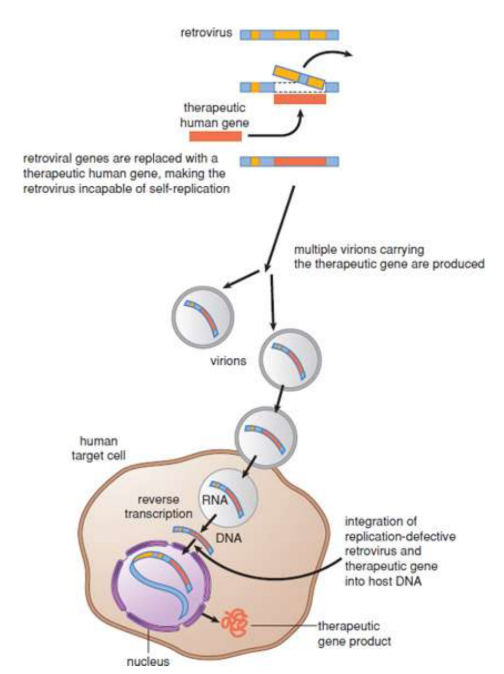
Gene therapy
potential cures for individuals with inherited diseases; By transferring a normal copy of the gene into the affected tissues, the pathology should be fixed; efficient gene delivery vectors must be used to transfer the cloned gene into the target cells’ DNA: modified viruses
ex SCID (severe combined immunodeficiency) - 1990
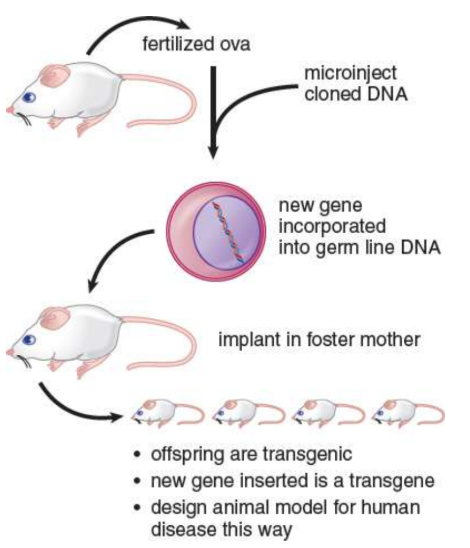
Transgenic mice
altered at their germ line by introducing a cloned gene into fertilized ova or into embryonic stem cells
germ line
cells that become/are gametes/embryos
transgene
cloned gene that is introduced into transgenic mice
knockout mice
a gene has been intentionally deleted to observe results
chimera
patches of cells, including germ cells, derived from each of the two lineages
Safety and Ethics
Ethical dilemmas associated with biotechnology
Read Frankenstein or something